Abstract
Human centromeres are specialized chromatin domains containing the centromeric histone H3 variant CENP-A. CENP-A nucleosomes are interspersed with nucleosomes containing histone H3 dimethylated at lysine 4, distinguishing centromeric chromatin (CEN chromatin) from flanking heterochromatin that is defined by H3 lysine 9 methylation. To understand the relationship between chromatin organization and the genomic structure of human centromeres, we compared molecular profiles of three endogenous human centromeres, defined by uninterrupted higher-order α-satellite DNA, with human artificial chromosomes that contain discontinuous blocks of higher-order α-satellite DNA and noncentromeric DNA. The underlying sequence did not correlate with chromatin states, because both higher-order α-satellite DNA and noncentromeric DNA were enriched for modifications that define CEN chromatin, euchromatin, and heterochromatin. Human artificial chromosomes were also organized into distinct domains. CENP-A and heterochromatin were assembled over noncentromeric DNA, including the gene blasticidin, into nonoverlapping domains. Blasticidin transcripts were enriched at sites of CENP-A binding but not at H3 methylated at lysine 9, indicating that formation of CEN chromatin within a repetitive DNA environment does not preclude gene expression. Finally, we tested the role of centric heterochromatin as a centromeric boundary by increasing CENP-A dosage to expand the CEN domain. In response, H3 lysine 9 dimethylation, but not trimethylation, was markedly decreased at all centromeres examined. We propose that human centromere regions normally exist in a dynamic state in which a regional boundary, defined by H3 lysine 9 dimethylation, separates CEN chromatin from constitutive heterochromatin.
Keywords: α-satellite, CENP-A, centromere, heterochromatin, histone
The centromere is the locus that mediates chromosome segregation in mitosis and meiosis (1). In multicellular eukaryotes, centromere identity and kinetochore formation are influenced by primary DNA sequence and epigenetic factors (2). The histone H3 variant CENP-A is a conserved marker at centromeres and is assembled into nucleosomes by replacing H3 (1, 3). Interspersed subdomains of CENP-A and histone H3 nucleosomes collectively produce centromeric chromatin (CEN chromatin), which is the structural foundation of the kinetochore (4). Given the importance of histone modifications in regulating chromatin dynamics, modification of H3 within CEN chromatin may impact centromere assembly, perhaps by recruiting CENP-A or marking sites for incorporation of newly synthesized CENP-A. In fact, H3 nucleosomes within CEN chromatin in humans and Drosophila are dimethylated at K4 (H3K4me2) (5). As a domain, CEN chromatin (containing both CENP-A and H3K4me2) is continuous, meaning that it is uninterrupted or not interspersed with other types of chromatin. Heterochromatin, defined by H3-K9 dimethylation and trimethylation (H3K9me2 and H3K9me3), flanks CEN chromatin (2, 6). Thus, CEN chromatin is structurally and functionally distinct from heterochromatin (2, 7).
Domain organization of centromere regions is highly conserved (6). CEN chromatin and heterochromatin are each required for chromosome segregation and de novo chromosome assembly (8–10). How heterochromatin contributes to structural attributes of the kinetochore is unclear, as is the nature of CEN chromatin itself. In Drosophila, heterochromatin is thought to prevent the spread of CENP-ACID (11) into noncentromeric DNA (12, 13). Separation of euchromatic and heterochromatic domains is often maintained by balancing different histone-modifying enzymes and chromatin components (14–16). For instance, overexpression of Su(var)3–9, the histone methyltransferase in Drosophila that trimethylates H3-K9 (17, 18), triggers expansion of repressive heterochromatic domains (14). A prediction of this model is that components of CEN chromatin and heterochromatin are similarly regulated.
