Abstract
Receptor-induced Ca2+ signals are key to the function of all cells and involve release of Ca2+ from endoplasmic reticulum (ER) stores, triggering Ca2+ entry through plasma membrane (PM) “store-operated channels” (SOCs). The identity of SOCs and their coupling to store depletion remain molecular and mechanistic mysteries. The single transmembrane-spanning Ca2+-binding protein, STIM1, is necessary in this coupling process and is proposed to function as an ER Ca2+ sensor to provide the trigger for SOC activation. Here we reveal that, in addition to being an ER Ca2+ sensor, STIM1 functions within the PM to control operation of the Ca2+ entry channel itself. Increased expression levels of STIM1 correlate with a gain in function of Ca2+ release-activated Ca2+ (CRAC) channel activity. Point mutation of the N-terminal EF hand transforms the CRAC channel current (ICRAC) into a constitutively active, Ca2+ store-independent mode. Mutants in the EF hand and cytoplasmic C terminus of STIM1 alter operational parameters of CRAC channels, including pharmacological profile and inactivation properties. Last, Ab externally applied to the STIM1 N-terminal EF hand blocks both ICRAC in hematopoietic cells and SOC-mediated Ca2+ entry in HEK293 cells, revealing that STIM1 has an important functional presence within the PM. The results reveal that, in addition to being an ER Ca2+ sensor, STIM1 functions within the PM to exert control over the operation of SOCs. As a cell surface signaling protein, STIM1 represents a key pharmacological target to control fundamental Ca2+-regulated processes including secretion, contraction, metabolism, cell division, and apoptosis.
Keywords: calcium signaling, calcium channel, patch-clamp, mast cells, T lymphocytes
Cytosolic Ca2+ signals control a wide array of cellular functions ranging from short-term responses such as contraction and secretion to longer-term regulation of cell growth and proliferation (1, 2). Ca2+ signals generated in response to receptors involve two closely coupled components: rapid, transient release of Ca2+ stored in the endoplasmic reticulum (ER), followed by slowly developing extracellular Ca2+ entry (1, 3–6). The initial Ca2+ release phase mediated by inositol 1,4,5-trisphosphate is well understood. The resulting depletion of Ca2+ stored within the ER lumen serves as the primary trigger for a message that is returned to the plasma membrane (PM), resulting in the activation of store-operated channels (SOCs), which mediate Ca2+ entry (3–6). The activation of SOCs is relatively slow (10–100 sec), and the Ca2+ entry phase of Ca2+ signals serves to mediate longer-term cytosolic Ca2+ elevations and provides a means to replenish intracellular stores (4–6). In certain cell types, including hematopoietic cells, SOCs carry a highly Ca2+-selective, nonvoltage-gated, inwardly rectifying current, termed the Ca2+ release-activated Ca2+ (CRAC) current (ICRAC) (3). The mechanism of coupling of depleted ER Ca2+ stores to activate SOCs and the nature of the channels mediating Ca2+ entry remain crucial but unresolved questions.
Recent screening approaches have revealed that the single membrane-spanning protein STIM1 plays an essential role in the activation of SOCs (7, 8). Reduction in the expression of STIM1 reduced SOC activation in response to store depletion (7, 8). The STIM1 protein is proposed primarily as a sensor of Ca2+ within stores. Thus, a single EF-hand Ca2+-binding motif located near the intraluminal N terminus of STIM1 may sense the decrease in free Ca2+ within stores (8, 9). The question of how this information is passed to the PM Ca2+ entry channels is important. Upon store emptying, it was observed that STIM1 undergoes a profound redistribution within the cell (8, 9). Thus, whereas labeled STIM1 normally appears to be uniformly distributed on the ER, store depletion leads to the appearance of densely labeled puncta, indicating that STIM1 becomes reorganized into spatially discrete areas after Ca2+ release from stores (8). Evidence suggested that, although these puncta were close to the PM, the STIM1 protein was not inserted into the PM (8). However, it was subsequently reported that store emptying results in a significant increase in the surface labeling of STIM1, suggesting that the protein may become translocated to the PM (9). Here we have determined that, in addition to being an ER sensor, STIM1 plays a crucial functional role in the PM, exerting control over the operation of SOCs.
Results and Discussion
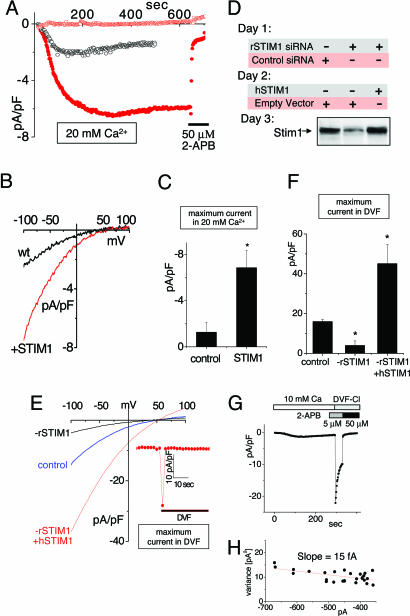
The STIM1 protein has been shown to be required for the function of SOCs and has been suggested to act in the ER as a sensor for luminal Ca2+ and, hence, to trigger the activation of Ca2+ entry channels (7–9). How STIM1 functions and what its role is in the activation of SOCs in the PM remain unresolved. Our studies addressed the question of whether STIM1 may have a direct role in controlling SOCs in the PM. Although SOC-mediated Ca2+ entry occurs in most cells, ICRAC is the only well characterized SOC current and is clearly operational in hematopoietic cells (3, 5, 6). Using Jurkat T cells, typical development of endogenous ICRAC in response to 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate (BAPTA)-induced store depletion is shown in Fig. 1A (black circles). The current becomes maximal at ≈100 sec, correlating with the slow BAPTA-driven depletion of stores The current–voltage (I–V) profile (Fig. 1B) reveals inward rectification and a reversal potential of approximately +50 mV, typical of this highly selective Ca2+ channel (3, 6). Overexpression of STIM1 in Jurkat T cells resulted in a large increase (average of 5.7-fold) in inward current density with the same I–V properties (Fig. 1 A–C). The increased channel activity had typical CRAC pharmacology being blocked by 50 μM 2-aminoethoxydiphenyl borate (2-APB) with the familiar current overshoot (Fig. 1A) (10, 11).
