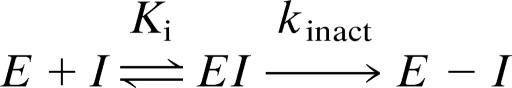Abstract
Resorcylic acid lactones containing a cis-enone are susceptible to Michael addition reactions and are potent inhibitors of several protein kinases. A structural-bioinformatics analysis identified a conserved Cys residue in the ATP-binding site of the kinases reported to be inhibited by cis-enone resorcylic acid lactones but absent in those that are not. Mining of the kinome database revealed that a subset of some 46 kinases contained this Cys residue. Screening a panel of 124 kinases with the resorcylic acid lactone hypothemycin showed that 18 of 19 targets containing the conserved Cys were inhibited. Kinetic analyses showed time-dependent inhibition, a hallmark of covalent inactivation, and biochemical studies of the interaction of extracellular signal-regulated kinase (ERK)2 with hypothemycin confirmed covalent adduct formation. Resorcylic acid lactones are unique among kinase inhibitors in that they target mitogen-activated protein (MAP) kinase pathways at four levels: mitogen receptors, MAP kinase kinase (MEK)1/2 and ERK1/2, and certain downstream ERK substrates. Cell lines dependent on the activation of Tyr kinase mitogen receptor targets of the resorcylic acid lactones were unusually sensitive toward hypothemycin and showed the expected inhibition of kinase phosphorylation due to inhibition of the mitogen receptors and/or MEK1/2 and ERK1/2. Among cells without mitogen receptor targets, those harboring an ERK pathway-activating B-RAF V600E mutation were selectively and potently inhibited by hypothemycin. Hypothemycin also prevented stimulated activation of the p38 cascade through inhibition of the Cys-containing targets MEK3/6 and TGF-β-activated kinase 1 and of the JNK/SAPK (c-Jun N-terminal kinase/stress-activated protein kinase) cascade through inhibition of MEK4/7.
Keywords: B-RAF V600E, hypothemycin, irreversible kinase inhibitor, Michael adduct
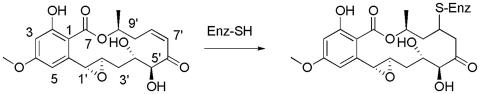
Resorcylic acid lactones (RALs) are polyketide natural products with a large macrocyclic ring fused to resorcylic acid. Some RALs contain an α,β-unsaturated ketone in the macrocycle, as exemplified by the cis-enone RALs hypothemycin, 5Z-7-oxozeaenol, and L-783,277 (Fig. 1). The cis-enone RALs have been shown to inhibit mammalian cell proliferation and tumor growth in animals (1–3). Furthermore, several reports have indicated that cis-enone RALs inhibit certain protein kinases, such as mitogen-activated protein (MAP) kinase (MAPK) kinase (MEK)1 (4), TGF-β-activated kinase 1 (TAK1) (5) and platelet-derived growth factor receptor (PDGFR) (6), but not others, such as RAF, PKA, PKC, endothelial growth factor receptor (EGFR), FGF receptor, ZAP70, MEK kinase 4, and lymphoid-specific Tyr kinase p56kk (LCK) (3, 4, 6). Where tested, targets inhibited by the cis-enone RALs were not affected by trans-enone RALs or RAL analogs lacking the α,β-unsaturated ketone (4, 5).
Fig. 1.
Structure of hypothemycin and its Michael addition product to a Cys thiol of a protein kinase. L-783,277 and 5Z-7-oxozeanol have carbon–carbon single and double bonds, respectively, instead of the 1′,2′ epoxide.
L-783,277 was shown to be a potent in vitro inhibitor of MEK1, competitive with ATP, that became even more potent upon preincubation (4). The apparent time-dependent inhibition caught our attention because it is a hallmark of affinity-directed covalent bond formation between enzyme and inhibitor, and the α,β-unsaturated ketone moiety is an effective Michael acceptor of protein nucleophiles, particularly Cys thiolate.
Off-target inhibition by a kinase inhibitor is usually unpredictable, and assessment of complete specificity usually requires screening of the entire kinome (7). In the present work, we identified a Cys residue that is conserved in the ATP site of kinase targets reported to be inhibited by cis-enone RALs but absent from those that are not. Furthermore, a structure–bioinformatics approach revealed that this Cys is present in a subset of some 46 protein kinases in the kinome that include such important targets as mitogen receptor Tyr kinases, MEK, and ERK. We show that hypothemycin forms stable covalent adducts with this Cys residue and is highly efficacious in inhibiting growth of cells dependent on the target kinases, in particular, cells dependent on mitogen receptor RAL targets or harboring B-RAF V600E mutations that drive the ERK pathway.
Results
Bioinformatics.
Sequence alignment of the kinases reported to be inhibited by cis-enone RALs revealed a conserved Cys residue (Cys-166 in human ERK2) adjacent to the completely conserved Asp that is involved in binding the Mg2+ complexed to ATP; kinases that were reportedly not inhibited by a RAL had no Cys residue at that position. Interrogation of the human kinome sequence database revealed that some 46 of 510 identified kinases contained the target Cys and were therefore considered candidates for RAL inhibition (Table 1).
Table 1.
Kinases containing the target Cys for RALs (Cys166 in human ERK2)
| Branches of human kinome | Kinases |
|---|---|
| TK | FLT1 (VEGFR1), KDR (VEGFR2), FLT4 (VEGFR3), FLT3, KIT, PDGFRα, PDGFRβ |
| TKL | TAK1, TGF-BR2, ZAK |
| CMGC | CDKL1, CDKL2, CDKL3, CDKL4, CDKL5, GSK3α, GSK3β, ERK1, ERK2, MAPK15, NLK, PRP4K |
| CAMK | MAPKAP5, MNK1, MNK2, RSK1*, RSK2*, RSK3*, RSK4*, PKD1, PKD2, PKD3, SPEG† |
| STE | MEK1, MEK2, MEK3, MEK4, MEK5, MEK6, MEK7, NIK1 |
| Other | AAK1, BMP2K, GAK, STK36, TOPK |
*The target Cys is in the more C-terminal of two kinase domains.