Human centromeres are genomically defined by α-satellite, a 171-bp monomeric repeat arranged into tandem, higher-order arrays that form de novo centromeres when introduced into human cells (19, 20). The size (≤4 Mb) and repetitive nature of human centromeres have impeded assembly of molecular maps and limited comprehensive functional analyses. Here, we report histone modification patterns at human centromeres and on human artificial chromosomes by using chromatin immunoprecipitation (ChIP) with a panel of antibodies that recognize specific methylated lysine residues on histone H3. We also used extended chromatin fibers to compare the arrangement of CEN chromatin at endogenous centromeres and on de novo human artificial chromosomes (21, 22) that contain interrupted blocks of α-satellite sequences. Finally, we show that CEN chromatin is assembled on noncentromeric sequences and does not silence gene expression within the context of a functional centromere and that centromere domain organization is disrupted when the dosage of CENP-A is altered. We conclude that CEN chromatin and constitutive heterochromatin in humans exist as distinct domains that are separated by variable amounts of chromatin defined by H3K9me2. These results provide insights into CENP-A chromatin and strengthen the emerging model that CEN chromatin is neither exclusively heterochromatic nor euchromatic.
Results and Discussion
Histone Modifications Are Conserved at α-Satellite DNA Arrays.
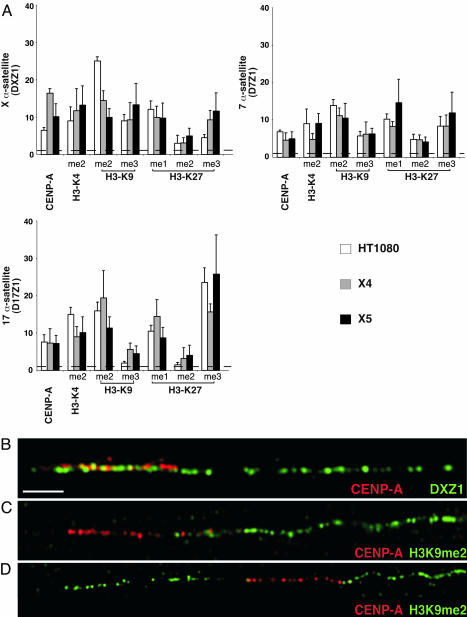
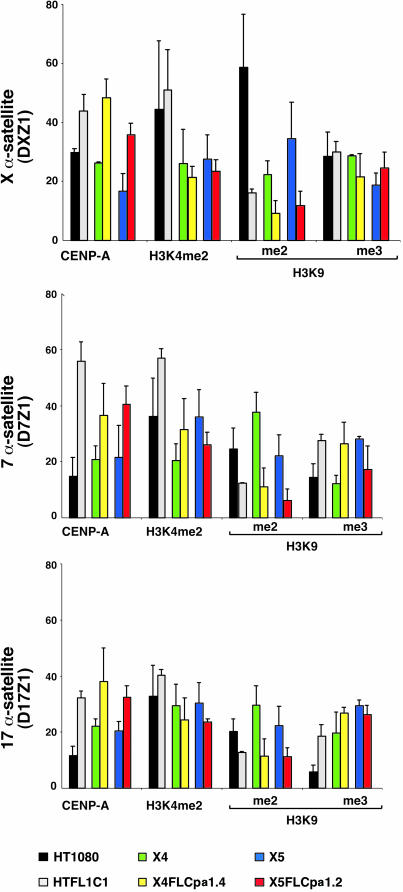
Human centromeres contain homogenous arrays of higher-order α-satellite DNA, but other smaller arrays of higher-order α-satellite and stretches of divergent monomeric α-satellite are also located in or near the primary constriction (22, 23). Noncoding RNAs transcribed from monomeric α-satellite sequences participate in RNA interference-mediated heterochromatin assembly (24), implying that heterochromatic histones are excluded from higher-order α-satellites at which kinetochore proteins and H3K4me2 nucleosomes are assembled (2). We analyzed the distribution of CENP-A and mono-, di-, and trimethylated H3 at higher-order α-satellite DNA from human chromosomes 7 (D7Z1), 17 (D17Z1), and X (DXZ1) by using ChIP and semiquantitative PCR (see Methods). As expected, D7Z1, D17Z1, and DXZ1 were enriched for CENP-A in all cell lines studied (Fig. 1). ChIP-PCR analysis also supported cytological data that α-satellite DNA is enriched for H3K4me2, which defines euchromatin (2, 25) (Fig. 1A). Using chromatin fibers to optically map entire centromere regions, we observed that CEN chromatin occupied 30–50% of a specific α-satellite array (Fig. 1B) (26). Given the close proximity of H3K9me to CEN chromatin and the limited extent of CENP-A on α-satellite DNA (Fig. 1 B–D), we hypothesized that higher-order α-satellite DNA also contributes to heterochromatin assembly.
