Abstract
Identifying sequence determinants of fibril-forming proteins is crucial for understanding the processes causing >20 proteins to form pathological amyloid depositions. Our approach to identifying which sequences form amyloid-like fibrils is to screen the amyloid-forming proteins human insulin and β2-microglobulin for segments that form fibrils. Our screen is of 60 sequentially overlapping peptides, 59 being six residues in length and 1 being five residues, covering every noncysteine-containing segment in these two proteins. Each peptide was characterized as amyloid-like or nonfibril-forming. Amyloid-like peptides formed fibrils visible in electron micrographs or needle-like microcrystals showing a cross-β diffraction pattern. Eight of the 60 peptides (three from insulin and five from β2-microglobulin) were identified as amyloid-like. The results of the screen were used to assess the computational method, and good agreement between prediction and experiments was found. This agreement suggests that the pair-of-sheets, zipper spine model on which the computational method is based is at least approximately correct for the structure of the fibrils and suggests the nature of the sequence signal for formation of amyloid-like fibrils.
Keywords: fibrils, structure
Amyloid fibrils may be defined as elongated, unbranched protein fibrils that accumulate in the extracellular space of various tissues in connection with disease and that bind Congo red (CR) with characteristic birefringence (1). For example, the intact globular proteins (2) lysozyme and β2-microglobulin (β2m) are deposited as amyloid in hereditary systemic amyloidosis and dialysis-related amyloidosis, whereas type II diabetes is associated with the peptide known as islet amyloid polypeptide (3) and injection amyloidosis with deposits of insulin (4). Human insulin and β2m are two of the smaller amyloid-forming proteins, and here we have searched systematically for hexameric segments of these proteins that form fibrils or related needle-like microcrystals in isolation from the rest of the protein.
The premise of our work is that small segments of proteins are capable of forming amyloid-like fibrils. In fact, there are numerous examples of small peptides (5–9), some as short as three or four residues, that form fibrils (10, 11). How such short peptides can form fibrils was illuminated by the crystal structures of the amyloid-like peptides of sequence NNQQNY and GNNQQNY (12). That study found that the protofibril is built from a pair of β-sheets, whose side chains intermesh at a dry “steric zipper.” That is, the protofibril is an elongated pair of β-sheets, with the β-strands running perpendicular to the fibril axis. Thus a fibril-forming segment need be only as long as the β-sheet is wide.
Amyloid-forming proteins are not in general homologs of each other, and in fact it has been challenging to find any strong sequence signal for the formation of amyloid-like fibrils. Short peptides offer the opportunity for systematic investigation of the sequence determinants of fibril formation, as in the work of Lopez de la Paz and Serrano (6), who varied residues at each position of a known hexameric fibril-forming peptide. Here, we take an alternative strategy of studying which hexameric peptides from the amyloid-forming proteins human insulin [INS_HUMAN (P01308) and β2m (B2MG_HUMAN (P61769)] themselves form fibrils. These hexamers, which cover all noncysteine residues of these two proteins, are varied in sequence (see Table 2, which is published as supporting information on the PNAS web site). Because some of these segments are fibril-forming and many others are not, our study is consistent with the idea that there is a strong sequence signature for fibril formation. By comparing our experimental observations to predictions with the 3D profile algorithm of ref. 13, we are able to draw conclusions about the nature of this sequence signature.
Results
Definition of Amyloid-Like.
We characterized peptide hexamers by EM, the binding of CR and thioflavin T (ThT), and x-ray diffraction. We consider hexamers to be amyloid-like only if they form straight, unbranched fibrils, 5–15 nm in diameter, or if they form needle-like microcrystals (Fig. 1D and F) that display the characteristic cross-β diffraction pattern (see Fig. 3, which is published as supporting information on the PNAS web site). Although we characterized every peptide by colorimetric and fluorimetric assays with CR or ThT (Table 3, which is published as supporting information on the PNAS web site), the correlation of these assays with the visible formation of fibrils and microcrystals was not high. Also the correlation of predictions from the 3D profile method with these assays was not as strong as with fibril and crystal formation. For these reasons, we do not discuss the detailed results of our colorimetric and fluorimetric assays.