†Expected to include at least two splice variants (24), each containing two kinase domains with the target Cys in the N-terminal domain.
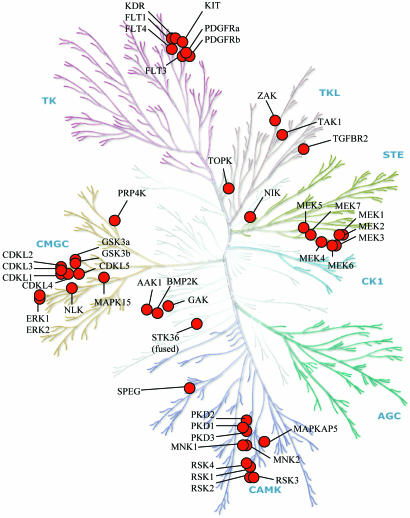
Of the 46 Cys-containing RAL targets, 38 exist in 8 evolutionarily related clusters, 7 of which lie within 5 major branches of the human kinase tree (Fig. 2) (8). The largest branch of ≈90 Tyr kinases contains a cluster of 7 RAL targets, all in the subbranch composed of the 12 split kinases associated with mitogen-stimulated cell proliferation; these include all three vascular EGFRs (VEGFRs) and four of five PDGFR-type kinases. In the branch containing the 74 calmodulin-dependent protein kinases and related kinases, 10 of the 11 RAL targets are concentrated in 2 clusters. Seven RAL targets are in a group of 11 MAPK substrates and consist of the 4 ribosomal subunit kinase (RSK) isoforms, two MAPK-interacting kinase 1/2 isoforms, and MAPK-activating protein 5; the remainders are the three protein kinase D members and SPEG (striated muscle preferentially expressed gene). In the 61-membered CMGC branch, three clusters and one isolate make up 12 of the Cys-containing RAL targets; these include all five CDKL (cyclin-dependent kinase-like) isoforms, four members of the MAPK branch, including ERK1/2, the two glycogen synthase kinase (GSK)3 isoforms, and the isolate PRP4K (pre-mRNA processing factor 4 kinase). The 47 STE kinases include 8 targets: one cluster of 7 MEK isoforms and 1 isolate. Among the 43 Tyr kinase-like kinases are three isolates, including TAK1 and TGF-β receptor type 2. Target kinases not belonging to the seven major branches include a cluster of three in the AAK1/BIKE/GAK (adaptor-associated kinase 1/bone morphogenetic protein 2-inducible kinase/cyclin G-associated kinase) group and three isolates. Two large branches of the tree, the 63 AGC and the 12 CK1 (casein kinase 1) kinases, are free of Cys-containing RAL targets.
Fig. 2.
Dendrogram of the kinase family relationships showing the location of the RAL target kinases. [Figure adapted with permission from Manning et al. (8) (Copyright 2002, Cell Signaling Technology).]
We also attempted to address whether other kinases and ATP-binding proteins might have the target Cys; however, most of these have different crystallographic folds than the Ser/Thr/Tyr-protein kinases, making predictions of RAL sensitivity difficult. However, there are at least three homology families [ATM/PIK3C/PIK4C (ataxia telangiectasia mutated/phosphatidylinositol kinase 3C/phosphatidylinositol kinase 4C), IPK (inositol polyphosphate kinase), and CHK/EK (cholin kinase/ethanolamine kinase) of human kinases that have a similar fold to that of the standard protein kinases, and none of these has a Cys at the equivalent position as in the putative RAL targets.
Kinase Screen.
Hypothemycin was screened against a panel of 124 kinases (Upstate Cell Signaling Solutions, Dundee, U.K.) containing 19 putative Cys-containing RAL targets (see Table 4, which is published as supporting information on the PNAS web site). After an initial screen with 0.20 μM hypothemycin, the Cys-containing targets and enzymes inhibited ≥20% were rescreened with 2.0 μM hypothemycin. With the exception of GSK3α, all of the putative Cys-containing targets were inhibited by 60–100% in one or both screens. Three kinases, cSRC, TRKA, and TRKB, lacking the target Cys residue, were also inhibited.
Enzyme Kinetics.
Affinity-directed covalent enzyme inactivation involves formation of a reversible complex with dissociation constant Ki, followed by a first-order loss of enzyme activity with rate constant kinact. Analogous to kcat/Km, the kinact/Ki is the second-order rate constant for covalent bond formation under dilute conditions (i.e., [I] and [E] ≪ Ki) and reflects a “specificity constant” for the reaction of the inhibitor with the enzyme (Scheme 1) (9).
Scheme 1.
A sensitive, continuous fluorometric assay that measures ADP formation‡ was used to determine kinetic parameters of a variety of kinases (see Table 5, which is published as supporting information on the PNAS web site); progress curves of the reaction in the presence of various concentrations of the inhibitor were generated and analyzed to assess inhibition kinetics (see Fig. 3, which is published as supporting information on the PNAS web site) (10). Table 2 shows the inhibition constants for hypothemycin against a number of kinases, including, where relevant, kinact and t1/2 values for time-dependent inactivation.
Table 2.
