Abstract
Epsins are endocytic proteins with a structured epsin N-terminal homology (ENTH) domain that binds phosphoinositides and a poorly structured C-terminal region that interacts with ubiquitin and endocytic machinery, including clathrin and endocytic scaffolding proteins. Yeast has two redundant genes encoding epsins, ENT1 and ENT2; deleting both genes is lethal. We demonstrate that the ENTH domain is both necessary and sufficient for viability of ent1Δent2Δ cells. Mutational analysis of the ENTH domain revealed a surface patch that is essential for viability and that binds guanine nucleotide triphosphatase-activating proteins for Cdc42, a critical regulator of cell polarity in all eukaryotes. Furthermore, the epsins contribute to regulation of specific Cdc42 signaling pathways in yeast cells. These data support a model in which the epsins function as spatial and temporal coordinators of endocytosis and cell polarity.
Keywords: actin, endocytosis, polarity
Endocytosis is an essential mechanism for internalizing extracellular material and controlling the composition of the plasma membrane; this is critical for cellular homeostasis, including down-regulation of signaling receptors and recycling of transmembrane proteins such as v-SNAREs that reside transiently at the plasma membrane (1). Many cytosolic proteins that contribute to the mechanisms and regulation of endocytosis have been identified, but assigning precise functions to each protein has been more challenging (2, 3). Some of these proteins may also participate in multiple steps or pathways (4, 5), either related to or independent from endocytosis, further complicating the elucidation of their function(s). Additionally, roles for the actin cytoskeleton in regulating or effecting specific stages of endocytosis are another active area of investigation (2). One goal is to identify multifunctional proteins that coordinate these various cellular processes.
The epsin proteins are proposed to function as endocytic clathrin adaptors for ubiquitinated cargo (6, 7). They are found in all eukaryotes and have an N-terminal phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2]-binding epsin N-terminal homology (ENTH) domain, two ubiquitin interaction motifs, and several peptide ligands that bind components of the endocytic machinery (7). In addition to putative adaptor roles, it has been shown previously that mammalian epsin binds RalBP1/RLIP76, a GTPase-activating protein (GAP) for Cdc42 and Rac1 (8). RalBP1 has been implicated in endocytosis, because it binds the plasma membrane clathrin adaptor AP-2 (8). The Cdc42 and Rac GTPases are key regulators of the actin cytoskeleton (9), thus suggesting that this complex links signaling, endocytosis, and actin cytoskeleton regulation.
The budding yeast Saccharomyces cerevisiae has two epsins, Ent1 and Ent2; deleting either alone leads to no detectable phenotype, but a double deletion is lethal (10). Here we show that the ENTH domain of yeast epsin is necessary and sufficient for viability of ent1Δent2Δ cells (ΔΔ). The essential function requires a patch of residues, conserved only in the ENTH domains of epsin proteins, that interacts with Cdc42 GAPs. Our results indicate that this interaction regulates levels of activated Cdc42 in vivo, and that an [epsin·GAP] complex plays a critical physiological role together with specific Cdc42 effectors. Furthermore, Cdc42 regulation via epsins is likely to be conserved in higher eukaryotes. Based on these data, we propose a model in which the epsins link the endocytic and cell polarity machineries to coordinate membrane and cytoskeletal pathways in response to polarized signals.
Results
The ENTH Domain Is Sufficient for Viability.
To determine which regions of epsin contribute to its essential role, we used ΔΔ containing a wild-type ENT1 or ENT2 plasmid and a marker for counterselection on a toxic compound; this allows for shuffling of mutated epsin plasmids with a different selection marker and evicting the original one. Cells will not grow on plates containing the toxic compound [5-fluoroorotic acid or 5-fluoroanthranilic acid/alcohol (5-FAA)] if the new plasmid encodes a protein that cannot fulfill the essential function of the wild-type epsin; this assay is used throughout. The ENTH domain of either Ent1 or Ent2, with or without an N-terminal hemagglutinin epitope tag and expressed from either endogenous or methionine (Met)-regulated MET25 promoters, was both necessary and sufficient for ΔΔ viability (Fig. 1A and data not shown). The ENTH domain of the related proteins Ent3 or Ent4 did not complement ΔΔ (Fig. 1A and data not shown). Thus, the epsin ENTH domain is required for cell viability.
Fig. 1.
Epsin ENTH domain essential residues. (A) The ENTH domain of Ent1 is necessary and sufficient for cell viability. Schematic of Ent1 constructs; the ENTH domain, ubiquitin-interaction motifs (UIMs), Asn-Pro-Phe tripeptides (NPF), and clathrin-binding motif (CBM) are indicated. ΔΔ with an ENT2 TRP1 plasmid and a second URA3 plasmid, empty or encoding the proteins or domains indicated (Right), were grown on plates lacking uracil and tryptophan (Left) or containing 5-FAA to evict the ENT2 TRP1 plasmid (Center) at 30°C for 3 days. The presence of an N-terminal hemagglutinin tag, plasmid copy number, and promoter used are also indicated. (B) ENTH domain-specific residues, Y100, T104, and E137, are solvent-exposed in a 3D model. Residues that are conserved in ENTH domains, different in nonepsin ENTH/AP180 N-terminal homology domains, and exposed to the solvent in a 3D model are shown [Y100 (green), T104 (red), and E137 (magenta)]. Lipid-binding residues (blue) and the N- and C-termini are indicated. (C) ENTH1Y100R domain is a hypomorph. ΔΔ cells containing an ENT2 TRP1 plasmid and a second high copy or single copy URA3 plasmid encoding ENTH1WT or ENTH1Y100R from endogenous or MET25 promoter were assayed as in A.
