Abstract
A genome-wide phenotype screen was used to identify factors and pathways that induce proliferation of human umbilical vein endothelial cells (HUVEC). HUVEC proliferation is a recognized marker for factors that modulate vascularization. Screening “hits” included known proangiogenic factors, such as VEGF, FGF1, and FGF2 and additional factors for which a direct association with angiogenesis was not previously described. These include the kinase TBK1 as well as Toll-like receptor adaptor molecule and IFN regulatory factor 3. All three proteins belong to one signaling pathway that mediates induction of gene expression, including a mixture of secreted factors, which, in concert, mediate proliferative activity toward endothelial cells. TBK1 as the “trigger” of this pathway is induced under hypoxic conditions and expressed at significant levels in many solid tumors. This pattern of expression and the decreased expression of angiogenic factors in cultured cells upon RNA-interference-mediated ablation suggests that TBK1 is important for vascularization and subsequent tumor growth and a target for cancer therapy.
Keywords: cancer, cDNA/libraries, expression cloning, human umbilical vein endothelial cells, screening
The availability of the human genome map and sequence, combined with information regarding genetic factors and/or disease-associated aberrations or variations, has accelerated the identification of disease-associated genes (1, 2). For example, high-throughput expression profiling (e.g., by chips) or bioinformatics analyses of databases with genetic or clinical information are currently successfully applied to identify disease associations and target genes (3–6).
Despite progress made by these procedures, there is still a gap between the large amount of sequence information on one side and the limited experimental information for most genes. To circumvent this bottleneck, we “inverted” the usual process of target identification by initiating the search for new targets, not with an in silico screen or profiling but, instead, with a high-throughput functional screen. For this screen, we developed a robotics platform that performs transfection of single cDNAs into mammalian cells, followed by phenotype determination (7, 8). The phenotype chosen was vascularization, assayed by determination of human umbilical vein endothelial cells (HUVEC) proliferation.
Proteins that drive tumor angiogenesis contribute to cancer progression (9–13). Examples include VEGF or other growth factors produced or induced by tumor cells. Therefore, we set up our genomics platform to detect secreted factors that control vascularization by the phenotype of transfected cells.
Here, we describe the identification and characterization of a set of genes (TANK-binding kinase 1 (TBK1), Toll/IL-1 receptor (TIR)-domain-containing adaptor-inducing IFN-β (TRIF), and IFN regulatory factor 3 (IRF3) that induce HUVEC proliferation and constitute a signaling pathway that controls gene expression through the transcription factor IRF3.
Results
Functional Genomics Screen for Proangiogenic Activities.
An automated high-throughput screening platform was applied to identify factors that stimulate HUVEC proliferation. Human embryonic kidney (HEK)293 cells were transfected individually with 251,000 defined plasmids from various cDNA expression libraries (35,000 clones from a cDNA collection, 66,000 poly-ribosomal library, 86,000 metabolic library, and 99,000 placenta library, see Methods). These “producer cells” were incubated for 48 h, and cell culture supernatants containing factors produced by the transfected cells were harvested. These factors are either encoded by the transfected cDNAs or produced as a consequence of their expression.
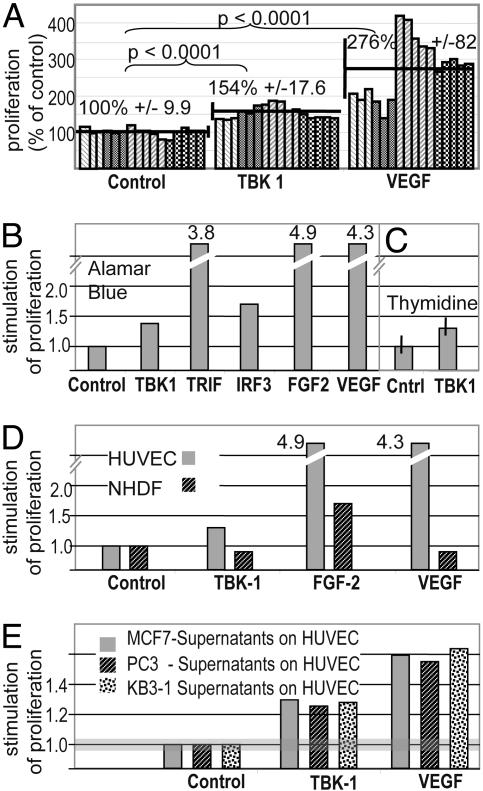
Exposure of HUVEC to these conditioned media revealed “hits” that cause proliferation. This activity was observed for only 0.02% of all screened supernatants with hit rates ranging from 0.0045% (polyribosomal library) to 0.037% (cDNA collection). The observed activities of the screening hits were clearly associated with specific cDNAs (e.g., independent library plasmids with the same insert cause the same phenotypes). Examples of the reproducibility of the assays are shown in Fig. 1A and B. Known proangiogenic factors, e.g., VEGF or FGF1 and FGF2, were found as multiple independent hits in independent libraries (e.g., VEGF in all cDNA sources, four independent clones alone in the metabolic library), or reproducibly in multiple experiments using the cDNA collection (FGF1 and FGF2, Fig. 1B). The importance of screening diverse cDNA sources was also confirmed by the fact that another known angiogenesis-associated factor, follistatin (placental protein), was observed twice in the placenta library but not in any other library. These screening data, which are available in Table 2, which is published as supporting information on the PNAS web site, validate the specificity of the screening system. Hits may not be only secreted factors with direct activity. Alternatively, the transfected cDNA can induce cellular pathways. Examples of this mode of action are cDNAs encoding the proteins TBK1, Toll-like receptor adaptor molecule (TRIF), and IRF3. These proteins were identified as inducers of HUVEC proliferation activities as repeated hits in independent libraries: TBK1 was reproducibly found in the cDNA collection in independent experiments, IRF3 was found hit in the polyribosomal library as well as in the metabolic library, and TRIF was identified once each in the cDNA collection and placenta library and also as four independent hits in the metabolic library (Table 2 and Fig. 1).