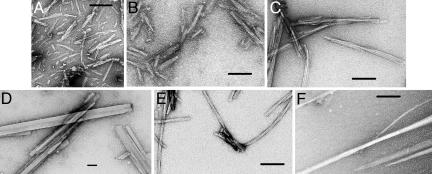
Fig. 1.
Electron micrographs showing fibrils (A–C and E) and needle-shaped microcrystals (D and F) formed by insulin and β2m and hexameric peptides from their sequences. (A) β2m91:KIVKWD. (B) β2m83:NHVTLS. (C) Full-length β2m. (D) β2m62:FYLLYY. (E) Full-length insulin. (F) IB12:VEALYL. The number following the protein name is the position in the protein sequence of the first residue. The sequence of the hexameric peptide follows. The length of the calibration bars is 100 nm.
Solution Conditions for the Screen.
Because both insulin and β2m form fibrils at low pH (14, 15), we chose pH 2.5 for the fibril formation screen. Fibril formation of both insulin and β2m is affected by the ionic strength and type of the salt present (14, 16). Insulin usually forms fibrils at low ionic strength (16). β2m fibril formation requires salt concentrations >100 mM (14). To have uniform conditions for the fibril screen, we chose to use 150 mM NaCl consistent with physiological conditions.
Systematic Survey of Amyloid Formation with Hexameric Segments from Insulin and β2M.
The one pentameric and 16 hexameric peptides from the insulin sequence and the 43 hexameric peptides from the β2m sequence were dissolved and inspected for fibrils or microcrystals (see Table 3). Three of the peptides from insulin and five of the peptides from β2m form fibrils or elongated microcrystals and qualify as amyloid-like by the definition of this article (Fig. 1 and Table 1).
Table 1.
The 18 six-residue peptides from insulin (I) and β2m predicted by the 3D profile method to form fibrils
| Protein, hexamer first residue, and sequence | Amyloid-like fibrils or needles* | X-ray diffraction characterization | Comment‡ |
|---|---|---|---|
| I A12: SLYQLE | A 9-residue peptide spanning residues A12 to A20 forms fibrils | ||
| I A13: LYQLEN | Needles | Needle-like crystals grew in sample buffer† and gave cross-β pattern | Predicted at threshold −19 kcal/mol |
| I A14: YQLENY | A 9-residue peptide spanning residues A12 to A20 forms fibrils | ||
| I B1: FVNQHL | |||
| I B8: GSHLVE | |||
| I B9: SHLVEA | Overlaps needle-forming IB11, IB12 | ||
| I B10: HLVEAL | Overlaps needle-forming IB11, IB12 | ||
| I B11: LVEALY | Needles | Needle-like crystals not characterized by x-ray diffraction | |
| I B12: VEALYL | Long fibrils and needles | Needle-like crystals grew in sample buffer† and gave cross-β pattern | A 9-residue peptide spanning residues B11 to B19 forms fibrils |
| β2m26: YVSGFH | |||
| β2m48: KVEHSD | |||
| β2m52: SDLSFS | |||
| β2m54: LSFSKD | Overlaps fibril-forming β2m58 | ||
| β2m58: KDWSFY | Short/medium fibrils | Needle-like crystals grew in conditions C7 and C11 and gave cross-β pattern | A 13-residue peptide spanning β2m residues 59–71 forms fibrils (18) |
| β2m62: FYLLYY | Needles | Needle-like crystals grew in sample buffer† and gave cross-β pattern | |
| β2m64: LLYYTE | Needles | Needle-like crystals grew in sample buffer† and gave cross-β pattern | Predicted at threshold −19 kcal/mol |
| β2m83: NHVTLS | Short fibrils | Needle-like crystals grew in condition E8 and gave cross-β pattern | |
| β2m91: KIVKWD | Short fibrils | Needle-like crystals grew in conditions B11, C5, and E10 and gave cross-β pattern |
*As observed by electron micrography. Short, medium, and long indicate short, medium, and long amyloid-like fibrils (as shown in Fig. 1 A, B, and E). Needles indicates needle-like crystals (as shown in Fig. 1 D and F).
†Sample buffer used for polymerization assays was 150 mM NaCl/50 mM phosphate, pH 2.5.
‡Notice that two of the needles were predicted only with the permissive threshold of −19 kcal/ mol rather than the standard threshold of −21 kcal/mol.