To identify candidate regions of epsin ENTH domains that could mediate essential functions, we compared the sequences of S. cerevisiae Ent1/2 ENTH domains with the homologous domains from Drosophila melanogaster (liquid facets) and Xenopus laevis (MP90) epsin, which also complement ΔΔ (ref. 11 and our unpublished results). We found three residues (Y100, T104, and E137) that were both conserved exclusively in epsin ENTH domains and also different in all other nonessential ENTH and AP180 N-terminal homology domain-containing proteins encoded by the yeast genome (Ent3, Ent4, Ent5, Yap1801, Yap1802, and Sla2). These three candidate essential residues were predicted to be on the surface of a model for the folded domain (Fig. 1B); moreover, T104 had been found previously as a critical residue in epsin function (10).
To test their functional importance, we individually mutated these three residues to a corresponding residue found in a noncomplementing ENTH/AP180 N-terminal homology domain (Y100R, T104D, and E137G) and asked whether the mutant ENTH domains could fulfill the essential domain function. ENTH1Y100R and ENTH1T104D domains did not complement ΔΔ when expressed from the endogenous promoter (Fig. 1C and Fig. 7A, which is published as supporting information on the PNAS web site). Western blotting confirmed that wild-type or mutant ENTH domain proteins was stable (data not shown). Purified recombinant wild-type and mutant ENTH domains had nearly identical structures, based on fluorescence emission of two tryptophan residues (W70 and W94; Fig. 8A, which is published as supporting information on the PNAS web site). Differential scanning calorimetry showed that the ENTH1 domains were properly folded, their denaturation being centered at 52.5°C (data not shown). ENTH domain lipid-binding activity was also preserved in the ENTH1Y100R domain; it bound IP6, which mimics the head group of phosphatidylinositol-4,5-bisphosphate (12), with a binding affinity and enthalpy identical to the ENTH1WT domain (Kd = 39 ± 6 mM, ΔH = −2.2 ± 0.9 kcal/mol; Fig. 8B). Thus, the inviability associated with the mutation of Y100 or T104 is unlikely to be caused by gross misfolding of the mutant domains. These results also suggest that the essential function of the ENTH domain mediated by the Y100/T104 patch is independent of lipid binding. ENTH domains with lipid-binding mutations partially complemented ΔΔ, further suggesting independent functions for lipid binding and the Y100/T104 patch (Fig. 8C).
ENTH1Y100R Phenotypes.
Although ENTH1Y100R could not complement ΔΔ when expressed from the endogenous promoter, cells with higher-level expression from the regulated MET25 promoter were viable, although they grew more slowly than ENTH1WT cells (Fig. 1C). In contrast, even high-level expression of ENTH1T104D could not support ΔΔ viability (Fig. 7B). This suggests that ENTH1Y100R acts as a hypomorphic allele. Thus, we used plasmids encoding ENTH1WT or ENTH1Y100R domains expressed from the MET25 promoter to study in vivo the effect of mutating the Y100/T104 essential patch. Cells with these plasmids grown in 0–0.02 mM Met had the highest protein levels; those grown in higher Met concentrations (0.2–2 mM) had lower protein levels (Fig. 9, which is published as supporting information on the PNAS web site). ENTH1Y100R-expressing cells grown in 1 mM Met (low protein levels) exhibited dramatically slow growth, inviability at 37°C, and greatly enlarged, fragile, and often broken cells (Fig. 2A and B).
Fig. 2.
ENTH1Y100R phenotypes. (A) ENTH1Y100R cells are larger and more fragile than ENTH1WT cells. ΔΔ cells expressing ENTH1WT or ENTH1Y100R domains from the MET25 promoter were grown in selective liquid media + 1 mM Met and visualized by differential interference contrast microscopy. (Scale bar, 10 μm.) (B) ENTH1Y100R cells are sensitive to high temperature and hyposmolarity. Serial dilutions of ΔΔ cells expressing ENTH1WT or ENTH1Y100R domains from the MET25 promoter were grown on YPD or 0.5xYPD plates at 30°C or 37°C for 3 days. (C) ENTH1Y100R cells have a depolarized actin cytoskeleton. ΔΔ cells expressing ENTH1WT or ENTH1Y100R domains from the MET25 promoter were grown in liquid media plus 1 mM Met. Fixed cells were labeled with rhodamine-phalloidin and DAPI and visualized by confocal microscopy. Arrowheads highlight small buds where actin patches should concentrate. (Scale bar, 5 μm.) (D) ENTHY100R cells show no defect on endocytosis of GFP-Ste3. ENTH1WT (Left) or ENTH1Y100R (Right) cells expressing a Ste3-GFP fusion protein were grown in liquid media plus 1 mM Met and visualized by confocal microscopy.
Sites of cell wall synthesis, secretion, and endocytosis in yeast correlate with the polarized locations of F-actin-containing structures (13, 14). Actin cables deliver newly synthesized secretory material to sites of growth (e.g., buds and necks) (15), whereas cortical patches are dynamic structures in the process of endocytosis (16). Unlike the normal actin organization observed in ENTH1WT cells, cortical patches were not polarized in the buds and few, if any, actin cables were visible in ENTH1Y100R cells (Fig. 2C). Thus, mutation of the essential patch of the ENTH domain altered cell growth, morphology, and polarity. These data also demonstrate that a wild-type ENTH domain is the minimal functional unit of the epsin molecule required to maintain proper actin organization/polarity (Fig. 2C). Interestingly, under the conditions of our experiments, the Ste3 mating pheromone receptor is endocytosed normally in both ENTH1WT and ENTH1Y100R cells (Fig. 2D; see Discussion).