Fig. 1.
Human α-satellite DNA is assembled into CEN chromatin, euchromatin, and heterochromatin. (A) ChIP-PCR of histone modifications at α-satellite DNA from chromosomes X, 7, and 17 in cell lines HT1080, X4, and X5. Bar graphs show relative enrichment (>1). Baseline enrichment was set at 1 (dashed line) (see Methods). ChIP-PCR was performed three times; averages are shown with standard deviation bars. (B–D) Optical mapping on extended chromatin fibers shows that CENP-A (red) occupies a fraction of the α-satellite array (DXZ1) of the human X chromosome. H3K9me2 (green) is closely located at one (C) or both (D) sides of the CEN chromatin domain marked by CENP-A (red), suggesting that α-satellite DNA is also assembled into heterochromatin. (Scale bar, 10 μm.)
By ChIP-PCR, α-satellite DNA was enriched for H3K9me2 and, to a lesser extent, H3K9me3. H3K27me1 and H3K27me3, but not H3K27me2, were also present at α-satellite DNA (Fig. 1A). We conclude that human centromeres containing homogeneous arrays of higher-order α-satellite DNA are assembled into three types of chromatin, CEN chromatin, euchromatin, and heterochromatin. Distinctive enrichment for certain modifications among three different centromeres, such as increased H3K9me2 and H3K27me3, was observed on chromosomes 17 and X, compared with chromosome 7 (Fig. 1A). Chromosome-specific variability in histone modifications may reflect underlying genomic differences of human centromeric DNAs.
CEN Chromatin Assembly over Noncentromeric DNA on Human Artificial Chromosomes.
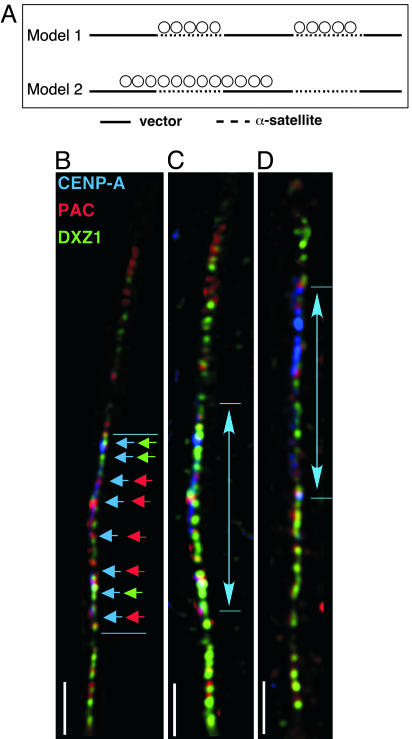
Unlike endogenous human centromeres that contain uninterrupted arrays of α-satellite DNA, human artificial chromosomes are organized as α-satellite arrays interspersed with noncentromeric vector [bacterial artificial chromosome (BAC)/P1 artificial chromosome (PAC)] sequences and multiple copies of a selectable marker gene (Fig. 2A) (27). Cytologically visible artificial chromosomes result from multimerization of input constructs (27), and centromere organization on artificial chromosomes might represent one of two models (Fig. 2A). CEN chromatin may be assembled only at blocks of α-satellite, whereas heterochromatin is located on noncentromeric sequences (1). Alternatively, CEN chromatin on artificial chromosomes could be organized as a single domain of CENP-A/H3K4me2 that is flanked, and not interrupted, by heterochromatin (2, 28).
Fig. 2.
CENP-A spreads over noncentromeric DNA on human artificial chromosomes. (A) Positions of primer pairs on human artificial chromosome input constructs for cell lines X4 and X5. (B) Proposed models for sequence-dependent (Model 1) and sequence-independent (Model 2) assembly of CENP-A on human artificial chromosomes. (C) IF-FISH on chromatin fibers of DXZ1-derived human artificial chromosomes shows localization of CENP-A (blue), PAC vector DNA (red), and α-satellite DNA (green). Colored arrowheads show overlap between CENP-A/PAC DNA and CENP-A/α-satellite DNA. (Scale bar, 15 μm.) Arrowed lines denote CENP-A staining (blue), illustrating that CEN chromatin containing CENP-A and H3K4me2 is organized as a single domain and not as multiple blocks along the entire artificial chromosome.