Kinetic parameters for inhibition of kinases by hypothemycin
| Kinase | Ki, M | kinact, sec−1 | t1/2, sec | kinact/Ki, M−1·sec−1 |
|---|---|---|---|---|
| MEK1* | 1.7 × 10−8 | 0.0020 | 347 | 1.2 × 105 |
| MEK2* | 3.8 × 10−8 | 0.0010 | 753 | 2.6 × 104 |
| ERK1 | 8.4 × 10−6 | 0.0050 | 139 | 6.0 × 102 |
| ERK2 | 2.4 × 10−6 | 0.0043 | 161 | 1.8 × 103 |
| PDGFRα | 1.5 × 10−6 | 0.0020 | 347 | 1.3 × 103 |
| PDGFRβ | 9.0 × 10−7 | 0.0020 | 347 | 2.2 × 103 |
| FLT-3 | 9.0 × 10−8 | 0.0045 | 154 | 5.0 × 104 |
| FLT-1 (VEGFR1) | 7.0 × 10−8 | 0.0050 | 139 | 7.1 × 104 |
| KDR (VEGFR2) | 1.0 × 10−8 | 0.0046 | 151 | 4.6 × 105 |
| PKD1 | 1.2 × 10−6 | 0.0012 | 578 | 1.0 × 103 |
| MAPKAP5 | 1.3 × 10−5 | 0.0020 | 347 | 1.6 × 102 |
| TRKA | 2.2 × 10−6 | — | — | — |
| TRKB | 3.7 × 10−7 | — | — | — |
| SRC | 5.1 × 10−6 | — | — | — |
| GSK3α† | >6.3 × 10−4 | — | — | — |
| GSK3β | 3.7 × 10−4 | 0.0020 | 347 | 5.4 |
Kinases were analyzed by using progress curve analysis obtained from a continuous fluorometric assay. The Ki and kinact values derived from Kapp and kobsd values as described in Materials and Methods are averages of at least two determinations; SEs were 7–69% for Ki and 4– 52% for kinact; t1/2 values are from ln2/kinact. Enzyme sources, KmATP values, and protein or peptide substrates are provided in Table 5. —, Not applicable (no time-dependent inactivation observed).
*Assay monitored ATPase activity in absence of peptide/protein substrate.
†Estimated assuming 5% inhibition at 100 μM inhibitor and 100 μM ATP.
GSK3β, one of the putative hypothemycin targets, was poorly inhibited in the initial screen and showed a high Ki and consequent low kinact/Ki. The fact that kinact is as rapid as for most other kinases tested suggests that once the weak reversible complex is formed, the acceptor enone and nucleophilic Cys are suitably juxtaposed for covalent bond formation. GSK3α is the single Cys-containing target studied that did not show time-dependent inhibition by hypothemycin.
Although cSRC, TRKA, and TRKB showed inhibition by hypothemycin in the single-point screening assay described above, they do not contain the target Cys for Michael adduct formation. When assayed and analyzed by the more rigorous kinetic method, hypothemycin showed competitive inhibition with respect to ATP but did not show time-dependent inactivation. Likewise, hypothemycin showed reversible but not time-dependent inhibition of ERK2 C166A, in which the target Cys is replaced with Ala (data not shown).
Interaction of Hypothemycin with ERK2.
In addition to enzyme kinetics, several other approaches verified an affinity-directed mechanism for covalent adduct formation between hypothemycin and the Cys-containing ERK2.
Reversible binding and thiol reactivity are necessary for potent inhibition.
The small thiol-reactive reagents cyclohex-2-enone and 5,6-dihydro-2H-pyran-2-one react with thiols 18- and 4-fold faster, respectively, than hypothemycin (see below) but do not inhibit ERK2 at 1 mM over a period of 60 min, indicating that reversible binding is required for effective covalent inactivation. Disruption of the cis-enone system by reduction of the double bond to give 7′8′-dihydro-hypothemycin gave a reversible inhibitor with Ki of 100 μM that did not show time-dependent inhibition of ERK2, demonstrating the necessity of a Michael acceptor for potent inhibition.
Hypothemycin adduct at Cys-166.
Unactivated ERK2 and activated ERK2 were individually treated with hypothemycin to form the covalent complex. The free enzyme controls and their complexes were alkylated with iodoacetamide and digested with trypsin, and the peptides were analyzed by MALDI-TOF MS. The control enzymes showed a mass peak of 951 corresponding to the smallest tryptic peptide (ICDFGLAR) containing the carboxamidomethyl Cys-166 residue. The tryptic digests of the unactivated and activated forms of ERK2 previously treated with hypothemycin showed a mass of 1,273, corresponding to the target Cys peptide plus hypothemycin.
Stability of covalent adducts.
Covalent complexes of [3H]hypothemycin with inactivated and activated ERK2 could be isolated by SDS/PAGE. To measure the rates of dissociation, unactivated and activated ERK2–[3H]hypothemycin complexes were treated with a 100-fold excess of unlabeled hypothemycin. Aliquots were removed at various times, and the remaining covalent complex was determined after denaturation by SDS/PAGE. Dissociation of the native unactivated and activated ERK2–[3H]hypothemycin complexes were first-order, with t1/2 values of 40 h.
Reactivity of Hypothemycin with Thiols.
As a model for nonspecific reactions, hypothemycin and certain analogs were treated with various concentrations of thiols at pH 7.5, and aliquots were analyzed by HPLC over time for remaining hypothemycin. Second-order rate constants of thiolate anion with hypothemycin were 3.6 ± 0.1 M−1·s−1 for 2-mercaptoethanol (pKa = 9.5) and 6.6 ± 0.1 M−1·s−1 for glutathione (pKa = 8.75). Second-order rate constants for reaction of cyclohex-2-enone and 5,6-dihydro-2H-pyran-2-one with 2-mercaptoethanol thiolate showed rate constants of 65 and 14.2 M−1·s−1, respectively.
Inhibition of RAL Targets in Cells.