Fig. 1.
Factors that specifically induce HUVEC proliferation. (A) TBK1 induces proliferation of HUVEC. Proliferative activities were detected by incubating HUVEC for 5 days with supernatants of transfected HEK293 producer cells [four independent experiments (different shades) with multiple samples each (individual bars)]. Increased signals of Alamar blue assays indicate proliferation. Supernatants of HEK293 cells producing VEGF (positive controls) confirm that the processing machinery releases secreted/shed factors in sufficient concentrations. For normalization of results, the mean value of the controls was set to 100% for each experiment; individual values for each experiment were calculated relative to that. Indicated are also the mean values ± SD of each group, and the significances of the signal increases (Student’s t test). (B–D) Representative single experiments. (B) Proliferative activities were detected as described in A. Increased signals of Alamar blue assays (fold increase relative to vector control = 1) indicate proliferation. Proliferation was also mediated by intracellular proteins through induction of signaling pathways. One pathway is defined by activities of TBK1, TRIF, and IRF3, which stimulate cells to release proliferative activities. (C) Endothelial cell proliferation accompanied by DNA replication: TIME cells were analyzed, applying a [3H]thymidine-incorporation assay. Increased signals (fold increase relative to control = 1) indicate DNA synthesis and cell proliferation (shown with SD). (D) Specificity. Supernatants were applied in parallel to endothelial cells (HUVEC, gray) and human fibroblasts (NHDF, black). TBK1, VEGF, and FGF2 cause HUVEC proliferation. Only FGF2 mediates proliferation of NHDF, confirming the specificity for endothelial cells of TBK1-induced activity. (E) HUVEC proliferation induced by supernatants of transiently transfected cancer cell lines.
Specific Induction of Endothelial Cell Proliferation by TBK1, TRIF, and IRF3.
Our primary screen does not distinguish activities that are specifically directed toward endothelial cells from general proliferative activities (e.g., FGF2). Therefore, positive supernatants were also analyzed by incubating them with normal human dermal fibroblasts (NHDF). Fig. 1D shows TBK1-generated activities that stimulate the proliferation of HUVEC but not NHDF. Thus, the specificity of TBK1-derived supernatants resembles that of VEGF. Supernatants from TRIF-expressing producer cells showed the same specificity (data not shown). Does TBK1-mediated proliferation of endothelial cells depend on the producer cell line that was used for our screen (HEK293) or a more general phenomenon? We analyzed the proliferation of endothelial cells by supernatants of TBK1-transfected MCF-7, PC3, and KB3–1 cancer cells. Fig. 1E shows that TBK1 expression in all three lines generates supernatants that promote the proliferation of endothelial cells. Thus, the proliferative TBK1 phenotype is observed in various cancer cell lines.
To confirm that the specificity of TBK1-induced supernatant activities is directed toward capillary endothelial cells (and not just restricted to vein endothelial cells), telomerase-immortalized microcapillary endothelial (TIME) cells were also tested. For these experiments, thymidine-incorporation assays were used. The data in Fig. 1C show that TBK1 expression generates activities that induce DNA syntheses in microcapillary endothelial cells.
These results indicate that the TRIF/TBK/IRF3 pathway leads to the induction of activities/secreted factors that specifically stimulate proliferation of endothelial cells.
Induction of Angiogenesis-Associated Factors as a Consequence of IRF-Pathway Activation by TBK1.