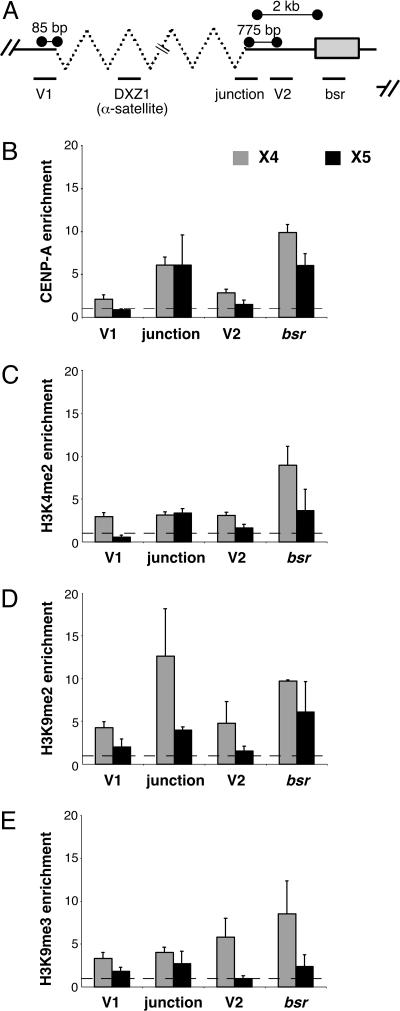
To test these models and to investigate the role of genomic structure in centromere organization, we studied two different human artificial chromosomes, X4 and X5, generated from X-chromosome-derived α-satellite sequences (DXZ1) (21, 22). Extended chromatin fibers from X4 and X5 were stained with antibodies to CENP-A. CENP-A localized to a single continuous region on each artificial chromosome, assembling across both PAC vector DNA and X α-satellite DNA (Fig. 2 B–D). To confirm these analyses, we examined CENP-A assembly on noncentromeric sequences using ChIP-PCR (Fig. 3). Primers specific for the α-satellite/vector junction distinguished DXZ1 on the artificial chromosome X from the endogenous X centromere (Fig. 3A) (see Methods). CENP-A was present at the junction between X α-satellite and the PAC vector, on vector sequences (V1, V2) flanking the X α-satellite array and on the bsr gene (Fig. 3B). Heterochromatic and euchromatic modifications were also detected on these same sequences (Fig. 3 C–E). The bsr gene on both X4 and X5 was enriched for H3K4me2, H3K9me2, and H3K9me3 (Fig. 3 C–E), implying that some copies of the gene are transcriptionally active and others are silenced. Chromatin organization on X4 and X5 was largely consistent, but there were some differences in enrichment for H3K4me2, H3K9me2, and H3K9me3 at V1 and V2 on X5 (Fig. 3 B–E). These discrepancies may reflect size differences between X4 and X5 that correlate with the amount of heterochromatin (29). Collectively, these results indicate that distinctive types of chromatin (CEN chromatin, heterochromatin, and euchromatin) are assembled on human artificial chromosomes (Fig. 2B, Model 2). Although they lack homogenous α-satellite arrays, human artificial chromosomes contain a single CEN chromatin domain that is not interrupted by blocks of heterochromatin, a configuration that resembles centromeres in fission yeast, flies, and humans (2, 6, 9). We also conclude that CENP-A can spread in cis over both centromeric (α-satellite) and noncentromeric DNA, including a gene. These findings emphasize the importance of conserved centromere organization (6, 30), despite differences in underlying DNA sequence.
Fig. 3.
Centromeric organization on human artificial chromosome is similar to endogenous centromeres. ChIP-PCR identified histone modifications on human artificial chromosomes X4 and X5. (A) PCR primers recognized specific regions (V1, V2, junction, and bsr gene) of the input subunit of the artificial chromosome. Average relative enrichments for CENP-A (B) and each histone modification (C–E) are shown as bar graphs.
Transcription of a Gene Occurs in CEN Chromatin.