Table 3 shows growth inhibitory properties of hypothemycin and two other kinase inhibitors, BAY 43–9006 and SU11248, toward cancer cell lines dependent on mutants of kinases that activate MAPK pathways. SKOV-3, which overexpresses ERB2, a MAPK pathway activating kinase that is not targeted by the inhibitors, does not show high sensitivity toward any of the tested inhibitors. Cell lines carrying mutations in the mitogen receptors c-KIT, FLT3, and PDGFRα, which are targets for all three inhibitors, were very sensitive to hypothemycin with IC50 values of 370, 6, and 0.4 nM, respectively, which in each case are comparable with the those of BAY43–9006 and SU11248. Inhibition of the target kinases was also investigated in these cell lines. Hypothemycin treatment resulted in complete inhibition of autophosphorylation of the receptor kinases c-KIT, FLT3 and PDGFRα in P815, MV4–11, and EOL1 cells, respectively, as well as downstream phosphorylation of MEK1/2 and ERK1/2 in MV4–11 and EOL1 (see Fig. 4, which is published as supporting information on the PNAS web site); with P815, phosphorylated MEK (P-MEK) and ERK (P-ERK) levels were too low to detect. As with SU11248 (11), MV4–11 (an AML cell line with the FLT3-ITD mutation) was >100-fold more sensitive to hypothemycin than an AML cell line with wild-type FLT3 (IC50 = 0.72 μM). Hypothemycin was also approximately equipotent with either BAY43-9006 or SU11248 in growth inhibition of the VEGFR-dependent human umbilical vein endothelial cell line; Western analysis with VEGF-stimulated human umbilical vein endothelial cells demonstrated complete inhibition of VEGFR, MEK1/2, and ERK1/2 phosphorylation.
Table 3.
Hypothemycin inhibition of cells dependent on activating kinase mutations and VEGFR
| Cell line | Ref. | Cancer | Activating mutation/RAL target | IC50, μM |
||
|---|---|---|---|---|---|---|
| Hyp | BAY43–9006 | SU11248 | ||||
| SKOV-3 | (25) | Ovarian | HER2 | 7.0 | 5.5 | 6.7 |
| A459 | (26) | NSCLC | KRAS | 6.0 | 5.5 | nt |
| COLO829 | (12) | Melanoma | BRAF V600E | 0.05 | 6.0 | 7.1 |
| MV-4–11 | (27) | AML | FLT3-ITD | 0.006 | 0.003 | 0.012 |
| P815 | (28) | Mastocytoma | c-KIT D814Y | 0.37 | 0.31 | 0.29 |
| EOL1 | (29) | EOL | FIP1L1-PDGFRα | 0.0004 | 0.0002 | 0.0015 |
| HUVEC | — | — | VEGFR | 0.07 | 0.23 | 0.05 |
nt, not tested; HUVEC, human umbilical vein endothelial cell; —, non-cancer cell line (growth depends on VEGF).
Analyses of the NCI60 cell line and gene mutation data (12, 13) show that seven of the eight cell lines most sensitive to hypothemycin (National Cancer Institute Developmental Therapeutics Program no. NSC#354462) harbor the activating B-RAF V600E mutation. To extend these results, six additional B-RAF V600 mutants were tested (Table 3; see also Table 6, which is published as supporting information on the PNAS web site) and found to be ≈15- to 300-fold more sensitive to hypothemycin than were A549 and SKOV-3 cells that have an active ERK pathway and wild-type B-RAF. Furthermore, hypothemycin was on average ≈85-fold (range 15- to 200-fold) more potent against B-RAF V600E mutants than the RAF inhibitor BAY43-9006, although the inhibitors are equipotent against A549 and SKOV-3 cells. Treatment of cells with hypothemycin for 1 h resulted in a concentration-dependent depletion of P-ERK1/2, an expected consequence of MEK1/2 inhibition. With the B-RAF V600E mutants, HT29 and COLO829, IC50 values for P-ERK1/2 depletion were 20 and 10 nM hypothemycin, respectively, which were significantly lower than the values of 300 and 600 nM for A549 and SKOV3, respectively (data not shown).
The time necessary for inhibition of the ERK pathway was determined by incubation of cells with 1 μM hypothemycin and analysis of extracts for P-ERK1/2 at various times. With the non-B-RAF mutants SKOV-3 and A549, it took 30–60 min before significant depletion of P-ERK1/2 was observed, whereas with the B-RAF V600E mutant cell lines HT29 and COLO829, P-ERK1/2 levels were depleted within 10 min (see Fig. 5, which is published as supporting information on the PNAS web site). We also examined the duration of ERK pathway inhibition after removal of the drug. Cells were treated with 1 μM of hypothemycin or the reversible MEK inhibitor U0126 for 1 h to deplete cells of P-ERK1/2, washed to remove free drug, and P-ERK1/2 levels monitored with time. With the reversible MEK inhibitor U0126, P-ERK1/2 levels in both B-RAF V600E mutant HT29 and B-RAF wild-type cell lines A549 recovered to control levels by the first time point examined (3 h) after removal of the drug. In contrast, with hypothemycin, there was prolonged depletion of P-ERK1/2 after treatment: In the B-RAF wild-type A549 cell line, P-ERK1/2 was easily detectable 3 h after drug removal and recovered by ≈6 h; in HT29, a B-RAF V600E mutant cell line, P-ERK1/2 was barely detectable even 24 h after drug removal (see Fig. 5).
Because hypothemycin has a long duration of action, we examined whether a brief daily exposure was sufficient to inhibit B-RAF V600E-dependent cell growth. Approximately 106 HT29 or COLO829 cells were exposed daily for 1 h to 2 μM hypothemycin or 10 μM U0126, and viable cells were counted on day 4. Whereas cells in the UO126-treated and control samples grew to equal numbers, hypothemycin-treated cells remained the same as originally seeded.
