Viruses, which infect a myriad of cell types from bacteria to mammalian cells, are the ultimate parasites. Upon infection of a host cell, these entities replicate their genetic material, express the proteins required to form mature viral particles, and exit the cell to initiate a subsequent round of infection. At all of these stages, viruses rely heavily on host resources and factors. It is, therefore, not surprising that the study of viral life cycles has contributed enormously to our understanding of basic cellular physiology. Furthermore, virus-induced diseases are perhaps the most prevalent pathological states in human beings; thus, understanding virus interactions with the human host has had an important medical impact. Much of our understanding of viral reproduction comes from genetic studies of virus–host interactions. For example, genetic analysis has proven to be a powerful tool for studying virus (phage)–host interactions in bacteria. Mutants of both phage and their host bacteria have allowed detailed understanding of the functions of phage genes, the functions of host factors hijacked by the phage, and the host defenses that inhibit phage reproduction (1). Genetic analysis has been similarly informative in analyzing the interactions between viruses and their plant hosts (2). The story is significantly different in animals, however, where genetic studies of virus–host interactions have generally been one-sided, focusing principally on the viral genome. Although human and murine mutations affecting various aspects of virus–host interactions have been described (3), a systematic, unbiased genetic approach to studying such interactions in these systems has not been possible. Natural viruses that attack the fruit fly Drosophila melanogaster exist, and a systematic genetic dissection of Drosophila C virus replication in cultured cells has been published recently (4); however, although some of the requirements for C virus to infect WT flies have been characterized (5), replication of these viruses in living animals has not been extensively studied. A natural virus for the nematode Caenorhabditis elegans has not been described; thus, until recently, this organism was not considered an appropriate model organism for studying virus–host interactions. However, the situation may be about to change. A series of articles published over the past few months, including an article by Liu et al. (6) in this issue of PNAS, now demonstrates that viruses that infect mammalian cells can infect, replicate, and assemble within C. elegans cells (7–9). These advances suggest that C. elegans could become an important system for understanding basic aspects of virus–host interplay.
Why would C. elegans be worth investigating as a host for virus reproduction? One answer lies in the powerful resources available for genetic analysis in this organism. Random mutagenesis to generate C. elegans strains resistant or hypersensitive to pathogens can be, in principle, easily accomplished (10–12) and detailed genetic and physical maps of C. elegans make the identification of mutant genes a relatively simple endeavor. RNA interference (RNAi) can also be used to inhibit gene function (13) and could be used to identify genes that cannot be targeted by using mutagenesis-based approaches. Indeed, genome-scale screens using RNAi are now commonplace (14, 15). Using these genetic strategies, several groups have identified interactions between C. elegans and pathogenic bacteria and fungi. These studies have led to the identification of innate immune mechanisms similar to those described in Drosophila and mammals and have also revealed novel mechanisms used by C. elegans to control pathogenesis (10–12).
The small cell number (959 somatic cells) and the transparency of its cuticle also make C. elegans attractive for studying viral reproduction. These properties allow the movements of subcellular particles to be tracked in living animals, a feat difficult to achieve in other model metazoans. Thus, for example, appropriately tagged viral proteins could be followed within infected cells in vivo to understand cell biological aspects of viral reproduction.
Although whole-animal and genetic studies are useful, using single cells to follow the viral life cycle offers important advantages, including the ability to examine the process at higher resolution and in biochemical detail. Most studies of mammalian viruses use cell lines to characterize viral reproduction and host resistance. Recently, simple methods for culturing primary embryonic cells from C. elegans have been developed that make studies of viral reproduction within isolated cells possible (16, 17). Using this technology, primary cells derived from C. elegans mutants resistant or hypersensitive to virus (identified, for example, by using the genetic strategies described above) could be studied in detail to reveal the stage of viral reproduction affected in the mutants. Although immortal C. elegans cell lines do not yet exist, these lines may not be necessary because primary cells may recapitulate the in vivo cellular environment more accurately than transformed cells.
Finally, it turns out that two key antiviral pathways operating in mammalian cells have been extensively studied, in other contexts, in C. elegans. The first involves cellular responses to dsRNA, an intermediate produced during the life cycle of many viruses. In mammalian cells, dsRNA is a potent trigger of IFN-mediated responses, which include the activation of nucleases that degrade the viral genome (18). In addition, recent studies also suggest that viral RNA may trigger the RNAi pathway (19), which also promotes cleavage of viral messages. The second pathway leads to the programmed cell death of virus-infected cells and is induced either by intracellular triggers or by the immune system (20). Many mutant strains of C. elegans exist that are defective in these pathways, and their genetics are understood in unparalleled detail. Indeed, these pathways were discovered and characterized first in C. elegans (13, 21). Thus, virus–host interactions involving RNAi and cell death could be studied in C. elegans at high resolution.