Because DNA from the selectable marker on artificial chromosomes was immunoprecipitated by CENP-A antibodies, we investigated whether transcription occurred within human CEN chromatin. RNA-ChIP was used to detect transcription of the bsr gene. Unsurprisingly, bsr transcripts were most highly enriched for H3K4me3 (see Fig. 6, which is published as supporting information on the PNAS web site), a modification linked to active transcription (31) and enriched to a lesser extent for H3K4me2. Negligible bsr transcription was detected within chromatin containing H3K9me3, a marker that defines constitutive heterochromatin. H3K9me and the low level of bsr transcripts by RNA-ChIP indicate that at least one copy of the gene is silenced by heterochromatin assembly. However, there was a significant enrichment for bsr transcripts immunoprecipitated with CENP-A antibodies compared with H3K9me3 (P = 0.03) (Fig. 6). These data suggest that transcription occurs within chromatin containing CENP-A, and, unlike constitutive heterochromatin, CEN chromatin does not inhibit gene expression. Our results agree with the association of active genes and transcription at plant centromeres, human neocentromeres, and other artificial chromosomes (7, 32, 33).
Chromatin Domains at Human Centromeres Are Dynamically Regulated.
In model organisms, dosage change in euchromatic or heterochromatic modifiers has dramatic effects on gene expression, chromatin assembly, and chromosome stability (8, 14, 15). To investigate the relationship between CEN chromatin and flanking heterochromatin in humans and to specifically test whether heterochromatin restricts CEN chromatin (13), we increased CENP-A dosage and examined the effect on CEN chromatin assembly at endogenous human centromeres and on human artificial chromosomes. Multiple cell lines were isolated that stably expressed FLAG-tagged CENP-A, increasing the amount of chromatin-associated CENP-A by 50% over normal levels (see Methods; and see Fig. 7, which is published as supporting information on the PNAS web site). Under these conditions, which differ from transient expression or mistargeting assays (11, 34, 35), CENP-A is not incorporated into chromosome arms but remains localized within centromeric regions (data not shown).
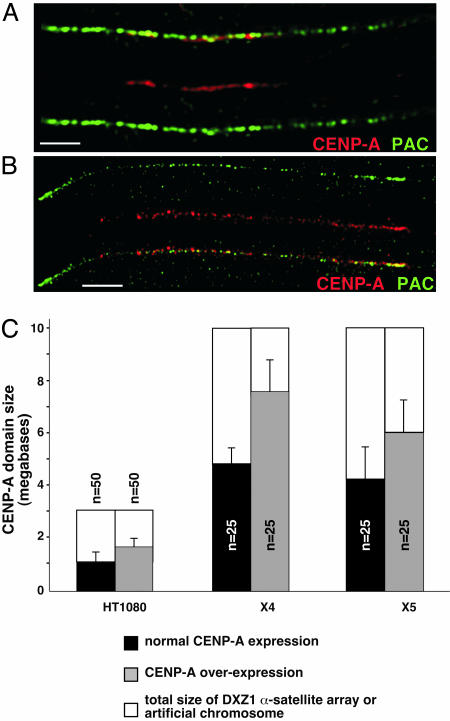
Arrays of α-satellite DNA from endogenous chromosomes X (DXZ1) and Y (DYZ3) were sized by pulsed-field gel electrophoresis (36) (data not shown). These two chromosomes are haploid in the cell lines studied and easily identified in long-range optical and molecular sizing analyses. The region occupied by CENP-A at endogenous centromeres and on artificial chromosomes was measured on chromatin fibers by colocalizing CENP-A antibodies and DNA probes specific for α-satellite DNA or PAC vector sequences. In FLAG-CENP-A lines, CENP-A covered 25–50% more of the α-satellite DNA array than in controls (Fig. 4). CEN size also expanded on artificial chromosomes X4 and X5, although less so on X5, the smaller artificial chromosome (Fig. 4 A–C).
Fig. 4.
Increased dosage of CENP-A causes the CEN domain to expand at endogenous centromeres and on human artificial chromosomes. (A and B) IF-FISH on chromatin fibers using CENP-A antibodies and α-satellite DNA probes spatially defined CEN chromatin domain on DXZ1 α-satellite array at endogenous centromeres and on human artificial chromosomes. (A) CENP-A (red) is present on ≈35% of the human artificial chromosome identified by PAC/α-satellite FISH probe (green). When CENP-A expression is increased (B), CENP-A (red) spreads over ≈80% of the artificial chromosome (green). (Scale bars, 10 μm.) (C) CEN chromatin, defined by CENP-A, also expands at the endogenous X centromere in HT1080 cells.