ENTH Domains Bind Cdc42 GAPs.
Because the lipid-binding activity of the ENTH domain appeared to be dispensable for the functions mediated by the Y100/T104 patch, we hypothesized that this essential region might bind proteins. A yeast two-hybrid screen with an ENTH1WT bait versus a yeast cDNA prey library identified fragments from all three known Cdc42 GAP proteins, one each for Rga1 and Bem3 and two for Rga2. Prey plasmids encoding each GAP interacted with baits for both ENTH1/2WT, as indicated by activation of the reporter genes ADE2, HIS3, and lacZ (Fig. 3A Top and data not shown). The interactions were confirmed to be both direct and specific by in vitro recombinant protein-binding assays. GST-Rga1/2 fusion proteins immobilized on glutathione-agarose beads were incubated with purified His6−-ENTH1/2; bound His6−-ENTH domains were detected by Western blotting with anti-His6− antibody (Fig. 3B Upper). Endogenous Ent1 from yeast lysates was also pulled down by GST-Rga2 immobilized on glutathione beads (data not shown). Isothermal titration calorimetry experiments gave an approximate affinity for the ENTH1–Rga2 interaction as a Kd of ≈0.6 mM (data not shown; see Supporting Text, which is published as supporting information on the PNAS web site). The ENTH1–Rga1 interaction appears to be considerably weaker.
Fig. 3.
The ENTH domain binds Cdc42 GTPase activating proteins. (A) ENTH domains interact with Cdc42 GAPs. Yeast two-hybrid cells with plasmids encoding GAL4 DNA-binding domain fusions to ENTH1WT, ENTH1Y100R, ENTH1T104D, ENTH1Y100R, T104D, ENTH2WT, or mouse p53 (mp53), and GAL4 activation domain fusions to Rga1, Rga2, Bem3, or SV40 T-large antigen (T-LAg) were grown on plates lacking histidine and adenine and containing 10 mM 3-amino-triazole. (B) In vitro interaction between ENTH domains and Rga1/2. GST, GST-Rga1, and GST-Rga2 were immobilized on glutathione-agarose beads and incubated with purified His-6-ENTH1, His-6-ENTH2, or His-6-ENTH1Y100R. Bound His-6-ENTH was detected by Western blot with anti-His-6 antibody. One percent of the input is shown. (C) Mapping the ENTH domain-binding site on Rga2. (Upper) GST-Rga2 fragments were used in His-6-ENTH1 pull-down experiments as in B. (Lower) Rga2 contains two LIM (Lin11, Isl-1, and Mec-3) domains, a coiled-coil (CC) region, and a RhoGAP domain. The dotted bar indicates the Rga2 minimal fragment from the two-hybrid screen. Solid bars represent the GST-Rga2 truncations used to map the EBS.
Directed two-hybrid experiments showed a reduced interaction when ENTH1Y100R bait was paired with the Cdc42 GAPs (Fig. 3A Middle). More severe impairment of the ENTH1–GAP interaction was seen with ENTH1T104D or ENTH1Y100R, T104D as bait (Fig. 3A Bottom). A role for the essential patch in the interaction was confirmed by in vitro binding assays showing reduced His-6-ENTH1Y100R–Rga2 interactions (Fig. 3B Lower). Together, our data indicate that the ENTH domain interacts with Cdc42 GAPs, and that this interaction depends upon the essential patch containing the Y100 and T104 residues. To complement these data, the minimal region of Rga2 that binds to ENTH1WT was mapped by using GST pull-down assays with Rga2 truncations (Fig. 3C). The ENTH-binding site (EBS) corresponded to Rga2604–786. Interestingly, Rga2710–721 shows sequence homology with the more divergent Cdc42 GAP, Bem3 (Bem3425–433). Preliminary data suggest that this homology region is involved in, but not sufficient for, ENTH domain recognition (data not shown).
In Vivo Requirement for ENTH Domain–Cdc42 GAP Interaction.
To address the role of Cdc42 GAP binding to the ENTH domain, we used several strategies predicted to influence [ENTH·GAP] complex formation in vivo.
Impaired ENTH–Cdc42 GAP interaction leads to decreased viability and cell polarity defects.
We affected the equilibrium of [ENTH·GAP] complex formation by manipulating each interaction partner (impairing the ENTH-binding affinity or decreasing the GAP dosage) or by adding a Cdc42 GAP competitor. First, mutations in the ENTH domain that affect its interaction with Cdc42 GAPs, yielding low levels of [ENTH·GAP] complex, led to impaired cell viability and cell polarity defects (Figs. 1C and 7A). Second, ΔΔ with lower levels of intracellular GAPs were obtained by deleting RGA1. rga1Δ in ΔΔ cells expressing ENTH1WT were temperature-sensitive, and the phenotypes were enhanced with rga1Δ in ΔΔ+ENTH1Y100R cells (Fig. 4A).
Fig. 4.