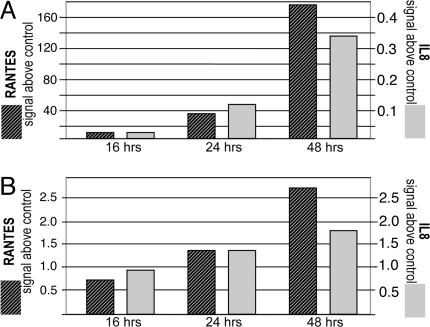
To elucidate the nature of the activities induced by the TBK1 pathway, RNAs of TBK1- or TRIF-transfected HEK293 were compared with control vector transfected cells by AffymetrixGeneChip analysis (14). We focused our analysis on secreted factors that could mediate TBK1-induced proliferative activity. Table 1 shows secreted factors that were up-regulated; these include RANTES and IL-8, both of which mediate proliferative activity on endothelial cells (15–17). Other factors, such as CXCL10, CXCL11, and β IFN were also induced. These factors are also known to modulate angiogenic processes (18–20). But not all of these factors are proproliferative. In fact, CXCL10, CXCL11, and IFNβ were described to modulate angiogenesis in a “negative” manner (18–20). Thus, expression of TBK1 generates a “mixture” of angiogenesis-modulating factors (Table 1). Exposure of endothelial cells to this mixture stimulates their proliferation. The induction of known proliferators of endothelial cells was also analyzed by quantitative (q)PCR. These analyses confirmed the induction of RANTES and IL-8 by TBK1 in HEK293 (Fig. 2A) as well as in MCF7 cells (Fig. 2B). The expression of individual factors, such as IL8 or RANTES, by themselves, does not result in HUVEC proliferation in our assay system. This indicates that the components of the mixture act in concert on the induction of endothelial cell proliferation.
Table 1.
Induction of secreted protein by TBK1 and TRIF
| Gene | Protein | Induction by TBK1 (SLR) | Induction by TRIF (SLR) |
|---|---|---|---|
| CCL5 | Chemokine (C-C motif) ligand 5, RANTES | 6.9 | 10.4 |
| IFNB1 | Interferon, β 1, fibroblast | 5.6 | 9.1 |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | 5.8 | 8 |
| G1P2 | Interferon, α inducible protein | 3 | 4.8 |
| CXCL10 | Chemokine (C-X-C motif) ligand 10 | 1.8 | 4.3 |
| IL8 | Interleukin 8 | 1.3 | 2.25 |
HEK293 were transfected with plasmids encoding TBK1 or TRIF, and expression levels of genes in comparison with empty-vector transfected controls were detected by Affymetrix-Chip analysis. Shown are all genes encoding secreted proteins (according to gene ontology) with signal log ratio (SLR) increases by >0.7 (= ≈1.6-fold) compared with the control cells. In agreement with the joined functionality of TRIF and TBK1 in signaling pathways, the set of induced secreted factors was identical in TRIF- and TBK1-expressing cells. Some induced factors modulate angiogenesis or are related to interferon response. In addition, we observed increased signals of NF-κB-dependent genes (e.g. NFKB1A).
Fig. 2.
TBK1 induces expression of secreted factors. The induction of RANTES and IL8 in recombinant TBK1-expressing cells was analyzed by qPCR using as reference the housekeeper G6PDH, which was tested in parallel in those experiments. (A) RNA of TBK1-transfected HEK293 cells was extracted at different time points after transfection, and mRNA levels were determined by qPCR. The relative signals (normalized to housekeeper expression) of the PCR reactions of RANTES and IL8 are different (RANTES, left axis; IL8, right axis), but both factors are induced with similar kinetics. (B) RANTES and IL8 induction by supernatants of transiently transfected cells is also observed in the MCF7 breast cancer cell line. Induction of the cytokines was detected as described in A.
TBK1-Induced Proangiogenic Activity: NF-κB vs. IRF3 Signaling.
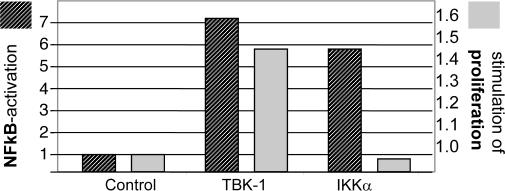
TBK1, TRIF, and IRF3 are components of the IRF3 pathway (21, 22), suggesting that signaling toward IRF3-dependent gene expression causes the observed phenotype. TBK1 is a member of the IKK family of protein kinases and activates not only IRF3 (and IRF7) but also NF-κB (23, 24). In agreement with that finding, induction of NF-κB activity through TBK1 occurred under our assay conditions (Fig. 3). Our Affymetrix GeneChip analyses indicated that NF-κB-dependent genes become induced by TBK1 expression (e.g., NFKBIA). Is the TBK1-mediated proliferative activity mediated by IRF or NF-κB signaling? We tested whether NF-κB activation, by itself, can generate proangiogenic phenotypes by transfecting TBK1 or IKKa into HEK293 cells. Fig. 3 shows that that both kinases activate NF-κB, as demonstrated by the induction of reporter constructs carrying an NF-κB promoter sequence. However, despite similar NF-κB activation, TBK1, but not IKKα, induced proliferative activity in supernatants of transfected HEK293 cells (Fig. 3). That IKKα-mediated NF-κB activation is not sufficient for induction of proliferative activity confirms the results of our primary screen, in which IKKα showed no phenotype. These data indicate that TBK1-mediated activation of IRF3, but not of NF-κB, plays the predominant role in the proangiogenic phenotype induced by TBK1.