We also tested whether hypothemycin would inhibit stimulated activation of the p38 and JNK/SAPK (c-Jun N-terminal kinase/stress-activated protein kinase) cascades, because p38 cascade contains the RAL targets TAK1 and MEK3/6, and JNK/SAPK cascade contains MEK4/7. Treatment of HT29 cells with 20 ng/ml phorbol 12-myristate 13-acetate (PMA) for 30 min resulted in high activation of p38. When cells were pretreated with 1 μM hypothemycin for 1 h, the PMA-stimulated phosphorylation of the MEK3/6 substrate p38 was prevented; similar pretreatment with the MEK1/2 inhibitor U01216 had no effect on p38 phosphorylation. Likewise, treatment of HeLa cells with 1 μM hypothemycin for 1 h inhibited phosphorylation of the TAK1 substrate IκB kinase β, induced by treatment with 10 ng/ml IL-1 for 15 min. Finally, treatment of A549 cells with 1 μM hypothemycin for 1 h completely prevented phosphorylation of the MEK 4/7 substrate JNK/SAPK induced by treatment with 20 ng/ml PMA for 30 min (see Fig. 4). As controls, 72-h exposure of all cell lines tested to 7′8′-dihydrohypothemycin showed IC50 values ≥100 μM, indicating requirement for the cis-enone moiety of hypothemycin for cytotoxicity and kinase inhibition.
Discussion
We identified a Cys residue that is conserved in the ATP site of Ser/Thr/Tyr protein kinases reported to be inhibited by cis-enone RALs but absent from those that are not. The target Cys (Cys-166 in human ERK2) is immediately N-terminal to the completely conserved Asp that binds Mg2+-ATP and is present in a subset of 46 of the ≈510 protein kinases. Because the enone moiety of RALs is susceptible to Michael addition, we speculated that the inhibition by RALs results from affinity-directed covalent adduct formation with the conserved Cys residue.
The 46 putative targets for cis-enone RALs represent ≈9% of the known Ser/Thr/Tyr kinases in the human kinome. Although this at first appears to be a large number of targets, it may be small compared with other kinase inhibitors of current interest. For example, BAY-43-9006 and SU11248, two multikinase inhibitors currently in clinical trials, have submicromolar Ki values on some 16% and 47%, respectively, of the ≈120 kinases assayed (7). Furthermore, in contrast to scattered profiles of kinases inhibited by leads obtained by random screening, the RAL targets are clustered in eight evolutionarily and functionally related groups. For example, in the branch of the some 90 Tyr kinases, all seven RAL targets are found in the subbranch containing the 12 mitogen receptor split kinases. Likewise, there are clusters of targets that encompass all of the MEK MAPK kinases, the ERK group, and certain groups downstream of ERK, such as the entire ribosomal subunit kinase group. In effect, the 46 individual RAL targets condense to major subsets of related targets that encompass multiple components of important pathways.
Initially, hypothemycin was screened as an inhibitor against a panel of some 124 kinases, 19 of which contained the conserved Cys. The RAL inhibited 18 of the 19 targeted Cys kinases and only three of the other kinases tested.
We investigated the kinetics of hypothemycin inhibition of 13 of the Cys-containing RAL targets. With the exception of GSK3α, all putative targets showed time-dependent inactivation consistent with initial formation of a reversible enzyme–inhibitor complex, followed by covalent Michael adduct formation. Additional evidence for the mechanism was obtained with experiments using ERK2, a kinase that has been notoriously resistant to inhibitor design. First, reagents that are more reactive toward thiols than hypothemycin but do not reversibly bind to ERK2 do not inhibit the enzyme. Second, the ERK2–[3H]hypothemycin complex was stable toward denaturation and isolable during SDS/PAGE. Third, dissociation of the ERK2–hypothemycin complex is extraordinarily slow (>24 h), a feature in accord with covalent inactivation. Fourth, trypsin digestion of ERK2–hypothemycin complexes yielded the expected active-site peptide containing the inhibitor attached to the target Cys-166. Finally, a crystal structure of the ERK2–hypothemycin complex showed covalent attachment of the 8′-carbon of the inhibitor to Cys-166 of the protein (our unpublished results).
The kinetic parameters for hypothemycin differed significantly for different Cys-containing target kinases that showed time-dependent inhibition: There was a ≈104-fold range in Ki for reversible binding, a ≈5-fold range in kinact, and a ≈105-fold range in kinact/Ki. The wide range of kinact/Ki values suggests that specificity of inhibition of certain kinases within the targeted subset might be achieved by controlling the exposure time; here, kinases such as MEK1 and the VEGFRs that have high kinact/Ki values would be inactivated by low concentrations of a RAL earlier than those with low reaction rates.
Curiously, the weak reversible complexes of hypothemycin with GSK3α and GSK3β had Ki values ≈102- to 104-fold higher than for the other targets examined. The peptide backbone between β-strand 4 and α-helix D of ERK2 contains Met-106 and is tightly packed against the aromatic ring of the inhibitor. In contrast, GSK3α and GSK3β have no corresponding Met but rather a Pro, and the region has six to nine additional residues not found in closely related kinases. We speculate that this region may serve as a selectivity filter (14) that accounts for reversible binding differences and that, within the weak GSK3α–hypothemycin complex, the cis-enone of the inhibitor is not suitably juxtaposed with the target Cys to form a covalent bond.
Kinetic analysis of the three kinases that were inhibited in the screen but do not contain the RAL targeted Cys residue (cSRC, TRKA, and TRKB) revealed that these enzymes are reversibly inhibited by hypothemycin but do not form covalent adducts although each contains a Cys residue in the ATP site that is remote from the RAL target Cys.
An important question is whether the RALs are sufficiently specific for their targets to be useful in a biological environment that contains free thiols and other thiol-containing proteins (15). At saturating concentrations of the inhibitor, covalent bond formation is limited by the first-order rates of inactivation. Depending on the enzyme, these rates occur with t1/2 values of 2–12 min, probably reflecting the positioning and proximity of the enone to the target Cys. At low concentrations of enzyme and inhibitor, kinact/Ki reflects a second-order rate constant for covalent adduct formation. The reaction is very fast, with most target kinases showing kinact/Ki values that exceed their kcat/Km and are some 105- to 107-fold greater than the second-order rate constants for the reaction of hypothemycin with thiols. Furthermore, kinases that contain Cys in different positions of their ATP sites than the target kinases, e.g., cSRC, TRKA, TRKB, PLK3, and NEK2, do not form covalent adducts with hypothemycin. Thus, the reaction of hypothemycin with most of its targets is significantly faster than with naturally occurring small thiols (e.g., glutathione) or, most important, other thiol-containing proteins.