In vivo analysis of the ENTH1–Cdc42 GAP interaction. (A) Cdc42 GAP RGA1 genetically interacts with the epsin ENTH domain. ent1Δent2Δrga1Δ cells with an ENT2 TRP1 plasmid and ENTH1 URA3 plasmid were grown on 5-FAA plates at 30°C for 3 days as in Fig. 1A. (B) Overexpressing the EBS phenocopies ENTH1Y100R. ENTH1WT cells overexpressing EBS from the MET25 promoter were stained for F-actin as in Fig. 2E. (Scale bar, 10 μm.) (C) Overexpressing Cdc42 GAPs alleviates ENTH1Y100R phenotypes. Serial dilutions of ENTH1Y100R cells with empty vector or overexpressing Rga1 or Rga2 were grown on 0.5xYPD plates as in Fig. 2B. (D) Cdc42·GTP levels are reduced in ENTH1Y100R cells. Lysates from wild-type, ENTH1WT, and ENTH1Y100R cells expressing GFP-Cdc42 grown in selective media plus 1 mM Met were incubated with GST-PAK (CRIB) beads that bind Cdc42·GTP. Bound GFP-Cdc42·GTP was detected by anti-GFP Western blotting. GTPγS-loaded lysates have equivalent GFP-Cdc42 in all samples. Levels of GFP-Cdc42·GTP (normalized relative to corresponding GTPγS-loaded GFP-Cdc42 signal) are calculated as a fraction of wild-type cell values.
We also used competitive inhibition of the ENTH–GAP interaction by overexpressing the Rga2 EBS. High levels of EBS expression in endogenous ENTH1WT cells yielded morphology and actin disorganization similar to ENTH1Y100R cells (Fig. 4B). These data indicate a dominant negative consequence of EBS expression consistent with competition for the endogenous interaction and suggest that the ENTH–Rga2 interaction is important for normal cell physiology.
Enhanced ENTH1Y100R–Cdc42 GAP interaction alleviates ENTH1Y100R phenotypes.
An in vivo role for the ENTH1–Cdc42 GAP interaction predicts that promoting the formation of [ENTH1Y100R·GAP] complexes by overexpressing either binding partner (ENTH1Y100R or GAP) should suppress the ENTH1Y100R phenotypes. Fig. 1C shows that increased levels of ENTH1Y100R improve ENTH1Y100R cell phenotypes. Conversely, overexpressing either Rga1 or Rga2 improved growth on hyposmotic media and at 37°C (Fig. 4C). Overexpression of Bem3 only mildly rescued ENTH1Y100R phenotypes (Table 1, which is published as supporting information on the PNAS web site; data not shown). In contrast, Bem2, a Rho GAP for both Rho1 (17) and Cdc42 (18), did not suppress ENTH1Y100R phenotypes when overexpressed (Table 1).
Cdc42·GTP Levels Depend on the ENTH Domain.
Our data indicate that impairing the ENTH–GAP interaction leads to cell viability and polarity defects, whereas promoting the formation of the [ENTH1Y100R·GAP] complex partially suppresses the ENTH1Y100R cell phenotypes. Because the newly identified ENTH interaction partners are Cdc42 regulators, we asked whether Cdc42 activation might be affected by the ENTH1Y100R mutation by measuring the levels of activated GFP-Cdc42 (Cdc42·GTP) in lysates from cells expressing ENTH1WT or ENTH1Y100R. The Cdc42/Rac interactive binding (CRIB) domain of the p21-activated kinase 1 binds specifically to the GTP-bound form of Cdc42 (19). A GST-CRIB fusion protein immobilized on glutathione beads was incubated with yeast lysates; bound GFP-Cdc42·GTP was visualized by Western blotting with anti-GFP antibody. Total Cdc42 present in the lysates was measured by Western blotting with anti-GFP (not shown) or by loading with nonhydrolyzable GTPγS and pulling down with CRIB beads as described above, as a measure of the total pool that can be activated (Fig. 4D). Amounts of GFP-Cdc42·GTP vs. GFP-Cdc42·GTPγS were quantified by densitometry after Western blotting. We found a 62% reduction in Cdc42·GTP in ENTH1Y100R cells relative to wild-type or ENTH1WT cells (Fig. 4D), indicating a dependence of Cdc42 regulation on ENTH1 function. Similar results were obtained for endogenous Cdc42 (data not shown). Attempts to demonstrate in vitro a direct regulation of the Cdc42 GAP activity by ENTH domain binding were unsuccessful. This suggests that full-length GAP, other proteins, posttranslational modifications, or localization-dependent events are required for the epsin-dependent effects observed in vivo. Thus, our data indicate that, at least in vivo, ENTH1Y100R mutants with reduced interaction with Cdc42 GAPs exhibit enhanced inactivation (or deficient activation) of Cdc42-regulated pathways.
The Cdc42-Dependent Gic1/2-Bem1 Pathway Depends on the ENTH Domain Essential Patch.
Next we asked whether overexpression of downstream Cdc42 effectors could bypass the deficiency in activated Cdc42 and rescue some of the ENTH1Y100R phenotypes. Only a subset of Cdc42 effectors (Gic1, Gic2, and to a lesser extent Bem1) allowed growth of ENTH1Y100R cells at 37°C, on hyposmotic medium at 30°C, or in 1 mM Met (Fig. 5A and data not shown). Overexpression of other Cdc42 effectors, including the protein kinases Ste20 and Cla4 or the formin Bni1, did not improve ENTH1Y100R cell growth on either hyposmotic media or at 37°C (data not shown). A complete list of all proteins tested, including other Cdc42 effectors and Rho1 effectors, is in Table 1.
Fig. 5.