Fig. 3.
NF-κB is not sufficient for induction of proliferative activity. TBK1 and IKKα were analyzed for NF-κB activation and induction of HUVEC proliferation. NF-κB activation (left axis): HEK293 were cotransfected with a NF-κB-promoter luciferase reporter construct and plasmids for expression of TBK1, IKKα, or empty vector. The relative signal (light units, fold above vector control) reflects NF-κB-dependent transcription. HUVEC proliferation (right axis): HEK293 cells were transfected (TBK1, IKKα, or vector control) and proliferative activities analyzed as described above. TBK1 and IKKα induce NF-κB-dependent transcription, but only supernatants of cells expressing TBK1 mediate HUVEC proliferation.
TBK1 is Induced in a Cell Culture Model for Hypoxia.
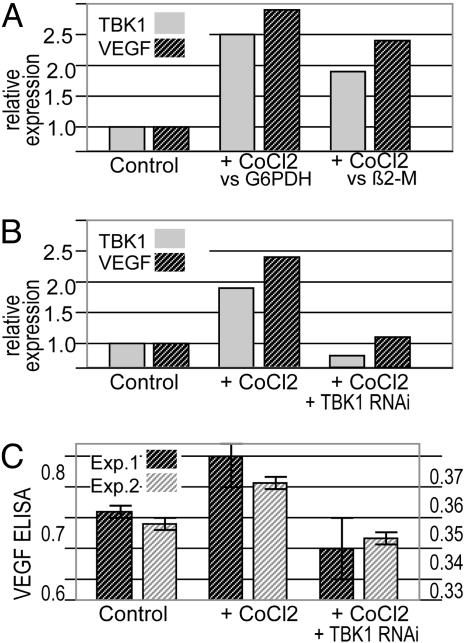
Hypoxia stimulates the induction of genes that counteract hypoxia through de novo stimulation of the vasculature (11–13). To analyze whether TBK1 is regulated by hypoxic stimuli, we compared its expression levels under hypoxic and nonhypoxic conditions. HEK293 cells were exposed to CoCl2, a model for hypoxia in which VEGF levels are elevated (13). Determination of VEGF levels (ELISA and qPCR) was therefore used as a control in our experiments. Fig. 4A shows that CoCl2 exposure induces both VEGF and TBK1 expression. Thus, hypoxia can induce the expression of TBK1. It is likely that this induction enables cells to use the TBK1/IRF3 pathway for generation of proangiogenic factors (Table 1).
Fig. 4.
TBK1 levels increase under hypoxic conditions and correlate with expression of VEGF. (A) Induction of TBK1 under hypoxic conditions. HEK293 cells were exposed to 50 mM CoCl2 for 24 h (chemical induction of hypoxia). Expression of VEGF and TBK1 was analyzed by qPCR (references were G6PDH or β2 microglobulin, relative values, control expression set to 1′). The housekeepers showed no evidence of nonspecific effects of RNAi; the presented data are all shown relative to the housekeeper. VEGF and TBK1 are induced by CoCl2 treatment. (B) RNAi-mediated reduction of TBK1. The effects of RNAi targeting TBK1 were analyzed by qPCR in CoCl2-treated cells. TBK1-RNAi reduces the expression of TBK1 and VEGF in CoCl2-treated cells. (C) Effects of RNAi against TBK1 on VEGF levels analyzed by ELISA. Hypoxia-induced HEK293 cells were exposed to TBK1-RNAi. VEGF levels were detected by ELISA (OD 492 nm) in two independent experiments (left axis, signal of experiment 1; right axis, experiment 2). Under our experimental conditions, VEGF expression is barely detectable without CoCl2 induction, and VEGF signals rise above background in CoCl2-treated cells and are reduced in TBK1-RNAi-treated cells.
Because the TBK1/IRF3 pathway is used for the generation of proangiogenic factors, we examined whether we could reduce the generation of such factors by RNA-interference-mediated depletion of TBK1. Fig. 4 shows that RNA interference reduced the levels of TBK1 by >90%. Even under hypoxic (CoCl2) conditions, cells exposed to TBK1 RNAi showed >2-fold reduction of TBK1 levels compared with cells under nonhypoxic conditions (Fig. 4B). The effect of RNAi-mediated reduction of TBK1 on the production of VEGF under hypoxic conditions is shown in Fig. 4 B and C: ELISA and qPCR analyses demonstrated that TBK1 depletion reduces VEGF expression in CoCl2 treated cells. This result indicates that it may be possible to reduce the generation of proangiogenic factors through inhibition of this pathway.
Elevated Expression Levels of TBK1 in Solid Tumors.