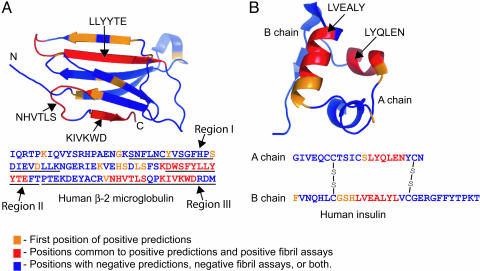
Of the three amyloid-like peptides from insulin, two (IB11:LVEALY and IB12:VEALYL) are overlapping peptides from the B chain. They are found in the native structure (Fig. 2) in an α-helix between the two intermolecular disulfide bonds (A7–B7 and A20–B19) and are predicted as fibril formers by the 3D profile algorithm. The third amyloid-like peptide from insulin has the sequence IA13:LYQLEN and resides in an α-helix on the A chain in the structure, also between the two intermolecular disulfide bonds. This peptide is not predicted to form fibrils with the standard energy threshold of −21 kcal/mol we adopt here, but is predicted to form fibrils with the more permissive threshold of −19 kcal/mol. Two other insulin peptides (IA12:SLYQLE and IA14:YQLENY) predicted to form fibrils at the threshold of −21 kcal/mol overlap with IA13:LYQLEN. In fact the nine-residue peptide (A12–A20) that spans all three of these six-residue peptides forms fibrils (data not shown). We conclude that there is good, but not perfect, agreement between predictions of fibril formation by the 3D profile method and our observations of fibril or needle formation. The extent of agreement mapped onto the 3D protein structures is shown in Fig. 2.
Fig. 2.
Ribbon diagrams of β2m (Protein Data Bank ID code 1LDS) (A) and insulin (Protein Data Bank ID code 1GUJ) (B) structures showing the positions of the predicted segments by the 3D profile method and the segments that were experimentally determined to form fibrils or needle-like crystals. Note that there is good, but not perfect, correlation between the predicted and experimentally determined segments, as discussed in the text.
Table 1 also shows that of the eight hexameric peptides from β2m predicted to be amyloid-like five were found to form fibrils or needle-shaped microcrystals. Earlier we found that fibrils and needle-shaped microcrystals grow under similar solution conditions and have similar structures (12). Also one of the four peptides predicted to form fibrils (β2m54:LSFSKD) but failing to do so overlaps with a peptide (β2m58:KDWSFY) that does form fibrils. Thus for β2m, as for insulin, there is good, but imperfect, agreement between predictions and observations, as shown graphically in Fig. 2.
It is noteworthy that six of the nine predicted fibril-forming peptides from β2m (Table 1) fall into three larger sequence regions of this protein, each of which has been found experimentally to form amyloid-like fibrils, as shown in Fig. 2. The peptide β2m26:YVSGFH is contained in the larger region from Ser-20 to Lys-41, found to form fibrils by Kozhukh et al. (17). The peptides β2m58:KDWSFY, β2m62:FYLLYY, and β2m56:LLYYTE are contained in the larger region Asp-59 to Thr-71 described by Jones et al. (18). And the peptides β2m83:NHVTLS and β2m91:KIVKWD are contained in the larger region found to form fibrils by Ivanova et al. (19).
Discussion
Hexameric Peptides Are Capable of Formation of Amyloid-Like Fibrils.
Our results reinforce earlier studies that have found that short protein segments can form amyloid-like fibrils (5–9, 18). A well studied example is the peptide GNNQQNY from the yeast prion Sup35, which forms fibrils with all well accepted biophysical properties of amyloid fibrils (5). At the same time, our findings are consistent with many earlier observations that not all short peptides form amyloid-like fibrils. Clearly amyloid formation requires some specific feature of sequence (12) or property of the peptide as a whole (20). We return below to the nature of this sequence signature for amyloid.
More than One Sequence Segment of a Protein Can Form Amyloid-Like Fibrils.