The ENTH domain is involved in Cdc42 signaling regulation. (A) Overexpressing the Cdc42 effectors Gic1, Gic2, and Bem1 partially rescues ENTH1Y100R phenotypes. ΔΔ expressing ENTH1WT or ENTH1Y100R with high-copy empty vector or encoding for the overexpression of the indicated proteins were tested for growth as in Fig. 4C. (B) Genetic interaction between ENTH1 and GIC1/2. Growth of ΔΔ with gic1Δ (Left) or gic2Δ (Right) cells expressing ENTH1WT (Upper) or ENTH1Y100R (Lower) was tested by plasmid shuffling as in Fig. 1A.
In agreement with the above results, deleting either GIC1 or GIC2 further reduced growth of ENTH1Y100R cells (Fig. 5B). Together, the results suggest a role for the ENTH domain in the Gic1/2-Bem1 specific branch downstream of Cdc42. Interestingly, a physical interaction between Rga1 and Gic2 has been reported (20), suggesting possible synergism between these two Cdc42·GTP-binding proteins. Moreover, both GAPs and Gics have been implicated in cell polarity regulation (21).
ENTH Domain Regulation of Cdc42 Is Conserved in Higher Eukaryotes.
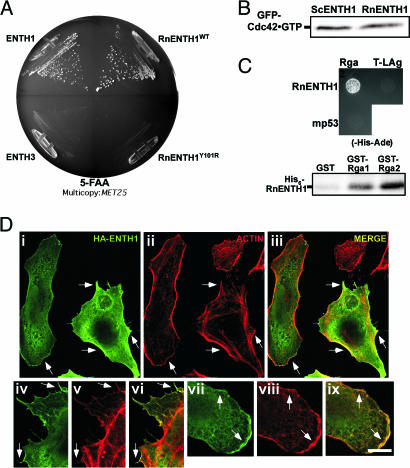
The Y100/T104/E137 patch is conserved in the ENTH domain of metazoans; thus, we asked whether the function of this patch is conserved in mammals. The rat epsin1 ENTH domain (RnENTH1), expressed from the MET25 promoter, complemented ΔΔ yeast cells (Fig. 6A). Importantly, these cells also had normal levels of activated Cdc42, by using the same GST-CRIB assay described above (Fig. 6B) and bound the Rga proteins in vitro (Fig. 6C). Individual mutation of the corresponding essential residues, Y101R and T105D, yielded nonfunctional ENTH domains (Fig. 6A and data not shown). Western blotting showed that each domain was stable (data not shown).
Fig. 6.
Epsin ENTH domain essential patch in higher eukaryotes. (A) Rat ENTH1 domain complements ΔΔ. ΔΔ with MET25 promoter plasmids encoding yeast ENTH1 and ENTH3 or RnENTH1WT and RnENTH1Y101R were grown on 5-FAA plates as in Fig. 1A. (B) RnENTH1 sustains normal levels of activated Cdc42. ΔΔ cells expressing RnENTH1 or yeast ENTH1 and GFP-Cdc42; levels of GFP-Cdc42·GTP were determined as in Fig. 4D. (C) RnENTH1 interacts with the Cdc42 GAPs. Binding of RnENTH1 domain to Cdc42 GAPs was assayed by two-hybrid (Upper) and by in vitro pull down (Lower) as in Fig. 3 A and B. (D) Yeast ENTH1 and ENTH2 domains localize to lamellipodia and filopodia in HeLa cells. HeLa cells transiently transfected with yeast hemagglutinin (HA)-ENTH1 (i–ix) were fixed, permeabilized, and incubated with mouse anti-HA antibody, Alexa 448-conjugated secondary antibody, and rhodamine-phalloidin. Representative cells showing ENTH domain localization (i, iv, and vii), actin structures (ii, v, and viii), and merge (iii, vi, and ix) are shown. (Scale bars, 10 μm.)
Additional evidence supporting functional conservation of ENTH domains was obtained by transfecting S. cerevisiae ENTH1 or ENTH2 into human HeLa cells. Immunofluorescence experiments showed significant colocalization between the yeast ENTH domains and peripheral F-actin-rich structures (Fig. 6D). Costaining was enriched in filopodia and lamellipodia, which are sites of Cdc42 and Rac activity, but not in stress fibers, which depend upon the GTPase Rho1 (22). In transfected HeLa cells, we also observed ENTH domain accumulation in Golgi structures (data not shown), which are enriched in activated Cdc42 (22). Interestingly, the mammalian Golgi-localized EpsinR protein shows potential conservation of the essential patch (Y106 and S110 residues), suggesting analogous Cdc42-related functions for this nonendocytic epsin-related protein at the Golgi apparatus of mammalian cells.
Discussion
It is well established that the epsins are key endocytic proteins; in yeast, they are also required for viability. Here, we showed that the ENTH domain is both necessary and sufficient for cell viability, identified an ENTH domain region in which the essential function resides, and showed its interaction with Cdc42 GAPs. Impairing this interaction in vivo (by mutating the ENTH domain, using dominant negative approaches with GAP fragments, or deleting GAP genes), led to reduced cell viability and defects in polarity and actin organization. ENTH domain mutations in this region are associated with decreased levels of Cdc42·GTP, a biochemical deficiency that explains some phenotypes of the epsin mutant cells.