When CENP-A is transiently overexpressed, it replaces H3 in chromosome arms (9, 34, 35). However, it is not known which H3 modifications are exchanged or whether any are resistant to replacement by CENP-A. We examined changes in histone modification patterns in cell lines expressing FLAG-CENP-A (Fig. 5). CENP-A enrichment clearly increased by 1.5- to 2-fold at D7Z1, D17Z1, and DXZ1 in each cell line (Fig. 5). H3K4me2 enrichment did not increase; however, the amount of H3K9me2 immediately flanking CEN chromatin decreased 2- to 3-fold (Fig. 5). We conclude that increased dosage of CENP-A causes CENP-A to spread along α-satellite arrays, displacing H3K9me2 but not H3K9me3 (see Fig. 8, which is published as supporting information on the PNAS web site). Decreasing pericentromeric H3K9me2 in this way did not affect chromosome segregation (10), because aneuploidy or segregation abnormalities were not detected (data not shown).
Fig. 5.
Overexpression of CENP-A correlates with reduction of H3K9me2 at human centromeres. ChIP-PCR of histone modifications at three human centromeres (D7Z1, D17Z1, and DXZ1) in CENP-A-overexpressing cell lines showed increased CENP-A enrichment at α-satellite from chromosomes 7 and X and concurrent decrease in H3K9me2. Depending on the centromere, H3K9me3 increased. % IP for histone modifications at α-satellite arrays was compared as relative ratios of each modification. Bar graphs are relative average ratios (% IP over input DNA) with standard deviations.
At some centromeres and on artificial chromosome X4, H3K9me3 enrichment increased at α-satellite DNA when CENP-A dosage increased. This finding may reflect a compensatory mechanism, similar to that described in mice, in which loss of H3K9me3 at pericentric heterochromatin causes redistribution of H3K27me3 to this region, preserving the repressive state (17). At human centromeres, H3K9me3 may spread to counteract potential disruption of the 3D metaphase kinetochore when H3K9me2 is decreased. Alternatively, expansion of H3K9me3 may reflect a passive response to the removal of a boundary. In Schizosaccharomyces pombe, specific structural elements physically separate heterochromatin from chromatin containing CENP-ACnp1 (6). However, boundaries between heterochromatin and CEN chromatin at human centromeres may be larger and less sequence-specific (22). Regional barriers, such as chromatin defined by H3K9me2, may, instead, demarcate distinctive centromeric domains (Fig. 8).
The centromeres studied here showed some differences in enrichment for specific histone H3 modifications. Variations in domain size or composition at each centromere may reflect differences in α-satellite array size or genomic structure and explain the different effects (i.e., no change or expansion) on H3K9me3 pericentric domains when H3K9me2 was reduced by CENP-A overexpression. Precise sizes of chromatin domains at individual centromeres remain to be determined as well as whether variations in histone modification enrichment are functionally significant. It will also be interesting to examine small, deleted, and rearranged human centromeres or centromeres of human chromosomes in rodent or other primate backgrounds for alterations in centromere organization and/or ratios of histone variants and modifications. Such studies will shed light on chromosome and centromere evolution and boundaries between functional chromatin domains.
Methods
Cell Culture.
Human HT1080 derivative cell lines X4 and X5 containing human artificial chromosomes assembled from X α-satellite DNA have been described in refs. 21 and 29. HTFL1c1, X4FLCpa1.4, and X5FLCpa1.2 are derivative clones that stably express FLAG-CENP-A (4).
ChIP.
Chromatin containing oligonucleosomes was prepared by micrococcal nuclease digestion (4). Soluble chromatin was immunoprecipitated overnight at 4°C with no antibody (mock), rabbit polyclonal (Upstate Biotechnology, Lake Placid, NY), or mouse monoclonal (Abcam, Cambridge, MA) antibodies to CENP-A and antibodies recognizing H3K4me2 (Abcam), H3K9me2, H3K9me3 (Abcam), H3K27me1, H3K27me2, and H3K27me3 (Abcam). For further details, see Supporting Methods, which is published as supporting information on the PNAS web site.