Fig. 1.
CRAC channel function correlates with altered expression of STIM1. (A) Development of ICRAC in Jurkat T cells overexpressing either STIM1 (red) or control empty vector (black). Current at −80 mV or +80 mV (red open circles) was measured in 20 mM extracellular Ca2+; addition of 50 μM 2-APB is indicated by the bar. (B) I–V profile determined at the time of maximal current in A. (C) Average maximal current at −100 mV in STIM1-overexpressing cells (n = 3) and control-expressing Jurkat T cells (n = 3). (D) Western analysis comparing STIM1 expression levels in RBL cells after knockdown and reexpression of STIM1. (E). The I–V profile for ICRAC was determined in DVF after maximal activation in Ca2+ex in control (blue), rSTIM1 knockdown (black), or rSTIM1 knockdown/hSTIM1-reexpressing RBL (red) cells. (Inset) Inactivation with overexpression of STIM1 WT in DVF. (F) Comparison of the average maximal current in DVF for control (n = 3), rSTIM1 knockdown (n = 3), and hSTIM1-reexpressing RBL (n = 4) cells. (G) ICRAC measured in rSTIM1 knockdown RBL cells reexpressing exogenous hSTIM1. Fifty micromolar 2-APB, but not 5 μM 2-APB, blocks the current in DVF. (H) Nonstationary noise analysis of the current in DVF from G to determine single-channel conductance (see Materials and Methods).
We also used rat basophilic lymphoma (RBL) cells to examine both short interfering RNA sequence (siRNA)-induced knockdown of endogenous rat STIM1 (rSTIM1) and reexpression of human STIM1 (hSTIM1) (Fig. 1 D–F). We recorded the I–V relationships after maximal activation by switching to divalent cation-free solution (DVF) to maximize the CRAC current (Fig. 1E). Treatment with rat-specific STIM1 siRNA reduced both CRAC current density and STIM1 protein levels by ≈75% (Fig. 1 D and E). In this rSTIM1 knockdown background, we were able to reexpress hSTIM1 and mutants thereof. hSTIM1 reexpression resulted in STIM1 protein and CRAC channel levels ≈10-fold above the knockdown WT (Fig. 1 D–F). Whereas CRAC current density was increased with overexpressed STIM1 in RBL cells, CRAC channel properties remained unchanged. Thus, removal of divalent cations induced a large, rapidly inactivating current, inhibited by 50 μM 2-APB but not 5 μM by 2-APB (Fig. 1G). Nonstationary noise analysis of the current traces during inactivation revealed a linear variance vs. current dependence (Fig. 1H), the slope giving a single-channel current of ≈15 fA, close to the reported value for endogenous ICRAC (6).
These gain-of-function results reveal an intriguing correlation between CRAC current and STIM1 expression. From Ca2+ imaging studies, Roos et al. (7) reported that high STIM1 overexpression resulted in only small additional Ca2+ entry. This was taken as evidence against STIM1 functioning as a component of the channel itself (7). However, in imaging studies, steady-state Ca2+ measurements reflect a combination of Ca2+ influx and efflux, and increased CRAC-mediated Ca2+ entry is efficiently removed by Ca2+ pumping (12). Hence, only current measurements reflect actual channel activity. Our data indicate that overexpression of STIM1 results in a substantial increase in CRAC channel activity, suggesting that STIM1 may be increasing the number of channels or their open probability. Hence, STIM1 may be more intimately linked with expression and/or function of channels in the PM, as opposed to playing only a sensing role in the ER. The earlier clear evidence that STIM1 is localized to the PM as well as the ER (13, 14) provides further support for the concept that it may play a role in the PM. This view is strengthened by the recent evidence that STIM1 can be translocated into the PM after store depletion (9).