Our results show that there are at least two segments of insulin, one in the A chain and one in the B chain, that form amyloid-like fibrils. Similarly, there are at least three segments of β2m that are capable of amyloid-like fibril formation. When these entire proteins are converted to fibrils, it is possible that only a single short segment from each molecule is incorporated into the cross-β spine of the fibril. Alternatively, more than one segment from each molecule may participate in the spine. Structural tools have not yet favored one of these alternatives over the other. In the case of insulin, the B-chain segments appear more prone to form fibrils than the A-chain segments, in that fibril conformations of B-chain peptides have lower energies than those of the A-chain peptides (Table 1). It may be that one segment, such as the B-chain segment in insulin, joins the zipper spine first, interfering with the ability of the second type of segment to join the fibril.
The Sequence Signature of Amyloid-Like Fibril Formation Is Dictated by the Structure of the Steric Zipper.
Our results, summarized in Table 1 and Fig. 2, show good agreement between our experiments on fibril formation of hexameric peptides from the sequences of insulin and β2m. This level of agreement can be quantified by a P value, which gives the probability that our predictions of fibril formation would match our experiments by chance. This P value is the probability that the agreement is no better than random and is given by
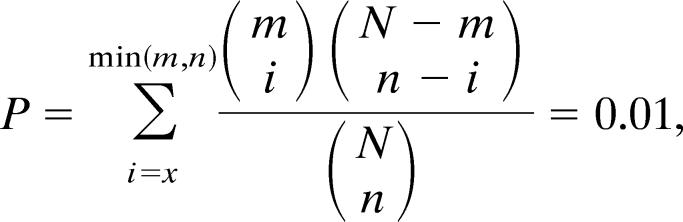 |
in which N = 59, the total number of hexameric peptides tested; m = 8, the total number of fibril-forming peptides; n = the number of predictions with energies below the threshold value; and x = the number of hexamers predicted to form fibrils that are observed to form fibrils.
This finding means that there is one chance in 100 that our results could have been observed by chance.
There are several ways that our computational and experimental methods could be optimized, which would be expected to further lower the P value. Our current computational model (13) assumes a rigid peptide backbone in the extended conformation, whereas there could be variations in the backbone dihedral angles from their most extended values. Also in generating the ensemble of templates, the variations are discrete steps, which could be more fine-grained. And we consider only parallel, in register β-sheets as seen in the structures of GNNQQNY and NNQQNY (12), rather than also antiparallel β-sheets. A model adopting these additional variations could improve predictions. At the same time, our conditions for observing fibrils may be too restrictive to capture all fibrils that could form. For example, our experiments have been carried out at a single pH and salt concentration, whereas other conditions might favor fibril formation more strongly.
The good correspondence between predictions and observations of fibril formation for hexameric peptides suggests that the sequence dependence of fibril formation is determined by constraints imposed by the pair-of-sheets, steric zipper structure seen in the structure of GNNQQNY (12). The reason is that the predictions are made by assuming that each hexamer assumes the pair-of sheets structure, or a structure with minor variations from it, as represented in the ensemble of templates. Table 1 shows neither massive overprediction nor underprediction of observed fibril-forming segments. There are five predicted peptide segments in Table 1 for which no overlapping fibrils or needle-like microcrystals were observed. One of these (β2m48:KVEHSD) is highly charged, which may offer problems for our energy-based algorithm. There are also two segments whose energies qualify as amyloid formers only at the permissive threshold but not the standard threshold. In practice, someone predicting fibril formation would note the possibility of fibril formation at the permissive threshold. On the whole, the level of agreement between prediction and experiment suggests that the underlying structural model of a pair-of-sheets structure with a dry steric zipper is at least moderately correct for these fibrils.
In short, the sequence signature for formation of amyloid-like fibrils appears to be a short, self-complementary sequence that is compatible with the dry steric zipper interface. The three residues of a hexamer that point into the interface are most important in achieving this self-complementarity. Charged, inward-pointing residues at positions 3 and 5 in the hexapeptide would be expected to be rare, because charged residues would be stacked close to one another, leading to electrostatic destabilization. Charged residues at position 1 are relatively solvent-exposed, particularly if the two β-sheets are shifted out of register with one another in the direction of the β-strands. Consistent with this finding, Lopez de la Paz and Serrano (6) found that the first position of their sequence pattern is tolerant of residue type. Inward-pointing residues greatly differing in size, such as Gly and Trp, would be unlikely, because of the difficulty in achieving a tight, self-complementary interface. Low-complexity sequences, having residues of similar size, might be expected to be favored in fibril-forming sequences, such as NNQQNY.