We anticipated that hypothemycin would be a potent inhibitor of cancer cell lines dependent on ERK pathway-activating kinase mutants that are molecular targets of RALs given that it inhibits most mitogen receptor kinases and uniquely inhibits both the downstream MAPK kinases (MEK1/2) and the MAPKs (ERK1/2). Cell lines carrying activating mutations in the mitogen receptors c-KIT, FLT3, or PDGFRα (all RAL targets) were inhibited by hypothemycin as effectively as by the potent receptor kinase inhibitors BAY43-9006 and SU11248. As expected, growth inhibition by hypothemycin correlated with inhibition of autophosphorylation of the receptor kinase and with phosphorylation of the downstream MEK1/2 and ERK1/2.
The ERK pathway-activating B-RAF V600E mutation is found in ≈60% of melanomas, ≈20% of colon cancers, and ≈2% of breast cancers (14). As initially indicated in the NCI60 cell panel screen and expanded upon here, hypothemycin is selectively potent against all tested melanoma, colon, and breast cancer cell lines having the ERK pathway-activating B-RAF V600E mutation. As reported for the potent MEK inhibitors CI-1040 and PD0325901 (16), hypothemycin inhibits such cells more effectively than those having an active ERK pathway absent the B-RAF mutation (e.g., activating RAS mutants) or the RAF inhibitor BAY43–9006. Solit et al. (16) have suggested that selectivity of MEK inhibitors for B-RAF mutants may be due to the selective decline in cyclin D that occurs upon inhibition of the ERK pathway in such cells. The “ultrasensitive” cooperative response of the ERK cascade (17) may also contribute to hypothemycin’s potency for B-RAF V600E cells. It has been shown that, whereas RAF exhibits hyperbolic Michaelis–Menten kinetics, the MEK and ERK enzymes behave cooperatively with Hill-coefficients of ≈2 and 5, respectively; hence, the downstream effects of the pathway should be most profound by inhibition of ERK pathway enzymes in the order ERK > MEK > RAF (15). Therefore, it is anticipated that RAF inhibitors, such as BAY43-9006, should not be particularly effective against such cells, and, although MEK inhibitors are effective, dual MEK/ERK inhibitors such as the cis-enone RALs should be superior. Indeed, in the NCI60 screen, B-RAF V600E cells are on average ≈23-fold more sensitive to hypothemycin than cells with RAS mutations, whereas there is just a 3-fold difference with the specific MEK inhibitor PD98059. An additional attractive feature of the RALs for inhibition of B-RAF V600E cells is that the onset of intracellular inhibition of MEK is rapid (≈10 min), and the duration is long (≈24 h); in cells with an active ERK pathway absent the B-RAF mutation, the onset of inhibition is longer and the duration is shorter. As expected, in preliminary experiments hypothemycin showed excellent activity in inhibiting a B-RAF V600E melanoma xenograft (our unpublished results).
Hypothemycin also was a potent inhibitor of VEGFR, was approximately equipotent with BAY43-9006 and SU11248 in inhibiting growth of VEGF-dependent human umbilical vein endothelial cells (HUVEC), and inhibited phosphorylation of VEGFR, MEK1/2, and ERK1/2 in these cells. Finally, we showed that pretreatment with hypothemycin prevented stimulated phosphorylation of the MEK3/6 substrate p38, the MEK4/7 substrate JNK/SAPK, and the TAK1 substrate IκB kinase β.
The RALs join a small but growing number of kinase inhibitors that function by affinity-directed covalent inactivation that includes inhibitors of phosphatidylinositol 3-kinase (18), EGFR (19), and certain Cys-containing members of the ribosomal subunit kinase family (14). The common prejudice against irreversible inhibitors is belied by the fact that 19 of the 71 enzyme targets of marketed drugs are covalently modified by the drug (20). Indeed, provided that target selectivity in a biological environment can be achieved, such inhibitors offer several potential advantages over reversible binding agents. First, the onset of covalent inhibition is a function of time and inhibitor concentration, so selectivity within the target subset might be altered by duration of exposure. Second, the reaction is thermodynamically driven toward the covalent adduct, and high concentrations of inhibitor are not needed to achieve effective inhibition. Third, once covalently bound, such inhibitors do not readily dissociate from the enzyme, and restoration of enzyme activity requires new synthesis. Fourth, as shown for EGFR, irreversible inhibition can circumvent drug resistance (21). Finally, with sufficient exposure, they are infinitely potent inhibitors that exceed the efficacy of reversible inhibitors.
Materials and Methods
Materials.
Rat ERK2 was expressed in E. coli, purified and activated as reported (22). Other kinases were obtained from Upstate Cell Signaling Solutions or Invitrogen and experiments used the manufacturer’s specifications of purity and concentration. Hypothemycin and 7′,8′-dihydrohypothemycin were isolated from Hypomyces subiculosus (23). [3H]Hypothemycin was a gift from C. Reeves (Kosan Biosciences).
Bioinformatics.
Human kinases were from the tabulation by Manning et al. (8) and updated with resources at National Center Biotechnology Information, including ENTREZ-GENE and blast (www.ncbi.nlm.nih.gov). Sequence analysis also used macvector 7.2.3 (Accelrys, San Diego), including its implementation of clustalw. Publicly available kinase crystal structures were obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank at www.rcsb.org/pdb, and the NCI60 database was accessed through www.dtp.nci.nih.gov/docs/cancer/cancer_data.html.
Kinase Assays.