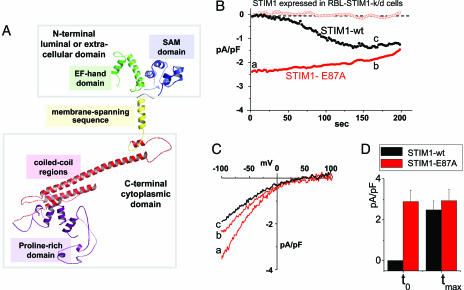
The STIM1 protein has several distinct functional domains, depicted in Fig. 2A. It exists as a membrane protein in both the ER and PM (13, 14) and may form multimers through interactions among the extensive cytoplasmic C-terminal coiled-coil regions (13, 15). The proposed ER Ca2+-sensing function seems to be mediated by the acidic 76–87 N-terminal consensus EF-hand Ca2+-binding sequence (8) within the ER lumen. Mutation of this region (D76A) was shown to cause a translocation of STIM1 from the ER toward the PM without store depletion, a movement exactly mimicking the translocation of the WT STIM1 protein in response to store emptying (8). In this study, there was a small but significant increase in Mn2+ influx that correlated with STIM1 translocation (8). In their recent study, Zhang et al. (9) revealed that expression of EF-hand mutants of STIM1 in Jurkat T cells resulted in a constitutive increase in Ca2+ entry. It was crucial to determine whether actual channel function was modified as a result of STIM1 EF-hand changes. We investigated whether the specific store-operated current, ICRAC, was altered by mutating the Ca2+-sensing EF hand of STIM1. We mutated E87A in STIM1 to reduce the negative charge on the EF hand and to lower its Ca2+-binding affinity. Expression of hSTIM1 E87A in the RBL rSTIM1 knockdown cells resulted in a profound alteration in ICRAC channel activation. After store depletion, ICRAC develops with the classical slow time course, requiring tens of seconds (3, 5, 6). This development is clearly shown in Jurkat T cells (Fig. 1A) and is similar in RBL cells reexpressing WT hSTIM1 (Fig. 2B, black circles). Importantly, in RBL cells expressing the hSTIM1 E87A mutant replacing endogenous rSTIM1, ICRAC was already fully activated at the moment of break-in, that is, without store depletion. The I–V profile for the E87A mutant immediately after break-in (Fig. 2C, time point a) or after 170 sec (Fig. 2C, time point b) was the same as for cells expressing WT hSTIM1 after maximal activation by store depletion at 170 sec (Fig. 2C, time point c). Maximal current density for CRAC for WT hSTIM1-reexpressing cells was not significantly different from maximal current in E87A-reexpressing cells (Fig. 2D).
Fig. 2.
Expression of the E87A STIM1 mutant with decreased EF-hand Ca2+-binding affinity leads to constitutively active ICRAC in RBL cells. (A) Predicted 3D structure of STIM1 (57–591) generated by using phyre software (www.sbg.bio.ic.ac.uk/∼phyre). The domain structures shown, EF-hand, sterile-α motif (SAM), transmembrane-spanning region, coiled-coil regions, and proline-rich N terminus, were determined by similarity to proteins with known structure. (B) Either WT hSTIM1 or the E87A mutant with predicted lowered EF-hand Ca2+ affinity were reexpressed in rSTIM1 knockdown RBL cells. In contrast to the slow-developing store depletion-dependent current observed with STIM1 WT (black circles), ICRAC was constitutively active at the time of break-in (to) with STIM1 E87A-reexpressing cells (−80 mV, red closed circles; +80 mV, red open circles). (C) I–V profiles of STIM1 WT or STIM1 E87A measured at the time points indicated by a, b, and c in B are similar to endogenous ICRAC, with high positive reversal potential in 10 mM extracellular Ca2+, reflecting Ca2+ selectivity. (D) Comparisons of average current observed at to reveal significant differences between STIM1 WT(n = 3) and STIM1 E87A (n = 3), but not in maximal activation (tmax).
This result provides direct evidence that the CRAC channel itself has undergone a fundamental change as a result of a mutation in STIM1 that removes its sensing of changes in ER luminal Ca2+. Indeed, with this result, the highly specific CRAC current has been shown to be fully and normally activated while Ca2+ stores are replete. The normal, slow time-dependence of ICRAC development may reflect the time required for reorganization of STIM1 in the ER and/or insertion into the PM. This would imply that the EF-hand mutant had already undergone the reorganization or translocation step.
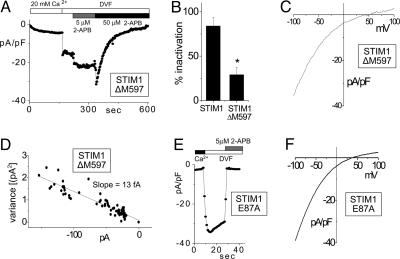
Considering the apparently close link between STIM1 and the function of PM CRAC channels, we investigated how intimately involved STIM1 is in CRAC channel function by examining the influence of mutations of STIM1 on either side of its transmembrane region. The cytoplasmic STIM1 C terminus contains a structurally significant proline-rich domain and a number of consensus phosphorylation sites (13). We expressed a C-terminal ΔM597 deletion mutant lacking a potential protein kinase C phosphorylation site (613 SSR) and two potential casein kinase II phosphorylation sites (620 SSPD and 660 SIGE). In Jurkat T cells, overexpression of ΔM597 caused a significant change in the CRAC channel function (Fig. 3A). The channel activated normally with passive store depletion in the presence of external Ca2+. Conductance increased with application of DVF; however, the rapid inactivation normally seen was substantially reduced (Fig. 3 A and B). This rapid inactivation in DVF is a well described parameter of CRAC channel function (6, 16); hence, this change represents a significant alteration in channel operation. Despite this, the pharmacology of the channel remained unchanged. The well described potentiation with 5 μM 2-APB and blockade with 50 μM 2-APB (10, 11) were clearly observed (Fig. 3A). Other CRAC channel properties remained unchanged, such as the I–V profile (Fig. 3C) and the single-channel conductance (Fig. 3D). Thus, the STIM1 ΔM597 deletion causes alteration in the operation of CRAC channels, suggesting that the STIM1 molecule is either functioning with the channel or modifying some regulatory component of the channel.