Methods
Polymerization Assays.
The 60 peptides listed in Table 2 (synthesized by Celtek Bioscience Peptides, Nashville, TN) were dissolved into 1 ml of sample buffer containing 150 mM NaCl/50 mM phosphate, pH 2.5 to a final concentration of 2 mM. Then this 1 ml was split into three and incubated at 37°C with shaking. All measurements (CR and ThT binding) were done immediately after dissolving the peptide, 10–12 and 40–45 days.
CR Binding Assay.
Spectroscopic assays, detecting a red shift upon binding of CR with peptides, were done as described by Klunk et al. (21). In short, a 5-μl sample was added to 75 μl of 15 μM filtered CR/150 mM NaCl/25 mM phosphate, pH 7.4. The contribution of the scatter was measured by adding a 5-μl sample to 75 μl of 150 mM NaCl/25 mM phosphate, pH 7.4. The CR alone was measured by adding a 5-μl sample buffer to 75 μl of 15 μM filtered CR/150 mM NaCl/25 mM phosphate, pH 7.4. All specimens were incubated at 37°C for 30 min before signal measurements, which were done at 407, 497, 540, and 550 nm.
ThT Binding Assay.
A 20-μl sample was added to 200 μl of 5 μM ThT/10 mM Tris, pH 8.0. Emission at 482 nm (2-nm slit width) was measured immediately with excitation at 444 nm (2-nm slit width) by using a Spex Fluorolog spectrofluorimeter (Jobin Yvan, Edison, NJ). Signal was corrected for scatter by subtracting the scatter at 482 nm (20-μl sample added to 200 μl of 10 mM Tris, pH 8.0) from the signal measured at 482 nm from the sample in ThT.
EM.
Sample was applied directly to hydrophilic 400-mesh carbon-coated formvar support films mounted on copper grids (Ted Pella, Inc., Redding, CA), allowed to adhere for 4 min, rinsed with distilled water, and stained for 1 min with 1% uranyl acetate (Ted Pella, Inc.). Grids were examined in a Hitachi (Tokyo) H-7000 electron microscope at an accelerating voltage of 75 kV.
X-Ray Diffraction from Fibrils.
Several hexamers that formed fibrils also formed needle-like crystals after screening with Index HT (HR2-134, Hampton Research, Riverside, CA).
As shown in Table 1, needle-like crystals grew in the following conditions: Index HT screen, B11, 2.1 M DL-Malic acid, pH 7.0, C5 60% (vol/vol) Tacsimate, pH 7.0; C7, 0.8 M KNa Tartrate tetrahydrate/0.1 M Tris, pH 8.5/0.5% (wt/vol) polyethylene glycol monomethyl ether 5000; C11, 1.0 M ammonium sulfate/0.1 M Hepes, pH 7.0/0.5% PEG 8000; E10, 0.1 M Bis·Tris, pH 6.5, 45% (vol/vol) PEG P 400; and Crystal Screen HT, E8, 1.5 M NaCl/10% (vol/vol) ethanol.
Clusters of needle-like crystals of hexamers were transferred to a 2-μl drop containing 30% glycerol. Then samples were exposed to copper Kα X-radiation from a Rigaku FR-D x-ray generator supplemented with Rigaku Blue Optics operating at 50 kV and 100 mA. Data were collected at 105 K for 5 min with 1° oscillations on a Rigaku IV++ imaging plate detector. X-ray photos were also taken on Beamline ID13 at the European Synchrotron Radiation Facility, Grenoble, France.
Supplementary Material
Acknowledgments
We thank Mari Gingery, Michael R. Sawaya, and Martin Phillips for advice on experiments and Melanie J. Bennett for discussion. This work was supported by the National Science Foundation, National Institutes of Health, and the Howard Hughes Medical Institute.