PCR Analysis.
Relative DNA enrichment for histone antibodies was determined by semiquantitative PCR using primers to amplify α-satellite DNA (37), α-satellite/pPAC4ΔoriP vector junction, bsr, and pPAC4ΔoriP vector sequences (Fig. 2A, primer sequences for test and control sequences are available upon request). PCR products were analyzed by agarose-gel electrophoresis and quantified by using the Alpha Imaging System (Alpha Innotech). Relative enrichment for CENP-A was calculated by using (%IP DNA/D7Z2 DNA)antibody/(%IP DNA/D7Z2 DNA)mock, where %IP is the percentage of immunoprecipitated DNA over input. Relative enrichment for H3K9me2, H3K9me3, and H3K27me2 was calculated by using (%IP DNA/5S rDNA)antibody/(%IP DNA/5S rDNA)mock. Relative enrichment for H3K4me2, H3K27 me1, H3K27me2, and H3K27me3 was calculated by using (%IP DNA/γ-satellite DNA)antibody/(%IP DNA/γ-satellite DNA)mock.
Immunofluorescence (IF)-FISH.
Extended chromatin fibers were generated as described in refs. 2 and 4. Chromosome-specific α-satellite DNA was identified by using PCR-cloned products (37). Vector DNA on human artificial chromosomes was detected by using pPAC4 or pPAC4ΔoriP (21, 38). Probes were labeled with biotin-16-dUTP (Roche), digoxygenin-11-dUTP (Roche), or AlexaFluor dUTPs (Molecular Probes). α-Satellite array sizes were estimated by pulsed-field gel electrophoresis and Southern blotting as described in refs. 36, 39, and 40 by using PCR-generated probes specific for DXZ1 and DYZ3 (37).
Microscopy and Image Analysis.
All images were acquired and analyzed by using the Deltavision Spectris Restoration Imaging System (Applied Precision) (4). The “measure distances” tool in the program softworx resolve 3d was used to calculate CENP-A domain size in control and FLAG-CENP-A lines. CENP-A antibody staining (in μm) was measured against the length of α-satellite FISH probe (37) and/or pPAC4 signal. α-Satellite FISH probe signal length represented array size that was determined by pulsed-field gel electrophoresis. CENP-A domain size was calculated from the ratio of the lengths of CENP-A antibody signal over the α-satellite FISH signal.
Supplementary Material
Acknowledgments
We thank Hunt Willard for comments on the manuscript. This work was supported by National Institutes of Health Grant GM069514 and March of Dimes Basil O’Connor Scholar Award #5-FY04-29 (to B.A.S.).
Abbreviations
- bsr
blasticidin
- CEN chromatin
centromeric chromatin
- ChIP
chromatin immunoprecipitation
- IF
immunofluorescence
- PAC
P1 artificial chromosome.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sullivan B. A., Blower M. D., Karpen G. H. Nat. Rev. Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan B. A., Karpen G. H. Nat. Struct. Mol. Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black B. E., Foltz D. R., Chakravarthy S., Luger K., Woods V. L., Jr., Cleveland D. W. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 4.Blower M. D., Sullivan B. A., Karpen G. H. Dev. Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner B. M. Nat. Struct. Mol. Biol. 2005;12:110–112. doi: 10.1038/nsmb0205-110. [DOI] [PubMed] [Google Scholar]
- 6.Partridge J. F., Borgstrom B., Allshire R. C. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 7.Nakashima H., Nakano M., Ohnishi R., Hiraoka Y., Kaneda Y., Sugino A., Masumoto H. J. Cell Sci. 2005;118:5885–5898. doi: 10.1242/jcs.02702. [DOI] [PubMed] [Google Scholar]