Fig. 3.
Expression of STIM1 mutants alters the pharmacology and inactivation properties of ICRAC. (A) Measured at a holding potential of −80 mV, ICRAC failed to inactivate in DVF in Jurkat T cells overexpressing the STIM1-ΔM597 deletion mutant, as distinct from endogenous ICRAC (see Fig. 4A Inset) or ICRAC measured after overexpression of hSTIM1 in RBL cells (Fig. 1G). In contrast, other ICRAC properties were similar to endogenous current: activation with passive Ca2+ store depletion, potentiation by 5 μM 2-APB, and inhibition by 50 μM 2-APB. (B) One minute after exposure to DVF, which followed maximal CRAC activation in 20 mM extracellular Ca2+, ICRAC showed ≈25% inactivation when STIM1-ΔM597 was overexpressed (n = 6), significantly different from the ≈80% inactivation observed in STIM1 WT-overexpressing cells (n = 3; P < 0.05). (C) The I–V curve for ICRAC in DVF was unaltered by overexpression of STIM1-ΔM597 (compared with STIM1 WT; Fig. 1E). (D) Assessment of the single-channel conductance by noise analysis (as in Fig. 1H) at −100 mV during inhibition by 2-APB in A revealed no differences from the single-channel current measured for STIM1 WT (Fig. 1H). (E) hSTIM1 E87A mutant was overexpressed after knockdown of rSTIM1 in RBL cells. Sensitivity of ICRAC to 2-APB was greatly altered: ICRAC was immediately inhibited by 5 μM 2-APB, as opposed to the normal potentiation seen in A. (F) Overexpression of STIM1 E87A did not result in any changes in the I–V curve of ICRAC in DVF as compared to STIM1 WT (see Fig. 1E).
Further examination of the STIM1 E87A EF-hand mutation revealed an important alteration in CRAC channel pharmacology. In rSTIM1 knockdown RBL cells overexpressing the E87A EF-hand point mutant, there was a significant modification in the action of 2-APB on CRAC channel function. As shown in Fig. 3E, the addition of 5 μM 2-APB caused a rapid block of CRAC channel activity, in clear contrast to the potentiation seen with 5 μM 2-APB in previous studies (10, 11) and the data in Fig. 3A. The mutation also caused a lower inactivation in DVF, as for the STIM1 ΔM597 truncation mutant, suggesting that multiple domains may contribute to the inactivation process. The I–V profile for CRAC in DVF with the EF-hand mutant (Fig. 3F) was similar to that with control cells (Fig. 1E). Although there seemed to be a slight decrease in the reversal potential, the significance of this change has not yet been determined. The alteration in 2-APB action indicates that the STIM1 mutation has modified the operation of the CRAC channel, again suggesting some more direct link between STIM1 and CRAC channel function. 2-APB is believed to modify CRAC channel activity by binding to a site on the outer surface of the PM (10). Although there is no evidence that 2-APB directly interacts with either the CRAC channel or STIM1, it is noteworthy that the EF hand is located on the outer surface when STIM1 is expressed in the PM; hence, it could be associated with the site of interaction of 2-APB.
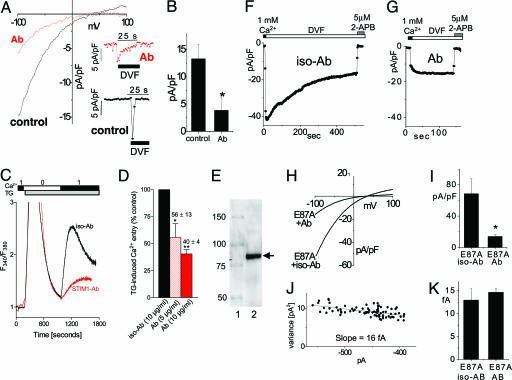
Many of the experiments described above provide circumstantial evidence for operation of the STIM1 protein either within or close to the PM, possibly even as a subunit of the SOC. This hypothesis derives further support from experiments using a STIM1 N-terminal Ab (Fig. 4). We used a purified N-terminal-specific STIM1 mAb raised against the 25–139 STIM1 sequence comprising mostly the helix–loop–helix EF-hand domain. Applied to the outside of intact Jurkat T cells, the Ab caused a substantial suppression of CRAC channel current (Fig. 4A). In this case, ICRAC was measured after activation in 20 mM Ca2+ followed by DVF to maximize current. Application of Ab (20 μg/ml) 30 min before break-in suppressed the maximal endogenous CRAC channel activity observed in DVF (Fig. 4A Inset), whereas the I–V profile was essentially unchanged (Fig. 4A). The Ab suppressed maximal current in DVF by an average of 70% (Fig. 4B). The matching mouse IgG2a isotype Ab at 20 μg/ml had no effect (data not shown). The Ab also seemed to reduce the rate of inactivation in DVF. A similar suppression of ICRAC by the same mAb was observed in the rSTIM1 knockdown RBL cells reexpressing human STIM1 E87A (see below). Whereas CRAC channels are observable only in hematopoietic cells, SOC activity is observed universally among cell types (5, 6). We assessed whether SOC-mediated Ca2+ entry is similarly modified by the STIM1 mAb in HEK293 cells. The data in Fig. 4C reveal that thapsigargin (TG)-induced Ca2+ entry in HEK293 cells was suppressed by a similar margin. At 10 μg/ml there was almost 70% inhibition of Ca2+ entry, the rate of entry being substantially slowed compared with isotype control (Fig. 4C), the latter having no effect on Ca2+ entry. As shown in Fig. 4D, the mAb effect was dose-dependent with almost 50% inhibition at 5 μg/ml. The specificity of the mAb for STIM1 seemed high: by Western analysis there was no obvious cross-reaction with other proteins, as shown, for example, in WT Jurkat T cells (Fig. 4E). When using HEK293 cells in which STIM1 was overexpressed, the effect of the STIM1 mAb was at least as effective (data not shown). We also examined the action of the STIM1 mAb on the STIM1 EF-hand mutant. As shown in Fig. 4 F and G, the mAb blocked a substantial portion of current in the E87A EF-hand mutant-expressing RBL cells. The EF-hand mutant still retained its high 2-APB sensitivity (Fig. 4 F and G). The I–V profile remained similar (Fig. 4H), and the average mAb block was ≈80% compared with the isotype control (Fig. 4I). The distinctive single-channel properties of CRAC channels remained the same with EF-hand mutant reexpression, measured as 16 fA at −100 mV (Fig. 4J), nor did Ab applied to the EF-hand mutant-reexpressing cells significantly alter the single-channel current (Fig. 4K). These data suggest that Ab blockade is more likely to reduce the number of channels rather than their open probability.