A continuous fluorimetric assay based on coupling ADP to resorufin formation was adapted from Dominguez et al.‡ Each well of a black, half-volume, 96-well plate was loaded to contain in 40 μl 25 mM Hepes (pH 7.4), 62.5 mM KCl, 12.5 mM MgCl2, 37.5 mM Na2HPO4, 0.125 mM EDTA, 0.125 mM Na3VO4, 1.25 mM phosphoenolpyruvate, 62.5 μM Amplex red, 0.125 μM FAD, 625 μM thiamine pyrophosphate, 1.25 mM ATP, 31.25 μM 2-mercaptoethanol, 5.0 units/ml pyruvate kinase, 5.0 units/ml pyruvate oxidase, 2.5 units/ml horseradish peroxidase, and ≈1 × 10−3 units/ml kinase. Reactions were initiated by adding a 10-μl solution of indicated amounts of kinase substrate (see Table 5) and various amounts of inhibitor (≈0.01–100 μM final concentration) in 10 mM Tris, pH 7.5. Fluorescence was monitored at 530:590 nm (excitation/emmission) in 30-sec intervals for 30 min at ambient temperature. Reversible competitive inhibition was analyzed by measuring changes in initial velocity with softmaxpro software. For time-dependent inactivation, product versus time curves were fitted (prism 4.0b, Graphpad, San Diego), to the equation [P] = (vi/kobs)·(1 − exp(−kobs·t)), where P is the product formed at time t, vi is the initial velocity, and kobs is the apparent first-order rate constant for enzyme inactivation (10). The data were then fitted to the equation kobs = kinact·[I]/(Kapp + [I]), where Kapp is the apparent dissociation constant of the reversible enzyme–inhibitor complex and kinact is the first-order rate constant for apparent irreversible conversion of the enzyme–inhibitor complex to covalently bound complex (10). Ki values were calculated from Ki = Kapp/(1 + [ATP]/KmATP) by using experimentally determined Km values for ATP (see Table 5).
MS of the ERK2–Hypothemycin Complex.
Approximately 70–100 μg of active and inactive ERK2 in 100 μl were dialyzed against 25 mM NH4HCO3, and half of each sample was incubated with 5 μl of 1 mM hypothemycin for 60 min at ambient temperature or left untreated (control). The proteins were reduced by adding 5 μl of 100 mM DTT at 50°C for 15 min, alkylated by adding 5 μl of 200 mM iodoacetamide at room temperature for 30 min, and then treated with 1 μl of a 0.4 μg/μl trypsin solution (Promega) for 18 h at 37°C. Samples were analyzed on a Voyager-DE STR BioSpectrometry Workstation (MALDI-TOF).
SDS/PAGE Binding Assay and Dissociation.
Reaction mixtures (180 μl) containing 1.1 μM [3H]hypothemycin at 10 Ci/mmol (1 Ci = 37 GBq) and 2.2 μM inactive or active ERK2 in binding buffer [20 mM Mops-KOH, pH 7.4/50 mM KCl/10 mM MgCl2/0.1 mM EDTA/2 mM tris(2-carboxyethyl)phosphine hydrochloride] were incubated at ambient temperature for 50 min. Twenty microliters of 1.0 mM unlabeled hypothemycin was added to give a final concentration of 100 μM. Aliquots of 20 μl were electrophoresed at various times by SDS/PAGE. The gel was dried, exposed to a Tritium storage phosphor screen, and scanned on a Typhoon imager (Molecular Devices).
Cytotoxicity Assays.
Cell lines were from American Type Culture Collection (Manassas, VA) or Deutsche Sammlung von Mikroorganismen Zellkulturen (Braunschweig, Germany) and grown according to the suppliers’ recommendations. Cells were plated into the 96-well microtiter plates at 4,000 cells per well, incubated overnight at 37°C, and then treated with various drug concentrations. After 72 h, cells were counted with the Cell Titer Glo Luminescent Cell Viability Assay kit (Promega).
Phosphoprotein Analysis.
Cell lines were grown to 70% confluency in recommended media supplemented with 10% FBS in 6-cm dishes. Inhibitors or DMSO (vehicle control) were added, and the plates were incubated at 37°C for the desired incubation time. For lysis of adherent cells, media was removed and cells were rinsed with PBS. Cells were lysed in 0.3 ml of supplemented radioimmunoprecipitation assay (RIPA) buffer (see Fig. 4) on ice and scraped. The lysate was passed through a 21-gauge syringe needle three times, and cell debris was removed by centrifugation at 15,000 × g for 10 min at 4°C. For nonadherent cells, cells were harvested by centrifugation at 193 × g for 10 min and washed with 5 ml of PBS. The cell pellet was lysed in 0.3 ml of RIPA buffer and treated as described above.
Protein concentrations were normalized to ≈1 mg/liter before addition of 4× protein sample buffer, boiling, and electrophoresis on 4–12% Bis-Tris gels (Invitrogen). Gels were blotted semidry to poly(vinylidene difluoride) membranes. Membranes were blocked and incubated with antibodies to visualize total and phosphorylated kinases by following the instructions of the manufacturers.
Supplementary Material
Acknowledgments
We thank C. Reeves for providing [3H]hypothemycin, A. Keatinge-Clay for performing the MS peptide analysis, K. Patel (Kosan Biosciences) for providing ERK C166A, the Kosan Process Science Department for hypothemycin and 7′,8′-dihydrohypothemycin, the Kosan Bioanalytical Facility for performing SDS/PAGE and thiol-reactivity experiments, and Cell Signaling Technology for permission to adapt their kinase dendrogram.
Abbreviations
- RAL
resorcylic acid lactone
- ERK
extracellular signal-regulated kinase
- P-ERK
phosphorylated ERK
- MAP
mitogen-activated protein
- MAPK
MAP kinase
- MEK
MAPK kinase
- P-MEK
phosphorylated MEK
- EGFR
endothelial growth factor receptor
- VEGFR
vascular EFGR
- GSK
glycogen synthase kinase
- PDGFR
platelet-derived growth factor receptor
- TAK1
TGF-β-activated kinase 1
- LCK
lymphoid-specific Tyr kinase p56kk.
Footnotes
Conflict of interest statement: No conflicts declared.