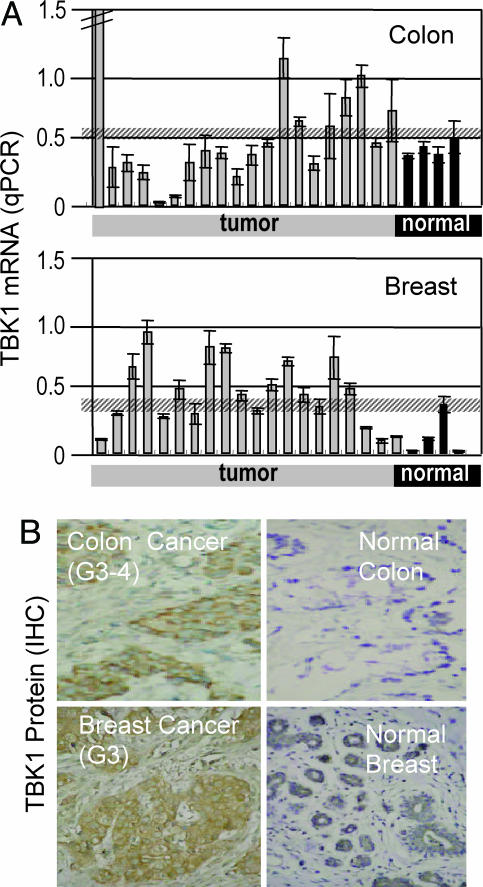
Because the TBK1/IRF3 pathway causes expression and release of proangiogenic factors in cultured cells, we analyzed the expression of TBK1 as a trigger of that pathway in tumors and normal tissues. Fig. 5A shows qPCR analyses which demonstrate increased expression of TBK1 compared with controls in colon and breast cancer samples.
Fig. 5.
Elevated expression levels of TBK1 in solid tumors. (A) RT-PCR (qPCR) for detection of TBK1 transcripts. RNAs from tumor and normal colon and breast were subjected to TBK1-specific qPCR. Each bar represents the qPCR-signal from one individual (y axis signal represents mRNA amount). The first (far left) colon tumor sample showed a very high expression (value, 21.6). The hatched horizontal lines represent the mean + SD of the expression of the nontumor samples. Thus, everything above these thresholds is increased expression. The mean values of expression were 0.43 and 0.12 for all normal colon or normal breast, respectively, and 1.52 or 0.40 for all colon or breast cancer samples, respectively. Preparations from tissues from control individuals display low signals, whereas many tumors show increased levels of the mRNA of TBK1. (B) Detection of TBK1 protein by immunohistochemistry. Exemplar shown is the analysis of tumor and adjacent normal tissue from colon and breast cancer. Both tumors show increased expression of TBK1 in malignant cells. IHC, immunohistochemistry.
To determine the cells in which TBK1 expression was elevated, we performed immunohistology with TBK1-specific antibodies. In agreement with the qPCR analyses, we observed increased TBK1 reactivity compared with nonmalignant tissue in colon cancer (Fig. 5B) and breast cancer (Fig. 5C). It is also evident that the reactivity is restricted to the malignant cells of the cancer. Thus, tumor-associated TBK1 is predominantly produced by the malignant cells. This enables tumor cells to activate the TBK1/IRF3 pathway and may, in consequence, induce the production of angiogenic factors.
Discussion
We show here, that identification of angiogenesis-associated factors can be achieved by a high-throughput functional genomics screening. This approach identified a signaling pathway that controls gene expression by TRIF, TBK1, and the transcription factor IRF3. This pathway controls not only the expression of “viral response” genes (21) but is also relevant for the production of a mixture of secreted angiogenesis-factors. These factors, in turn, stimulate the proliferation of endothelial cells. We also show that TBK1, a trigger of this pathway, is induced under hypoxic conditions and expressed at significant levels in many solid tumors, indicating that this pathway is important for tumor growth and/or vascularization.
High-Throughput Functional Genomics for Identification of Angiogenesis Pathways.
The screening platform is based on the gain-of-function principle because of transient expression of large numbers of individual cDNAs in mammalian cells. This technique, performed on a robotics platform, was applied to the identification of cDNAs that directly affect the phenotype of the transfected cells (7, 8). For the herein-described identification of factors that are associated with angiogenesis, the technology was adapted to identify activities in cell-culture supernatants. These activities, in turn, can induce phenotypes in nontransfected cells, such as endothelial cells. On the technical level, this task was achieved by harvesting the media of transfected producer cells at defined time points after transfection and analyzing the proliferative activity of these supernatants in the HUVEC-proliferation model. The challenge of such an assay is that the supernatants comprise “conditioned media” of cells that are exposed to stress (transfection procedure) and are incubated for extended periods of time for the development of phenotypes. The identification of specific activities above the background nonspecific effects represents a major challenge. The reproducible identification of positive controls (e.g., FGF, VEGF) demonstrates that, despite these challenges, the identification of a proangiogenic phenotype is possible. Also, the fact that activity is observed for just a small fraction of supernatants (0.02% of all screened supernatants, see Table 2), and that this activity is highly reproducible, indicates the high specificity of our screen.
The TRIF/TBK1/IRF3 Pathway Induces Proangiogenic Activity.