Fig. 4.
ICRAC and store-operated Ca2+ entry were inhibited by an anti-N-terminal STIM1 (25–139) mAb. Ab is depicted in red, and control (with or without isotype Ab) is depicted in black. (A–J) Jurkat T cells (A and B), HEK293 cells (C and D), or rSTIM1 knockdown RBL cells reexpressing hSTIM1 E87A mutant (F–K) were incubated for 30–60 min with STIM1 mAb or IgG2a isotype control Ab. (A) I–V curves for ICRAC were measured in DVF after maximal activation in Ca2+ in the presence or absence of 20 μg/ml mAb. Current inhibited by mAb was similar to control with respect to inward rectification and reversal potential. (Inset) Time course of ICRAC at −80 mV, measured after maximal activation. (B) Average maximal current measured at −100 mV was decreased by ≈70% in the presence of 20 μg/ml mAb compared with control (n = 3; P < 0.05). (C) SOC-mediated Ca2+ entry assessed in fura-2/acetoxymethyl ester-loaded HEK293 cells was similarly inhibited by the same mAb. No Ca2+ entry was observed with transient Ca2+ addition before 2 μM TG addition. Addition of Ca2+ after TG-induced store depletion revealed a reduction in SOC-induced Ca2+ entry in mAb-pretreated cells. (D) The effect of mAb on SOC was dose-dependent: ≈45% (P < 0.05) reduction at 5 μg/ml and ≈60% (P < 0.01) reduction at 10 μg/ml. (E) Western analysis of Jurkat T cell lysate using the same STIM1 mAb revealed only a single major band corresponding to the STIM1 protein. Lanes: 1, Size standards (kDa); 2, Jurkat T cell lysate. (F–K) ICRAC in hSTIM1 E87A-reexpressing RBL cells after rSTIM1 knockdown was inhibited by STIM1 Ab. (F) Incubation in isotype control Ab did not induce a change in ICRAC compared with control cells (Fig. 3E). (G) Incubation with mAb reduced ICRAC. Subsequent addition of 5 μM 2-APB resulted in complete blockade of the remaining current. (H) I–V curves of the cells shown in F and G in DVF were similar in inward rectification and reversal potential. (I) Average maximal current measured at −100 mV was decreased by ≈80% in the presence of mAb (n = 3; P < 0.05). (J) Noise analysis of the cell in G showed a preserved single-channel current of 16 fA, typical for ICRAC. (K) No change in the single-channel conductance was observed for the residual current after the mAb block (n = 3), compared with the isotype control (n = 3).
Overall, these data provide some perspectives on the role of STIM1 in SOC activation. Recent studies have indicated that STIM1 functions within the ER as a Ca2+ sensor and becomes translocated toward the PM (7–9). After store depletion, STIM1 is reorganized in the ER into vesicular structures, or puncta, close to the PM (8). Although it was first reported that there was no evidence of STIM1 insertion into the PM (8), subsequent biotinylation experiments revealed some STIM1 appearing at the cell surface after sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA) pump inhibition (9). An insertional model based on this information is depicted in Fig. 5A–D. The blocking effect of the STIM1 Ab shown here provides strong evidence that STIM1 has a functional presence in the PM. However, as yet there are no data to support the hypothesis that translocation of STIM1 into the PM is required for the activation of SOCs. Indeed, there has previously been little evidence to support any role for vesicle trafficking in the activation of SOCs (5, 6, 17). Importantly, the earlier studies of Dziadek and coworkers (14, 15) revealed that a significant proportion of STIM1 is in the PM in normal cells. Indeed, using N-terminal STIM1 Abs, our FACS analysis of STIM1 expression in normal and STIM1-overexpressing cells revealed a substantial presence of STIM1 in the PM (data not shown). Here, the use of the N-terminal mAb to block SOC function involved treating cells with Ab and then removing the Ab before measurement of CRAC or SOC activity. Thus, the Ab likely is targeting preexisting STIM1 in the PM. Compatibly with the insertional model, it is possible that Ab bound to preexisting PM STIM1 might have a dominant modifying action on the formation or function of the STIM1 multimers required for SOC activation. It is established that STIM1 can form multimers and that the C-terminal coiled-coil region mediates this interaction (15). However, without the information that STIM1 insertion is required for SOC activation, we could also consider an alternative model. Thus, although some insertion of STIM1 into the PM may be measurable upon store emptying, it is possible that this is not required for the activation of SOCs. Instead, as shown as an alternative model in Fig. 5 A, B, E, and F, channel activation may result from an “influence” of the reorganized ER STIM1 molecules on the PM, through interactions with preexisting PM STIM1 molecules and possibly the entry channel itself. Thus, through a conformational coupling process, aggregated STIM1 molecules on the ER may activate the entry channels without necessarily becoming inserted into the PM. Such a conformational coupling model would provide a simpler explanation of the STIM1 Ab results and would be compatible with much earlier speculation and evidence that close interactions, but not necessarily fusion, between the ER and PM are involved in the activation of SOCs (5, 6, 18–20).