Dominguez, J. M., Rodriguez, B., Zapatero, C., Vasquez, M. J., Magan, C., Martin, J. J. & Carranza, C., Annual Meeting of the Society for Biomolecular Screening, Sept. 22–26, 2002, The Hague, The Netherlands.
References
- 1.Camacho R., Staruch M. J., DaSilva C., Koprak S., Sewell T., Salituro G., Dumont F. J. Immunopharmacology. 1999;44:255–265. doi: 10.1016/s0162-3109(99)00085-5. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka H., Nishida K., Sugita K., Yoshioka T. Jpn. J. Cancer Res. 1999;90:1139–1145. doi: 10.1111/j.1349-7006.1999.tb00688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams D. H., Wilkinson S. E., Purton T., Lamont A., Flotow H., Murray E. J. Biochemistry. 1998;37:9579–9585. doi: 10.1021/bi972914c. [DOI] [PubMed] [Google Scholar]
- 4.Zhao A., Lee S. H., Mojena M., Jenkins R. G., Patrick D. R., Huber H. E., Goetz M. A., Hensens O. D., Zink D. L., Vilella D., et al. J. Antibiot. 1999;52:1086–1094. doi: 10.7164/antibiotics.52.1086. [DOI] [PubMed] [Google Scholar]
- 5.Ninomiya-Tsuji J., Kajino T., Ono K., Ohtomo T., Matsumoto M., Shiina M., Mihara M., Tsuchiya M., Matsumoto K. J. Biol. Chem. 2003;278:18485–18490. doi: 10.1074/jbc.M207453200. [DOI] [PubMed] [Google Scholar]
- 6.Giese N. A., Lokker N. 1998 U.S. Patent 5,728,726. [Google Scholar]
- 7.Fabian M. A., Biggs W. H., III, Treiber D. K., Atteridge C. E., Azimioara M. D., Benedetti M. G., Carter T. A., Ciceri P., Edeen P. T., Floyd M., et al. Nat. Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 8.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 9.Fersht A. Enzyme Structure and Mechanism. New York: Freeman; 1984. [Google Scholar]
- 10.Copeland R. A. Enzymes: A practical Introduction to Structure, Mechanism and Data Analysis. New York: Wiley; 2000. pp. 318–349. [Google Scholar]
- 11.O’Farrell A. M., Abrams T. J., Yuen H. A., Ngai T. J., Louie S. G., Yee K. W., Wong L. M., Hong W., Lee L. B., Town A., et al. Blood. 2003;101:3597–3605. doi: 10.1182/blood-2002-07-2307. [DOI] [PubMed] [Google Scholar]
- 12.Davies H., Bignell G. R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M. J., Bottomley W., et al. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 13.Garraway L. A., Widlund H. R., Rubin M. A., Getz G., Berger A. J., Ramaswamy S., Beroukhim R., Milner D. A., Granter S. R., Du J., et al. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 14.Cohen M. S., Chao Z., Shokat K. M., Taunton J. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight Z. A., Shokat K. M. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Solit D. B., Garraway L. A., Pratilas C. A., Sawai A., Getz G., Basso A., Ye Q., Lobo J. M., She Y., Osman I., et al. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C. Y., Ferrell J. E., Jr. Proc. Natl. Acad. Sci. USA. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker E. H., Pacold M. E., Perisic O., Stephens L., Hawkins P. T., Wymann M. P., Williams R. L. Mol. Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 19.Fry D. W., Bridges A. J., Denny W. A., Doherty A., Greis K. D., Hicks J. L., Hook K. E., Keller P. R., Leopold W. R., Loo J. A., et al. Proc. Natl. Acad. Sci. USA. 1998;95:12022–12027. doi: 10.1073/pnas.95.20.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson J. G. Biochemistry. 2005;44:5561–5571. doi: 10.1021/bi050247e. [DOI] [PubMed] [Google Scholar]
- 21.Kwak E. L., Sordella R., Bell D. W., Godin-Heymann N., Okimoto R. A., Brannigan B. W., Harris P. L., Driscoll D. R., Fidias P., Lynch T. J., et al. Proc. Natl. Acad. Sci. USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khokhlatchev A., Xu S., English J., Wu P., Schaefer E., Cobb M. H. J. Biol. Chem. 1997;272:11057–11062. doi: 10.1074/jbc.272.17.11057. [DOI] [PubMed] [Google Scholar]
- 23.Dombrowski A., Jenkins R., Raghoobar S., Bills G., Polishook J., Pelaez F., Burgess B., Zhao A., Huang L., Zhang Y., Goetz M. J. Antibiot. 1999;52:1077–1085. doi: 10.7164/antibiotics.52.1077. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh C. M., Fukumoto S., Layne M. D., Maemura K., Charles H., Patel A., Perrella M. A., Lee M. E. J. Biol. Chem. 2000;275:36966–36973. doi: 10.1074/jbc.M006028200. [DOI] [PubMed] [Google Scholar]
- 25.King B. L., Carter D., Foellmer H. G., Kacinski B. M. Am. J. Pathol. 1992;140:23–31. [PMC free article] [PubMed] [Google Scholar]
- 26.Valenzuela D. M., Groffen J. Nucleic Acids Res. 1986;14:843–852. doi: 10.1093/nar/14.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quentmeier H., Reinhardt J., Zaborski M., Drexler H. G. Leukemia. 2003;17:120–124. doi: 10.1038/sj.leu.2402740. [DOI] [PubMed] [Google Scholar]
- 28.Tsujimura T., Furitsu T., Morimoto M., Isozaki K., Nomura S., Matsuzawa Y., Kitamura Y., Kanakura Y. Blood. 1994;83:2619–2626. [PubMed] [Google Scholar]
- 29.Griffin J. H., Leung J., Bruner R. J., Caligiuri M. A., Briesewitz R. Proc. Natl. Acad. Sci. USA. 2003;100:7830–7835. doi: 10.1073/pnas.0932698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.