TBK1 is a Ser/Thr kinase activated by TRIF. Both proteins feed into a divided intracellular signaling pathway by their ability to serve two separate transcription factors, NF-κB and IRF3. Activation of TBK1 leads to IRF3 activation and expression of IRF3-dependent genes, important in response to viral infection. In addition, TRIF/TBK1 signaling activates NF-κB-dependent gene expression (22–24). NF-κB-dependent genes are also activated by IKKα or IKKβ that encode factors associated with inflammation. Our data presented here demonstrate that the IRF3 “arm” of the TBK1-signaling pathway is not only involved in the viral response but also plays a significant role in tumor angiogenesis. Interestingly, within this split pathway, only IRF3-dependent expression is sufficient to induce the production of a proangiogenic mixture of secreted factors (see below). Nevertheless, NF-κB-dependent gene expression may additionally contribute to the expression of proliferative factors in tumor angiogenesis. In fact, the importance of induction of proangiogenic factors by the IRF3 pathway, with a possible contribution of NFκB, is supported by recent works of Mizukami et al. (25). This group has demonstrated that, in addition to HIF1, other regulatory mechanisms contribute on a transcriptional level to tumor angiogenesis. Interestingly, again, IL8 was noted as being a significant factor produced by tumors to compensate for insufficient HIF1 activity under hypoxic conditions. This finding is in line with our observation of IL8 induction upon expression of TBK1. Mizukami et al. (25) suggest that induction of proangiogenic factors is associated with hypoxia-induced NFκB activation. Our findings that TBK1 is expressed under hypoxic conditions suggest that TBK1 activity could also contribute to the effects that were observed by Mizukami et al.
That activity toward HUVEC was detected in supernatants of cells transfected with TBK1, TRIF, or IRF3 indicates the induction of expression of proliferative secreted or shed proteins. Because TRIF, TBK1, and IRF3 are part of one signaling pathway, we conclude that this pathway causes the production of proangiogenic factors. However, TRIF/TBK1 signaling also activates NF-κB-dependent gene expression under our experimental conditions (Fig. 3; Table 1). For two reasons, we believe that the IRF3 pathway, and not NF-κB, is the major effector of the phenotype: (i) the expression of IKKα as inducer of NF-κB signaling generated no activity in our proliferation assay, indicating that activation of NF-κB alone is not sufficient to generate the activities; (ii) the presence of IRF3 as a screening hit in multiple cDNA sources and libraries shows that IRF3 by itself is sufficient for the phenotype.
The induction of proangiogenic activities by the IRF3 pathway, which is otherwise well known to participate in antiviral responses, indicates that angiogenesis can be promoted in the course of antiviral responses. Thus, angiogenesis may support defensive activities against viruses. Increased vascularization might also overcome local hypoxia that might be caused by the infection. It is interesting to note in this context, that certain viruses, (e.g., KS herpesvirus or B19 erythrovirus) induce replication under hypoxic conditions.
Specific Proangiogenic Mixture of Secreted Factors.
Upon TBK1 expression, a variety of secreted factors become induced that are associated with modulation of angiogenesis as well as inflammation or antiviral responses (14–19). These factors include RANTES and IL-8, for which induction of endothelial cell proliferation is described (15–17). Interestingly, certain factors that were induced by TBK1, e.g., RANTES or IL-8, were not found as hits in our screen. Nor did they cause HUVEC proliferation upon individual recombinant expression in our assay system. Thus, these factors probably constitute a mixture (of proliferative as well as modulating factors), which mediates the specific phenotype. One explanation for the observation that IL8 or RANTES alone showed no activity in our assay could be that the expression of the different receptors for signal transduction is strictly regulated (26). Thus, the mixture has better activity than single factors because it may simultaneously induce receptor expression as well as provide ligands for multiple receptors. The production of cocktails of growth factors that act in concert resembles the mechanism by which tumor cells induce and maintain angiogenesis.
Relevance of the TRIF/TBK1/IRF3 Pathway for Tumor Angiogenesis.
Increased expression of TBK1, which was observed in solid tumors, could be explained, in part, by inflammatory processes within the tumor and/or by infiltrating lymphocytes (27). However, immunohistochemistry shows unambiguously that the majority of TBK1 protein within tumors is found within the tumor cells (Fig. 5). This and the observation that, in cell culture, TBK1 expression is increased under hypoxic stimuli suggests that TBK1 in tumor cells takes part in the generation of factors that promote angiogenesis. Because TBK1 takes part in the generation of factors that promote angiogenesis and tumor growth, it should be possible to decrease the generation of such factors by inhibition of the TBK1/IRF3 pathway. The observation that RNAi inhibition of TBK1 decreases the synthesis of angiogenesis factors (even under hypoxia-induced conditions) supports this notion.
The proangiogenic phenotype of TBK1 and associated pathway genes, expression in human tumors, and the possibility to interfere with the generation of angiogenesis factors by inhibition of TBK1 makes this kinase a promising target for cancer therapy.