There are many independent observations supporting this role for the epsins in the Cdc42 pathway. (i) ENTH1Y100R cells have decreased levels of Cdc42·GTP in vivo. (ii) The ENTH domain interacts with the three known Cdc42 GAPs, all at least in part via the Y100/T104 patch. (iii) Y100/T104 patch mutants of the ENTH domain exhibit genetic interactions with the Cdc42 GAPs and a subset of Cdc42 effectors (Gic1 and Gic2). (iv) Cells with deletion of both GIC1 and GIC2 Cdc42 effectors phenocopy ENTH1Y100R cells. (v) Cdc42 GAPs and a subset of Cdc42 effectors (Bem1, Gic1, and Gic2), but not GAPs or effectors for other Rho proteins, were high-copy suppressors of the ENTH1Y100R phenotypes. (vi) The epsins localize to sites of polarized growth and to the bud neck of yeast, whereas in mammalian cells, the ENTH domain localizes to filopodia and lamellipodia (and not to Rho-dependent actin stress fibers), all sites of activated Cdc42 accumulation. (vii) Other authors’ and our unpublished data show that the yeast epsins bind Cdc24, the Cdc42 guanine nucleotide exchange factor.
The Essential Epsin Function May Be in a Cdc42-Gic1/Gic2/Bem1 Pathway.
Gene deletion and overexpression studies indicated that, despite the pleiotropic nature of Cdc42 signaling, the epsins are specifically involved in a Gic1/Gic2/Bem1-dependent Cdc42 pathway (23, 24). Genetic interactions between the ENTH1Y100R mutant and deletion of either GIC1 or GIC2 were more severe than those produced by GAP deletion, indicating a fundamental link between the epsins and these Cdc42 effectors. That gic1Δgic2Δbem1Δ triple deletion is lethal (23) further suggests an essential role of these effectors may be linked to an essential ENTH function on the Cdc42 pathway. Because Bem1 recruits the Cdc42 guanine nucleotide exchange factor to sites of polarized growth (25), its impaired function would also contribute to deficient Cdc42 activation.
It is also significant that a gic1Δgic2Δ double knockout strain exhibits phenotypes very similar to ENTH1Y100R cell phenotypes, including enlarged fragile cells with actin cytoskeleton depolarization. The absence of actin cables in ENTH1Y100R cells is compatible with deficient function of the yeast formin Bni1, which in turn depends on Gic1/2 activation (26).
The Cdc42 effectors Gic1/2 have also been proposed to act as scaffold proteins that stabilize the interactions of activated Cdc42 with yet other effectors (26). Interestingly, a similar effector complex-stabilizing role has been proposed for the Cdc42 GAPs (27). We postulate an effector-stabilizing role for Cdc42 GAPs that is mediated by a protein complex containing epsin, Cdc42 GAP, Cdc42·GTP, and the effectors Gic1/2-Bem1. Besides further supporting the hypothesis that the epsins are involved in Cdc42 signaling, this pathway selectivity suggests the interesting possibility that the epsins could specify activation of one particular branch among multiple Cdc42 pathways. Although we do not favor this alternative possibility, it could be that all Cdc42 pathways are equally affected in ENTH domain cells, but only some of these, such as the Gic1/2-Bem1 pathway, are required for viability.
Cdc42 Regulation Function Is Independent of the Endocytic Role of the Epsins.
Our results suggest independent functions for the epsins in endocytosis and cell polarity, because the endocytic determinants (UIMs, NPF tripeptides, CBMs) are dispensable for the epsin essential function (Fig. 1A), and ENTH1Y100R cells lack endocytic defects (Fig. 2D). We are investigating the lack of an endocytic defect in ENTH1Y100R cells, which may be explained by our preliminary evidence that the endocytic function of the epsins can be fulfilled by other proteins (L. Maldonado-Baez and B.W., unpublished work). Thus, we propose that dual functions in endocytosis and polarity allow epsins to coordinate these two processes; for example, endocytosis and recycling of Cdc42 and other proteins are part of a positive-feedback loop for establishment and maintenance of polarity (28–30).
Evolutionary Conservation of Epsin Regulation of Cdc42 Pathways.
We began our study by analyzing the sequences of the yeast ENTH/AP180 N-terminal homology domain-containing proteins and later found our conclusions to be valid in other species. Residues Y100 and T104 are conserved among the epsins from evolutionarily distant species and among epsin isoforms (including the tolerated Y100F substitution; Fig. 7).
We showed genetic and physical evidence that the rat ENTH domain functions like the yeast ENTH domain in nearly every respect (Fig. 6). Interestingly, an interaction between mammalian epsin and a Cdc42 GAP has been previously reported, although its physiological role was unclear (8). We also found that the yeast ENTH domain, expressed in human cells, colocalizes with F-actin structures rich in activated Cdc42 and not with Rho-dependent actin stress fibers. Thus, the essential function of the epsin ENTH domain in Cdc42 regulation and actin polarity may be conserved in mammalian cells.
Other ENTH Domain Interaction Partners.
The triple rga1Δrga2Δbem3Δ Cdc42 GAP deletion is viable, in contrast to epsin deletion, suggesting the existence of unidentified ENTH domain-interaction partners contributing to the epsin essential function. We believe that a main function of the [epsin·GAP] complex is to activate the essential Cdc42 effectors Gic1/2 and Bem1 (gic1Δgic2Δbem1Δ cells are inviable). Thus, the unidentified ENTH domain-binding proteins need not be Cdc42 GAPs but could be factors that contribute in other ways to signaling downstream of Cdc42·GTP. Candidates for such factors were also found in our two-hybrid screen, Fir1 and Nis1. Like the Cdc42 GAPs, both are localized at the bud neck (31). Nis1 also interacts with the bud neck-localized septins (32); the septins in turn interact with Gic1/2 and are also regulated by the Cdc42 GAPs (20, 33).