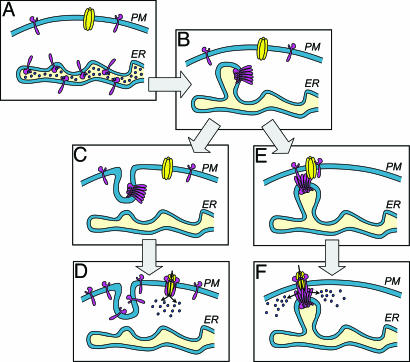
Fig. 5.
Models for the proposed actions of STIM1 in the activation of SOCs. (A) Cell at rest with ER stores filled with Ca2+. STIM1 molecules (pink) with Ca2+ bound are shown predominantly in the ER but also appear in the PM. The SOC (yellow) is closed. (B) Upon Ca2+ depletion of the ER, the ER STIM1 proteins become aggregated into puncta, shown as distinct regions of the ER close to the PM. (C) In the “insertional” model, the STIM1 protein is translocated and inserted into the PM. (D) The high content of STIM1 in the PM is sufficient to activate SOCs; this is shown as a hypothetical interaction between STIM1 and SOCs. (E) In the “influence” model, the aggregated STIM1 in puncta near the PM conformationally couples to STIM1 preexisting in the PM and causes association and reorganization of STIM1 in the PM. (F) This reorganization induces formation of a similar hypothetical complex between STIM1 and SOCs, leading again to channel activation.
Our data using the STIM1 EF-hand mutant provide direct evidence that STIM1, as a sensor for Ca2+ within the ER, is triggering the CRAC SOC itself. Our results also reveal that expression of STIM1 causes a gain of CRAC channel function. Moreover, mutations in STIM1 alter some significant characteristics of CRAC channel operation, including pharmacology and inactivation properties. These data, together with the information that STIM1 is functioning within the PM, suggest that STIM1 might operate as a regulatory component of the channel. Although we cannot exclude the possibility that it is the channel per se, the single transmembrane-spanning STIM1 protein does not resemble known channel proteins. However, its structure and role are reminiscent of the MinK-related peptides functioning as ancillary or β subunits of Kv channels (21). These single transmembrane proteins confer changes in the conductance, gating kinetics, and pharmacology of Kv channels and are crucial to normal channel function (21).
As an equivalent regulatory subunit of SOCs, STIM1 may lead to identification of other SOC components. Last, it seems that this single molecule, STIM1, may fulfill both ER sensing and PM channel activation functions. Thus, STIM1 would be an interesting example of a protein undertaking two crucial functions in two different membranes.
Materials and Methods
Reagents.
ATP and 2-APB were from Sigma. Fura-2/acetoxymethyl ester and stealth siRNA duplexes for rSTIM1 and hSTIM1 were from Invitrogen. Anti-STIM1 (25–139), IgG2a isotype control Ab, and enhanced yellow fluorescent protein (YFP) vector were from BD Biosciences (Mountain View, CA). TG was from EMD Biosciences (San Diego).
RNA Interference Design.
All siRNA sequences were designed by using Invitrogen block-it software. A mixture of two siRNA duplex sequences exclusively targeting rSTIM1, but not hSTIM1, were used: (i) start nucleotide 935, GCAUGGAAGGCAUCAGAAGUGUAUA, and (ii) start nucleotide 970, GGAUGAGGUGAUACAGUGGCUGAUU. The control double-stranded RNA sequence, GGUUCUCCACCUUUAUAGGUGGCUU, was used as negative control. Transfections were performed by electroporation using the Gene Pulser II electroporation system (Bio-Rad) at 350 V, 960 μF, and infinite resistance.
DNA Expression Constructs and Mutagenesis.
The 2.7-kb BamHI fragment of hSTIM1 (GenBank accession no. U52426) was subcloned into pIRESneo (Clontech) either as a WT cDNA fragment or after introduction of a premature stop codon (ΔPEST) as described (15), producing a 597-aa ΔPEST protein without the two proline-rich regions (residues 617–634 and 651–671) of STIM1 (STIM1-ΔM597). The hSTIM1 E87A mutation was introduced by using the QuikChange site-directed mutagenesis kit (Stratagene) and confirmed by sequencing. Cells were cotransfected with yellow fluorescent protein (YFP) plus vectors described by electroporation, and YFP-expressing cells were selected by fluorescence (22).