Materials and Methods
cDNA Expression Plasmids and Libraries.
A normalized cDNA library from human placenta mRNA in pCMVSport6 was obtained from Invitrogen (Placenta Library). For the normalized and full-length enriched {“metabolic” library (by GeneTrapper, custom generated by Invitrogen), cDNA of human liver, visceral fat, skeletal muscle, hypothalamus, and pancreas was cloned in pCMV6-XL, a modified pcDNA3. These tissues have high secretion rates and, thus, the library is enriched for secreted proteins. The “secreted” library was made from membrane-associated polyribosomal mRNA from four breast cancer cell lines, a prostate cancer cell line, and a normal breast cell line and was enriched for secreted factors and surface proteins (4). The collection of human cDNA expression plasmids with known full-length inserts consists of 35,000 clones. A total of 99,000 clones from the “placenta” library, 86,000 clones from the metabolic library, 66,000 clones from the secreted library, and 35,000 clones from our cDNA collection were screened. Because all of the clones from the 35,000 cDNA collection were unique, and because the redundancy in our libraries was generally <15%, transcripts of most human genes should have been covered by our screen.
Cell Cultivation and Transfection.
HEK293 cells were grown in DMEM/5% FCS, HeLa in DMEM/10% FCS, and HUVEC in EGM with supplements (PromoCell, Heidelberg), 2% FBS, 50 μg/ml gentamycin, 0.4 μg/ml amphotericin B, and 50 units/ml nystatin. HEK293 cells were transfected by calcium phosphate coprecipitation and HeLa with FuGene (Roche Diagnostics). TIME cells, derived from human dermal microvascular endothelial cells, were a kind gift from Dr. Martin McMahon (Cancer Research Institute, University of California, San Francisco) (28). TIME cells were cultured in endothelial cell basal medium MV2 with supplements (PromoCell), without antibiotics, on gelatin-coated dishes.
Functional Genomics Screen.
A robotics platform for DNA preparation, transfection, and phenotype identification (7), was adapted for the identification of biological activities in cell culture supernatants. Fifty to 70 plates per batch were simultaneously processed. Details of the screening procedures are available in Supporting Methods, which is published as supporting information on the PNAS web site. Briefly, HEK293 cells were transfected 24-h postseeding by using calcium phosphate coprecipitation (7), and cells were switched 4 h after transfection to nutrient-deficient DMEM to generate conditioned supernatants. Twenty-four hours later, supernatants were transferred to HUVEC, which were incubated with supernatants for 4 days. Metabolic activity of HUVEC was determined by using Alamar blue (BioSource International, Camarillo, CA). Thymidine-incorporation assays were applied to TIME cells.
Analytical Assays.
VEGF in cell culture supernatants was detected by ELISA, using the PromoKine human VEGF ELISA kit reagents (PromoCell).
qPCR analyses using the lightcycler (Roche) were applied to quantify the expression of TBK1, IL-8, RANTES, and VEGF: cDNA was synthesized from 1 μg of total RNA by using random hexamers and avian myeloblastosis virus (AMV)-RT (Roche). PCR contained 0.5 μM each sense and antisense primers, 3 mM MgCl2, 1× SYBR green MasterMix, and 2 μl of cDNA.
The PCR conditions and primer sequences are listed in Supporting Methods. References for quantification were β2M and G6PDH. Data were analyzed by using lightcycler analysis software.
Activation of NF-κB.
HEK293 cells were cotransfected with a constitutive active expression plasmid for renilla luciferase, a NF-κB-dependent firefly luciferase reporter construct, and expression plasmids for TBK1 or IKKα, or empty vector. Twenty-four hours after transfection, cells were lysed, and NF-κB-dependent firefly luciferase activity was measured. Activity of renilla luciferase was determined to normalize for transfection efficiencies.
RNAi.
HEK293 cells were transfected with TBK1 RNAi by using siPORT Amine according to the manufacturer’s protocol (Ambion). Reactions contained 100 nM each preannealed siTBK1 sense (GGAGACAACAACAAGACAUtt) and antisense (AUGUCUUGUUGUUGUCUCCtc). TBK1 mRNA levels were analyzed by qPCR. For the combination of RNAi and hypoxia-experiments, cells were incubated 24 h after transfection with medium containing 50 mM CoCl2.
Expression Analyses.
mRNA levels of TBK1 were quantified by qPCR (see above) with a LightCycler or by TaqMan (relative expression of TBK1 versus 18S rRNA, Cytomyx, Cambridge, U.K.). Immunostaining (Applied Phenomics, Tartu, Estonia) was performed with anti-TBK1 antibody (Calbiochem) on paraformaldehyde-fixed and paraffin-embedded material by using the DAKO Duet HRP kit and standard citrate/microwave pretreatment. Nonspecific binding of secondary reagents was prevented by biotin blocking. The results were evaluated by experts in immunohistochemistry and a pathologist (Applied Phenomics).