- 8.Ekwall K., Olsson T., Turner B. M., Cranston G., Allshire R. C. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 9.Blower M. D., Karpen G. H. Nat. Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernard P., Maure J. F., Partridge J. F., Genier S., Javerzat J. P., Allshire R. C. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 11.Henikoff S., Ahmad K., Platero J. S., van Steensel B. Proc. Natl. Acad. Sci. USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams B. C., Murphy T. D., Goldberg M. L., Karpen G. H. Nat. Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- 13.Maggert K. A., Karpen G. H. Genetics. 2001;158:1615–1628. doi: 10.1093/genetics/158.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebert A., Schotta G., Lein S., Kubicek S., Krauss V., Jenuwein T., Reuter G. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura A., Umehara T., Horikoshi M. Nat. Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 16.Meneghini M. D., Wu M., Madhani H. D. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 17.Peters A. H., Kubicek S., Mechtler K., O’Sullivan R. J., Derijck A. A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., et al. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 18.Rice J. C., Briggs S. D., Ueberheide B., Barber C. M., Shabanowitz J., Hunt D. F., Shinkai Y., Allis C. D. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 19.Willard H. F. Trends Genet. 1990;6:410–416. doi: 10.1016/0168-9525(90)90302-m. [DOI] [PubMed] [Google Scholar]
- 20.Harrington J. J., Van Bokkelen G., Mays R. W., Gustashaw K., Willard H. F. Nat. Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 21.Rudd M. K., Mays R. W., Schwartz S., Willard H. F. Mol. Cell. Biol. 2003;23:7689–7697. doi: 10.1128/MCB.23.21.7689-7697.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schueler M. G., Higgins A. W., Rudd M. K., Gustashaw K., Willard H. F. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- 23.Rudd M. K., Willard H. F. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 25.Schneider R., Bannister A. J., Myers F. A., Thorne A. W., Crane-Robinson C., Kouzarides T. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 26.Spence J. M., Critcher R., Ebersole T. A., Valdivia M. M., Earnshaw W. C., Fukagawa T., Farr C. J. EMBO J. 2002;21:5269–5280. doi: 10.1093/emboj/cdf511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mejia J. E., Willmott A., Levy E., Earnshaw W. C., Larin Z. Am. J. Hum. Genet. 2001;69:315–326. doi: 10.1086/321977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choo K. H. Dev. Cell. 2001;1:165–177. doi: 10.1016/s1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- 29.Grimes B. R., Babcock J., Rudd M. K., Chadwick B., Willard H. F. Genome Biol. 2004;5:R89. doi: 10.1186/gb-2004-5-11-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pidoux A. L., Allshire R. C. Chromosome Res. 2004;12:521–534. doi: 10.1023/B:CHRO.0000036586.81775.8b. [DOI] [PubMed] [Google Scholar]
- 31.Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 32.Nagaki K., Cheng Z., Ouyang S., Talbert P. B., Kim M., Jones K. M., Henikoff S., Buell C. R., Jiang J. Nat. Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 33.Saffery R., Sumer H., Hassan S., Wong L. H., Craig J. M., Todokoro K., Anderson M., Stafford A., Choo K. H. Mol. Cell. 2003;12:509–516. doi: 10.1016/s1097-2765(03)00279-x. [DOI] [PubMed] [Google Scholar]
- 34.Shelby R. D., Vafa O., Sullivan K. F. J. Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Hooser A. A., Ouspenski II, Gregson H. C., Starr D. A., Yen T. J., Goldberg M. L., Yokomori K., Earnshaw W. C., Sullivan K. F., Brinkley B. R. J. Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 36.Mahtani M. M., Willard H. F. Genomics. 1990;7:607–613. doi: 10.1016/0888-7543(90)90206-a. [DOI] [PubMed] [Google Scholar]
- 37.Warburton P. E., Greig G. M., Haaf T., Willard H. F. Genomics. 1991;11:324–333. doi: 10.1016/0888-7543(91)90139-6. [DOI] [PubMed] [Google Scholar]
- 38.Grimes B. R., Rhoades A. A., Willard H. F. Mol. Ther. 2002;5:798–805. doi: 10.1006/mthe.2002.0612. [DOI] [PubMed] [Google Scholar]
- 39.Mahtani M. M., Willard H. F. Genome Res. 1998;8:100–110. doi: 10.1101/gr.8.2.100. [DOI] [PubMed] [Google Scholar]
- 40.Floridia G., Zatterale A., Zuffardi O., Tyler-Smith C. EMBO Rep. 2000;1:489–493. doi: 10.1093/embo-reports/kvd110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.