Although a natural virus for C. elegans is not known, several groups have attempted to use viruses with promiscuous host specificities to examine whether these can infect, replicate, and assemble in C. elegans. Wilkins et al. (7) and Schott et al. (8) recently showed that cultured primary cells from C. elegans embryos could be infected with vesicular stomatitis virus (VSV), a negative-strand RNA virus able to infect both insects and mammals. They showed that VSV could enter C. elegans cells, express viral proteins, and produce new infectious viral particles. Remarkably, both groups showed that RNAi plays a key role in restricting viral reproduction in C. elegans. Specifically, mutations in the C. elegans rde-1 gene, an argonaute family member, or in the rde-4 gene, encoding a dsRNA-binding protein that interacts with the RDE-1 protein, resulted in a marked increase in virus production, suggesting that the RNAi pathway inhibits viral reproduction in these cells. Consistent with this observation, both groups demonstrated that mutations that enhance cellular RNAi responses reduced viral infectivity.
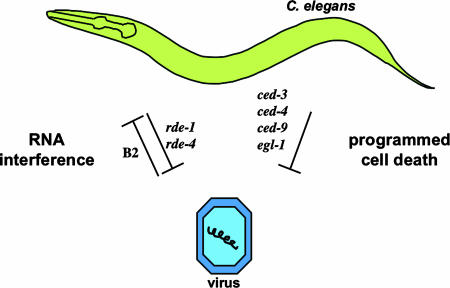
In a different set of experiments, Lu et al. (9) demonstrated a convincing genetic interaction between the C. elegans RNAi pathway and a viral protein that inhibits RNAi. These authors examined genome replication of the Flock house virus (FHV), a positive-strand RNA virus that has a large spectrum of hosts, including plants, yeast, insects, and mammalian cells. In these studies, FHV RNA was produced in living C. elegans from a transgene containing a heat-shock promoter driving expression of DNA encoding the viral genome. The authors showed that a point mutation disrupting the ORF of the viral B2 gene greatly reduced viral replication. Remarkably, inactivating RNAi by a mutation in rde-1 allowed the mutant FHV RNA to be replicated, suggesting that RNAi plays a key role in restricting FHV replication in C. elegans and that the B2 gene functions to inhibit the RNAi pathway (Fig. 1).
Fig. 1.
Genes of the RNA interference (RNAi) and programmed cell death pathways control nonnative viral reproduction in C. elegans. Gene names are indicated next to arrows. Blunt arrows indicate an inhibitory effect. A Flock house virus (FHV) protein, B2, can inhibit C. elegans RNAi defenses, as indicated by the arrow pointing from the virus to C. elegans. RNAi and cell death genes inhibit viral reproduction, as indicated by arrows pointing from C. elegans to the virus.
It makes sense for the cell to use RNAi to interfere with the replication of RNA viruses, because dsRNA is an integral part of the RNA virus life cycle. Although some DNA viruses also encode proteins that inhibit RNAi, suggesting that their hosts do mount an RNAi response, many DNA viruses encode proteins meant to inhibit other cellular defense mechanisms, including programmed cell death. To address whether C. elegans can be infected by DNA viruses and whether it is able to mount an appropriate response, Liu et al. (6) examined whether C. elegans could be infected with vaccinia virus (VV), an enveloped DNA poxvirus with a broad host range. They developed a method to permeabilize living animals by using polyethylene glycol (PEG), leaving ≈75% of animals alive and allowing successful VV infection. Interestingly, the authors showed that key components of the core programmed cell death machinery played roles in restricting VV replication. Specifically, loss-of-function mutations in the ced-3 caspase-encoding gene, or in the ced-4/Apaf-1 adapter gene, required for converting proCED-3 caspase to the mature form, resulted in a 2-fold increase in VV replication. Similarly, a gain-of-function mutation in ced-9, encoding an antiapoptotic Bcl-2-related protein, or loss-of-function mutations in the egl-1 gene, encoding a BH3-only protein that antagonizes the CED-9 protein, allowed increased VV replication. This apparent increase in replication could not be attributed to the presence of additional “undead” cells in these mutants, because mutants affecting cell division, which contained many additional cells, did not serve as better hosts for the virus.