Abbreviation
- β2m
β2-microglobulin
- ThT
thioflavin T
- CR
Congo red.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Westermark P. FEBS J. 2005;272:5942–5949. doi: 10.1111/j.1742-4658.2005.05024.x. [DOI] [PubMed] [Google Scholar]
- 2.Westermark P., Benson M. D., Buxbaum J. N., Cohen A. S., Frangione B., Ikeda S., Masters C. L., Merlini G., Saraiva M. J., Sipe J. D. Amyloid. 2002;9:197–200. doi: 10.3109/13506120209114823. [DOI] [PubMed] [Google Scholar]
- 3.Westermark P., Wernstedt C., Wilander E., Hayden D. W., O’Brien T. D., Johnson K. H. Proc. Natl. Acad. Sci. USA. 1987;84:3881–3885. doi: 10.1073/pnas.84.11.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dische F. E., Wernstedt C., Westermark G. T., Westermark P., Pepys M. B., Rennie J. A., Gilbey S. G., Watkins P. J. Diabetologia. 1988;31:158–161. doi: 10.1007/BF00276849. [DOI] [PubMed] [Google Scholar]
- 5.Balbirnie M., Grothe R., Eisenberg D. S. Proc. Natl. Acad. Sci. USA. 2001;98:2375–2380. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez de la Paz M., Serrano L. Proc. Natl. Acad. Sci. USA. 2004;101:87–92. doi: 10.1073/pnas.2634884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenidis K., Waldner M., Bernhagen J., Fischle W., Bergmann M., Weber M., Merkle M. L., Voelter W., Brunner H., Kapurniotu A. J. Mol. Biol. 2000;295:1055–1071. doi: 10.1006/jmbi.1999.3422. [DOI] [PubMed] [Google Scholar]
- 8.MacPhee C. E., Dobson C. M. J. Mol. Biol. 2000;297:1203–1215. doi: 10.1006/jmbi.2000.3600. [DOI] [PubMed] [Google Scholar]
- 9.Ventura S., Zurdo J., Narayanan S., Parreno M., Mangues R., Reif B., Chiti F., Giannoni E., Dobson C. M., Aviles F. X., Serrano L. Proc. Natl. Acad. Sci. USA. 2004;101:7258–7263. doi: 10.1073/pnas.0308249101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reches M., Porat Y., Gazit E. J. Biol. Chem. 2002;277:35475–35480. doi: 10.1074/jbc.M206039200. [DOI] [PubMed] [Google Scholar]
- 11.Tjernberg L., Hosia W., Bark N., Thyberg J., Johansson J. J. Biol. Chem. 2002;277:43243–43246. doi: 10.1074/jbc.M205570200. [DOI] [PubMed] [Google Scholar]
- 12.Nelson R., Sawaya M. R., Balbirnie M., Madsen A. O., Riekel C., Grothe R., Eisenberg D. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson M. J., Sievers S. A., Karanicolas J., Ivanova M. I., Baker D., Eisenberg D. Proc. Natl. Acad. Sci. USA. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McParland V. J., Kad N. M., Kalverda A. P., Brown A., Kirwin-Jones P., Hunter M. G., Sunde M., Radford S. E. Biochemistry. 2000;39:8735–8746. doi: 10.1021/bi000276j. [DOI] [PubMed] [Google Scholar]
- 15.Brange J., Andersen L., Laursen E. D., Meyn G., Rasmussen E. J. Pharm. Sci. 1997;86:517–525. doi: 10.1021/js960297s. [DOI] [PubMed] [Google Scholar]
- 16.Whittingham J. L., Scott D. J., Chance K., Wilson A., Finch J., Brange J., Dodson G. J. Mol. Biol. 2002;318:479–490. doi: 10.1016/S0022-2836(02)00021-9. [DOI] [PubMed] [Google Scholar]
- 17.Kozhukh G. V., Hagihara Y., Kawakami T., Hasegawa K., Naiki H., Goto Y. J. Biol. Chem. 2002;277:1310–1315. doi: 10.1074/jbc.M108753200. [DOI] [PubMed] [Google Scholar]
- 18.Jones S., Manning J., Kad N. M., Radford S. E. J. Mol. Biol. 2003;325:249–257. doi: 10.1016/s0022-2836(02)01227-5. [DOI] [PubMed] [Google Scholar]
- 19.Ivanova M. I., Gingery M., Whitson L. J., Eisenberg D. Biochemistry. 2003;42:13536–13540. doi: 10.1021/bi0301486. [DOI] [PubMed] [Google Scholar]
- 20.Dobson C. M. Philos. Trans. R. Soc. London B. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klunk W. E., Pettegrew J. W., Abraham D. J. J. Histochem. Cytochem. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.