Electrophysiology.
We used conventional whole-cell recordings as described (23). The cells were transferred into the recording chamber on poly(l-lysine)-coated coverslips before the experiment and allowed to settle. Immediately after establishment of the whole-cell configuration, voltage ramps of 50-msec duration spanning a voltage range from −100 to +100mV followed by a 500-msec step to −100 mV were delivered from a holding potential of 0 mV at a rate of 0.5 Hz. Currents were filtered at 6 kHz and sampled at 50-μsec intervals. 200-msec segment steps at −100 mV were used for nonstationary noise analysis (24, 25). We used the automatic capacitive and series resistance compensation of an EPC 10 amplifier (HEKA Electronics, Lambrecht/Pfalz, Germany). patchmaster, pulsetools, fitmaster, and origin software (HEKA Electronics) were used for acquisition and analysis. The temporal development of inward (−80 mV) and outward (+80 mV) currents were measured from the individual ramps at each time point. 2-APB was added to the external solution as indicated. STIM1 and isotype control Ab (20 μg/ml) were added 30–60 min before experiments. The intracellular solution contained the following (in mM): 145 CsGlu, 10 Hepes, 10 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate (BAPTA), 8 Na, 1 Mg, and 4 Mg-ATP (total 8 mM Mg) at pH 7.2. To inhibit TRPM7, 8 mM Mg and ATP were used (25). For extracellular solution A we used the following: 130 mM CsGlu, 20 mM CaGlu2, 2 mM MgCl2, and 10 mM Hepes at pH 7.4 or 160 mM NaGlu, 10 mM EDTA, and 10 mM Hepes at pH 7.4 (DVF). For extracellular solution B we used the following: 145 mM NaCl, 2.8 mM KCl, 10 mM CsCl, 10 mM Hepes, and 10 mM glucose at pH 7.4 with 2 mM EDTA (DVF-Cl) or 2 mM MgCl2 and CaCl2 (as indicated). We applied a 10-mV junction potential compensation for Cl-based external solutions and used a salt bridge for glutamate-based external solutions. All I–V profiles were filtered offline at 1 kHz. The I–V curves in DVF were derived from the current ramp at the time of maximum activation defined from the current–time dependence at −80 mV. All I–V curves were shown at the time of maximum activation. The current after channel inactivation or block by 2-APB (average of three to five traces) was used for leak subtraction. For all I–V curves in high Ca, with the exception of the EF-hand mutant (where a control cell was used), an average of the first three to five traces after establishment of whole-cell recording was used for leak subtraction. Anti-STIM1 Ab was preadded for 30 min at 22°C. The maximum CRAC currents at −100 mV were used for statistical analysis. Each statistical bar plot (Figs. 1 C and F, 2D, and 4 B and I) is an average of three or more experiments. The star on the bar plots represents significant difference from control (P < 0.05).
Western Blot.
Proteins were resolved on 6% SDS/PAGE gels (26) and electroblotted onto nitrocellulose membranes (Bio-Rad) (27). After transfer, nitrocellulose membranes were blocked (for l h at room temperature) in Tris-buffered saline/Tween 20 (TBST) (10 mM Tris·HCl, pH 8.0/150 mM NaCl/0.1% Tween 20) containing membrane-blocking agent (5%; Amersham Pharmacia), then incubated (overnight at 4°C) with mouse anti-hSTIM1 Ab (BD Biosciences). After washing, membranes were incubated (for 30 min) with secondary Ab (horseradish peroxidase-conjugated goat anti-mouse IgG; 1:2,500 in TBST) and washed. Peroxidase activity was visualized with enhanced chemiluminescence.
Ca2+ Measurements.
Cells grown on coverslips were placed in “cation-safe” medium free of sulfate and phosphate anions (107 mM NaCl/7.2 mM KCl/1.2 mM MgCl2/11.5 mM glucose/20 mM Hepes-NaOH, pH 7.2) and loaded with fura-2/acetoxymethyl ester (2 μM) for 30 min at 20°C. Cells were washed, and dye was allowed to deesterify for a minimum of 30 min at 20°C. Cells on coverslips were placed in cation-safe medium in the absence or presence of 1 mM CaCl2 (28, 29). Ca2+ measurements used an InCyt dual-wavelength fluorescence imaging system (Intracellular Imaging, Cincinnati). Fluorescence emission at 505 nm was monitored with excitation at 340 and 380 nm; intracellular Ca2+ measurements are shown as 340- to 380-nm ratios. Store-operated Ca2+ entry was induced by incubation with 2 μM TG. All traces are averages from multiple (20–50) cells and are representative of at least three separate experiments. Anti-STIM1 Ab was added at 4°C 30–40 min before the experiments.
Acknowledgments
We thank Drs. David J. Weber, Sean Tang, and Martin F. Schneider for helpful advice. This work was supported by National Institutes of Health Grants HL55426 and AI058173 (to D.L.G.) and the Interdisciplinary Training Program in Muscle Biology of the University of Maryland School of Medicine (to M.A.S.).