A Model for Epsin Regulation of Cdc42.
Our working model proposes that, by binding to sites of nascent endocytosis, the epsins stabilize or reinforce sites of Cdc42 activation to establish a spatial-temporal link between the endocytic and the cell polarity machineries. At these sites, the epsins will form complexes with the Cdc42 GAPs, Cdc42·GTP, and perhaps certain effectors (Bem1, Gic1, and Gic2). Although in vivo there might be an ENTH domain-dependent inhibition of the GAP activity, we think that the key function of the ENTH-GAP interaction is to allow proper effector selection and activation to trigger the appropriate signal transduction pathway. We speculate that other proteins (not epsin) selectively activate non-Gic1/2-Bem1-dependent Cdc42 pathways.
Epsin·Cdc42 GAP complexes may also promote the cycling of Cdc42 through iterative rounds of GTP hydrolysis. It has been proposed that assembly of septins requires this type of iterative Cdc42 activation (34). The ENTH1Y100R cells show septin mislocalization and cytokinesis defects (unpublished results), as predicted for cells with impaired Bem1 activation, and therefore defective Cdc42 guanine nucleotide exchange factor recruitment (35).
Motile chemotactic cells have enhanced endocytosis toward the leading edge, where secretion and polarized growth are directed (36). These processes also occur in metastatic cells as they migrate (37). We propose that the endocytic epsin protein links sites of endocytosis and polarized cell growth and, by interacting with Cdc42 regulators, epsin contributes to the stabilization of the position of cellular responses to internal or external signals.
Materials and Methods
Materials were purchased from Fisher Scientific or Sigma.
Yeast Culture Conditions and Transformation Procedures.
Yeast strains were grown in standard yeast extract–peptone–dextrose (YPD) or synthetic medium with dextrose and amino acids required for plasmid maintenance at 30°C or 37°C for 3–4 days. 0.5xYPD plates are equivalent to YPD plates but with half the concentration of each component.
For liquid culture assays, ≈105 cells were inoculated in 10 ml of selective media, incubated at 30°C for 48 h in the presence or absence different concentrations of Met, and the OD at 600 nm measured. Yeast were transformed by the Li-Acetate method in the Clontech yeast handbook.
Plasmid Shuffle.
ΔΔ expressing ENT1 or ENT2 from a plasmid containing an auxotrophic marker (TRP1 or URA3) were used for counterselection on 5-FAA or 5-fluoroorotic acid (5-FOA), respectively. Cells cotransformed with mutant URA3 and wild-type TRP1 plasmids were grown on 5-FAA plates (to evict the TRP1 plasmid) at 30°C for 3 days. Alternatively, cells cotransformed with mutant TRP1 and wild-type URA3 plasmids were grown on 5-FOA plates (to evict the URA3 plasmid) at 30°C for 3 days.
Plasmids, strains, and antibodies used and detailed experimental procedures are in Supporting Text. Detailed experimental procedures and plasmids, strains, and antibodies used can be found in Tables 2, 3, and 4, respectively, which are published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Erfei Bi (University of Pennsylvania, Philadelphia), Alan Hall (University College London, London), George Sprague (University of Oregon, Eugene), Daniel Lew (Duke University, Durham, NC), Elmar Schiebel (Zentrum für Molekulare Biologie, Heidelberg), Doug Johnson (University of Vermont, Burlington), Michael Hall (University of Basel, Basel), Yoshikazu Ohya (University of Tokyo, Tokyo), Maria Molina (Universidad Complutense de Madrid, Madrid), Kazuma Tanaka (Hokkaido University, Sapporo, Japan), Yoshimi Takai (Osaka University, Osaka), Keith Kozminski (University of Virginia, Charlottesville), Clarence Chan (University of Texas, Austin), Brian Kay (University of Illinois, Chicago), Jean François (Institut National des Sciences Appliquées, Toulouse, France), Dave Katzmann (Mayo Clinic, Rochester, NY), Michael Edidin (Johns Hopkins University, Baltimore), Ludwig Brand (Johns Hopkins University, Baltimore), and Sarah Boyle (Johns Hopkins University, Baltimore) for strains, plasmids, and other reagents. We also thank Erfei Bi for helpful discussions and David Katzmann and members of the Wendland laboratory for constructive comments on the manuscript. We are grateful to Catherine Sciambi and Keisha Meeks for excellent technical assistance, Michael Edidin and Sarah Boyle for tissue culture facilities, and Ludwig Brand for help with tryptophan fluorescence. This work was supported by grants from the National Institutes of Health (National Institute of General Medical Sciences Grant R01-60979), the Human Frontiers Scientific Program (to B.W.), and the National Science Foundation (Grant 0131241, to E.F.); by a Howard Hughes Medical Institute predoctoral fellowship (to J.D.S.); and by a Ford Foundation Thesis Fellowship (to H.A.W.).
Abbreviations
- Met
methionine
- ENTH
epsin N-terminal homology
- CRIB
Cdc42/Rac interactive binding
- EBS
ENTH-binding site
- RnENTH
rat epsin1 ENTH domain
- ΔΔ
ent1Δent2Δ cells
- GAP
GTPase-activating protein
- 5-FAA
5-fluoroanthranilic acid
- UIM
ubiquitin-interaction motif
- NPF
Asn-Pro-Phe tripeptides
- CBM
clathrin-binding motif.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
See Commentary on page 3953.