Although these results are consistent with programmed cell death playing an important role in VV restriction in C. elegans (Fig. 1), Liu et al. (6) were unable to show excess cell death in VV-infected animals. One possible explanation for these results is that the C. elegans cell death pathway restricts VV infection in a previously undescribed, cell death-independent manner. Alternatively, cell death induced by VV may use components of the canonical cell death pathway but may be morphologically or dynamically distinct from cell death normally seen in C. elegans and, therefore, would not be detected. Interestingly, >80% of VV-infected animals eventually died; however, death was independent of the programmed cell death pathway, suggesting that this pathway was not sufficient to generate a complete response against the virus.
The results of the studies discussed here are exciting and suggest that C. elegans may become an important system to study virus–host interactions; however, several issues still remain to be addressed. For example, although viral infections of C. elegans could be demonstrated for multiple viruses, all of these studies required high viral titers, and replication was modest in comparison to other well established systems. These problems highlight the somewhat contrived nature of these studies and suggest that identifying a virus for which C. elegans is a true host would be of significant importance. A second issue is that the systems currently available for studying viral reproduction in C. elegans do not yet allow utilization of the full power of genetic analysis in this animal. Specifically, unbiased genetic screens to identify mutant genetic backgrounds defective in VSV reproduction cannot be carried out easily in cell culture. Furthermore, a high proportion of PEG-treated animals either fail to take up VV or are not killed by its presence, precluding the isolation of rare mutants exhibiting defects in either viral replication or resistance to virus. However, the FHV transgenic strategy could be modified to allow large-scale genetic screens if appropriate reporters were engineered that correlated with levels of virus replication.
C. elegans has proven, perhaps surprisingly, to be an important organism for understanding basic cellular processes such as RNAi and programmed cell death. It may surprise us yet again in revealing the secrets of virus–host interactions.
Conflict of interest statement: No conflicts declared.
See companion article on page 4174.
References
- 1.Friedman D. I., Olson E. R., Georgopoulos C., Tilly K., Herskowitz I., Banuett F. Microbiol. Rev. 1984;48:299–325. doi: 10.1128/mr.48.4.299-325.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson R. S., Citovsky V. Plant Physiol. 2005;138:1809–1814. doi: 10.1104/pp.104.900167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrington M., O’Brien S. J. Annu. Rev. Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 4.Cherry S., Doukas T., Armknecht S., Whelan S., Wang H., Sarnow P., Perrimon N. Genes Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherry S., Perrimon N. Nat. Immunol. 2004;5:81–87. doi: 10.1038/ni1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W.-H., Lin Y.-L., Wang J.-P., Liou W., Hou R. F., Wu Y.-C., Liao C.-L. Proc. Natl. Acad. Sci. USA. 2006;103:4174–4179. doi: 10.1073/pnas.0506442103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkins C., Dishongh R., Moore S. C., Whitt M. A., Chow M., Machaca K. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 8.Schott D. H., Cureton D. K., Whelan S. P., Hunter C. P. Proc. Natl. Acad. Sci. USA. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu R., Maduro M., Li F., Li H. W., Broitman-Maduro G., Li W. X., Ding S. W. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan M. W., Ausubel F. M. Curr. Opin. Microbiol. 2000;3:29–34. doi: 10.1016/s1369-5274(99)00047-8. [DOI] [PubMed] [Google Scholar]
- 11.Pradel E., Ewbank J. J. Annu. Rev. Genet. 2004;38:347–363. doi: 10.1146/annurev.genet.38.072902.092528. [DOI] [PubMed] [Google Scholar]
- 12.Kurz C. L., Ewbank J. J. Nat. Rev. Genet. 2003;4:380–390. doi: 10.1038/nrg1067. [DOI] [PubMed] [Google Scholar]
- 13.Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 14.Sieburth D., Ch’ng Q., Dybbs M., Tavazoie M., Kennedy S., Wang D., Dupuy D., Rual J. F., Hill D. E., Vidal M., et al. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 15.Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., Ahringer J., Ruvkun G. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 16.Christensen M., Estevez A., Yin X., Fox R., Morrison R., McDonnell M., Gleason C., Miller D. M., 3rd, Strange K. Neuron. 2002;33:503–514. doi: 10.1016/s0896-6273(02)00591-3. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y., Ma C., Delohery T., Nasipak B., Foat B. C., Bounoutas A., Bussemaker H. J., Kim S. K., Chalfie M. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs B. L., Langland J. O. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 19.Lu S., Cullen B. R. J. Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomson B. J. Int. J. Exp. Pathol. 2001;82:65–76. doi: 10.1111/j.1365-2613.2001.iep0082-0065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis H. M., Horvitz H. R. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]