Abbreviations
- ER
endoplasmic reticulum
- PM
plasma membrane
- SOC
store-operated channel
- CRAC
Ca2+ release activating Ca2+
- ICRAC
CRAC current
- I–V
current–voltage
- 2-APB
2-aminoethoxydiphenyl borate
- RBL
rat basophilic lymphoma
- siRNA
short-interfering RNA sequence
- DVF
divalent cation-free solution
- TG
thapsigargin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Berridge M. J., Lipp P., Bootman M. D. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Berridge M. J., Bootman M. D., Roderick H. L. Nat. Rev. Mol. Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 3.Parekh A. B., Penner R. Physiol. Rev. 1997;77:901–930. doi: 10.1152/physrev.1997.77.4.901. [DOI] [PubMed] [Google Scholar]
- 4.Putney J. W., Jr., Broad L. M., Braun F. J., Lievremont J. P., Bird G. S. J. Cell Sci. 2001;114:2223–2229. doi: 10.1242/jcs.114.12.2223. [DOI] [PubMed] [Google Scholar]
- 5.Venkatachalam K., van Rossum D. B., Patterson R. L., Ma H. T., Gill D. L. Nat. Cell Biol. 2002;4:E263–E272. doi: 10.1038/ncb1102-e263. [DOI] [PubMed] [Google Scholar]
- 6.Parekh A. B., Putney J. W., Jr. Physiol. Rev. 2005;85:757–810. doi: 10.1152/physrev.00057.2003. [DOI] [PubMed] [Google Scholar]
- 7.Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., et al. J. Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Meyer T. Curr. Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S. L., Yu Y., Roos J., Kozak J. A., Deerinck T. J., Ellisman M. H., Stauderman K. A., Cahalan M. D. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prakriya M., Lewis R. S. J. Physiol. 2001;536:3–19. doi: 10.1111/j.1469-7793.2001.t01-1-00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma H. T., Venkatachalam K., Parys J. B., Gill D. L. J. Biol. Chem. 2002;277:6915–6922. doi: 10.1074/jbc.M107755200. [DOI] [PubMed] [Google Scholar]
- 12.Bautista D. M., Lewis R. S. J. Physiol. 2004;556:805–817. doi: 10.1113/jphysiol.2003.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams R. T., Manji S. S., Parker N. J., Hancock M. S., Van Stekelenburg L., Eid J. P., Senior P. V., Kazenwadel J. S., Shandala T., Saint R., et al. Biochem. J. 2001;357:673–685. doi: 10.1042/0264-6021:3570673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manji S. S., Parker N. J., Williams R. T., Van Stekelenburg L., Pearson R. B., Dziadek M., Smith P. J. Biochim. Biophys. Acta. 2000;1481:147–155. doi: 10.1016/s0167-4838(00)00105-9. [DOI] [PubMed] [Google Scholar]
- 15.Williams R. T., Senior P. V., Van Stekelenburg L., Layton J. E., Smith P. J., Dziadek M. A. Biochim. Biophys. Acta. 2002;1596:131–137. doi: 10.1016/s0167-4838(02)00211-x. [DOI] [PubMed] [Google Scholar]
- 16.Prakriya M., Lewis R. S. Cell Calcium. 2003;33:311–321. doi: 10.1016/s0143-4160(03)00045-9. [DOI] [PubMed] [Google Scholar]
- 17.Bakowski D., Burgoyne R. D., Parekh A. B. J. Physiol. 2003;553:387–393. doi: 10.1113/jphysiol.2003.055335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berridge M. J. Biochem. J. 1995;312:1–11. doi: 10.1042/bj3120001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson R. L., van Rossum D. B., Gill D. L. Cell. 1999;98:487–499. doi: 10.1016/s0092-8674(00)81977-7. [DOI] [PubMed] [Google Scholar]
- 20.Berridge M. J. Sci. STKE. 2004 Jul 27; doi: 10.1126/stke.2432004pe33. 10.1126/stke.2432004pe33. [DOI] [PubMed] [Google Scholar]
- 21.McCrossan Z. A., Abbott G. W. Neuropharmacology. 2004;47:787–821. doi: 10.1016/j.neuropharm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 22.Venkatachalam K., Zheng F., Gill D. L. J. Biol. Chem. 2003;278:29031–29040. doi: 10.1074/jbc.M302751200. [DOI] [PubMed] [Google Scholar]
- 23.Spassova M., Eisen M. D., Saunders J. C., Parsons T. D. J. Physiol. 2001;535:689–696. doi: 10.1111/j.1469-7793.2001.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinemann S. H., Conti F. Methods Enzymol. 1992;207:131–148. doi: 10.1016/0076-6879(92)07009-d. [DOI] [PubMed] [Google Scholar]
- 25.Prakriya M., Lewis R. S. J. Gen. Physiol. 2002;119:487–508. doi: 10.1085/jgp.20028551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli U. K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Towbin H., Gordon J. J. Immunol. Methods. 1984;72:313–340. doi: 10.1016/0022-1759(84)90001-2. [DOI] [PubMed] [Google Scholar]
- 28.Patterson R. L., van Rossum D. B., Ford D. L., Hurt K. J., Bae S. S., Suh P. G., Kurosaki T., Snyder S. H., Gill D. L. Cell. 2002;111:529–541. doi: 10.1016/s0092-8674(02)01045-0. [DOI] [PubMed] [Google Scholar]
- 29.van Rossum D. B., Patterson R. L., Sharma S., Barrow R. K., Kornberg M., Gill D. L., Snyder S. H. Nature. 2005;434:99–104. doi: 10.1038/nature03340. [DOI] [PubMed] [Google Scholar]