References
- 1.Sorkin A., Von Zastrow M. Nat. Rev. Mol. Cell Biol. 2002;3:600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 2.Engqvist-Goldstein A. E., Drubin D. G. Annu. Rev. Cell Dev. Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 3.Conner S. D., Schmid S. L. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 4.McLauchlan H., Newell J., Morrice N., Osborne A., West M., Smythe E. Curr. Biol. 1998;8:34–45. doi: 10.1016/s0960-9822(98)70018-1. [DOI] [PubMed] [Google Scholar]
- 5.Sever S. Curr. Opin. Cell Biol. 2002;14:463–467. doi: 10.1016/s0955-0674(02)00347-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen H., Fre S., Slepnev V. I., Capua M. R., Takei K., Butler M. H., Di Fiore P. P., De Camilli P. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- 7.Wendland B. Nat. Rev. Mol. Cell Biol. 2002;3:971–977. doi: 10.1038/nrm970. [DOI] [PubMed] [Google Scholar]
- 8.Rosse C., L’Hoste S., Offner N., Picard A., Camonis J. J. Biol. Chem. 2003;278:30597–30604. doi: 10.1074/jbc.M302191200. [DOI] [PubMed] [Google Scholar]
- 9.Ridley A. J. Traffic. 2001;2:303–310. doi: 10.1034/j.1600-0854.2001.002005303.x. [DOI] [PubMed] [Google Scholar]
- 10.Wendland B., Steece K. E., Emr S. D. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overstreet E., Chen X., Wendland B., Fischer J. A. Curr. Biol. 2003;13:854–860. doi: 10.1016/s0960-9822(03)00326-9. [DOI] [PubMed] [Google Scholar]
- 12.Ford M. G., Mills I. G., Peter B. J., Vallis Y., Praefcke G. J., Evans P. R., McMahon H. T. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 13.Sloat B. F., Adams A., Pringle J. R. J. Cell Biol. 1981;89:395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams A. E., Pringle J. R. J. Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schott D., Huffaker T., Bretscher A. Curr. Opin. Microbiol. 2002;5:564–574. doi: 10.1016/s1369-5274(02)00369-7. [DOI] [PubMed] [Google Scholar]
- 16.Kaksonen M., Sun Y., Drubin D. G. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 17.Wang T., Bretscher A. Mol. Biol. Cell. 1995;6:1011–1024. doi: 10.1091/mbc.6.8.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marquitz A. R., Harrison J. C., Bose I., Zyla T. R., McMillan J. N., Lew D. J. EMBO J. 2002;21:4012–4025. doi: 10.1093/emboj/cdf416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burbelo P. D., Drechsel D., Hall A. J. Biol. Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 20.Drees B. L., Sundin B., Brazeau E., Caviston J. P., Chen G. C., Guo W., Kozminski K. G., Lau M. W., Moskow J. J., Tong A., et al. J. Cell Biol. 2001;154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown J. L., Jaquenoud M., Gulli M. P., Chant J., Peter M. Genes Dev. 1997;11:2972–2982. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukata M., Nakagawa M., Kaibuchi K. Curr. Opin. Cell Biol. 2003;15:590–597. doi: 10.1016/s0955-0674(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki R., Fujimura-Kamada K., Toi H., Kato H., Tanaka K. Genes Cells. 2003;8:235–250. doi: 10.1046/j.1365-2443.2003.00629.x. [DOI] [PubMed] [Google Scholar]
- 24.Bi E., Chiavetta J. B., Chen H., Chen G. C., Chan C. S., Pringle J. R. Mol. Biol. Cell. 2000;11:773–793. doi: 10.1091/mbc.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gulli M. P., Jaquenoud M., Shimada Y., Niederhauser G., Wiget P., Peter M. Mol. Cell. 2000;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- 26.Jaquenoud M., Peter M. Mol. Cell. Biol. 2000;20:6244–6258. doi: 10.1128/mcb.20.17.6244-6258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith G. R., Givan S. A., Cullen P., Sprague G. F., Jr Eukaryot. Cell. 2002;1:469–480. doi: 10.1128/EC.1.3.469-480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irazoqui J. E., Howell A. S., Theesfeld C. L., Lew D. J. Mol. Biol. Cell. 2005;16:1296–1304. doi: 10.1091/mbc.E04-05-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valdez-Taubas J., Pelham H. R. Curr. Biol. 2003;13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Wedlich-Soldner R., Altschuler S., Wu L., Li R. Science. 2003;299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- 31.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O’Shea E. K. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 32.Iwase M., Toh-e A. Genes Genet. Syst. 2001;76:335–343. doi: 10.1266/ggs.76.335. [DOI] [PubMed] [Google Scholar]
- 33.Caviston J. P., Longtine M., Pringle J. R., Bi E. Mol. Biol. Cell. 2003;14:4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gladfelter A. S., Bose I., Zyla T. R., Bardes E. S., Lew D. J. J. Cell Biol. 2002;156:315–326. doi: 10.1083/jcb.200109062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bose I., Irazoqui J. E., Moskow J. J., Bardes E. S., Zyla T. R., Lew D. J. J. Biol. Chem. 2001;276:7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- 36.Rappoport J. Z., Simon S. M. J. Cell Sci. 2003;116:847–855. doi: 10.1242/jcs.00289. [DOI] [PubMed] [Google Scholar]
- 37.Schmitz A. A., Govek E. E., Bottner B., Van Aelst L. Exp. Cell Res. 2000;261:1–12. doi: 10.1006/excr.2000.5049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.