Supplementary Material
Acknowledgments
We thank Peter Vajkoczy for constructive suggestions and Theresia Walter and Angelika Waldschmidt for excellent technical assistance. This research was supported, in part, by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Abbreviations
- HEK
human embryonic kidney
- HUVEC
human umbilical vein endothelial cells
- IRF
IFN regulatory factor
- qPCR
quantitative PCR
- RNAi
RNA interference
- TBK1
TANK-binding kinase 1
- TIME
telomerase-immortalized endothelial
- TRIF
Toll-like receptor adaptor molecule.
Footnotes
Conflict of interest statement: C.K., H.G., R.S., K.K.-H., and U.B. were or are employees of Xantos Biomedicine.
References
- 1.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zoby M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. errata (2001) 411, 720 and (2001) 412, 565. [DOI] [PubMed] [Google Scholar]
- 2.Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., Smith H. O., Yandell M., Evans C. A., Holt R. A., et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. erratum (2001) 292, 1838. [DOI] [PubMed] [Google Scholar]
- 3.Bera T. K., Lee S., Salvatore G., Lee B., Pastan I. Mol. Med. 2001;7:509–516. [PMC free article] [PubMed] [Google Scholar]
- 4.Egland K. A., Vincent J. J., Strausberg R., Lee B., Pastan I. Proc. Natl. Acad. Sci. USA. 2003;100:1099–1104. doi: 10.1073/pnas.0337425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boguski M. S., Lowe T. M., Tolstoshev C. M. Nat. Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 6.Boon K., Osorio E. C., Greenhut S. F., Schaefer C. F., Shoemaker J., Polyak K., Morin P. J., Buetow K. H., Strausberg R. L., De Souza S. J., et al. Proc. Natl. Acad. Sci. USA. 2002;99:11287–11292. doi: 10.1073/pnas.152324199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koenig-Hoffmann K., Bonin-Debs A. L., Boche I., Gawin B., Gnirke A., Hergersberg C., Madeo F., Kazinski M., Klein M., Korherr C., et al. Int. J. Cancer. 2005;113:434–439. doi: 10.1002/ijc.20601. [DOI] [PubMed] [Google Scholar]
- 8.Zitzler J., Link D., Schafer R., Liebetrau W., Kazinski M., Bonin-Debs A., Behl C., Buckel P., Brinkmann U. Mol. Cell. Proteomics. 2004;3:834–840. doi: 10.1074/mcp.M400054-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Folkman J., Watson K., Ingber D., Hanahan D. Nature. 1989;339:58–61. doi: 10.1038/339058a0. [DOI] [PubMed] [Google Scholar]
- 10.Ferrara N. Nat. Rev. Cancer. 2002;2:795–803. doi: 10.1038/nrc909. [DOI] [PubMed] [Google Scholar]
- 11.Carmeliet P. Nat. Med. 2000;6:389–395. doi: 10.1038/74651. [DOI] [PubMed] [Google Scholar]
- 12.Gerwins P., Skoldenberg E., Claesson-Welsh L. Crit. Rev. Oncol. Hematol. 2000;34:185–194. doi: 10.1016/s1040-8428(00)00062-7. [DOI] [PubMed] [Google Scholar]
- 13.Liu X. H., Kirschenbaum A., Yao S., Stearns M. E., Holland J. F., Claffey K., Levine A. C. Clin. Exp. Metastasis. 1999;17:687–694. doi: 10.1023/a:1006728119549. [DOI] [PubMed] [Google Scholar]
- 14.Lipshutz R. J., Fodor S. P., Gingeras T.R., Lockhart D. J. Nat. Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 15.Adler E. P., Lemken C. A., Katchen N. S., Kurt R. A. Immunol. Lett. 2003;90:187–194. doi: 10.1016/j.imlet.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Bar-Eli M. Pathobiology. 1999;67:12–18. doi: 10.1159/000028045. [DOI] [PubMed] [Google Scholar]
- 17.Li A., Dubey S., Varney M. L., Dave B. J., Singh R. K. J. Immunol. 2003;170:3369–3376. doi: 10.4049/jimmunol.170.6.3369. [DOI] [PubMed] [Google Scholar]
- 18.Seville L. F., Mathiak G., Bagasra O. Cytokine Growth Factor Rev. 1997;8:207–219. doi: 10.1016/s1359-6101(97)00015-4. [DOI] [PubMed] [Google Scholar]
- 19.Lasagni L., Francalanci M., Annunziato F., Lazzeri E., Giannini S., Cosmi L., Sagrinati C., Mazzinghi B., Orlando C., Maggi E., et al. J. Exp. Med. 2003;197:1537–1549. doi: 10.1084/jem.20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fidler I. J. J. Natl. Cancer Inst. Monogr. 2001;28:10–14. doi: 10.1093/oxfordjournals.jncimonographs.a024251. [DOI] [PubMed] [Google Scholar]
- 21.Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 22.Sato S., Sugiyama M., Yamamoto M., Watanabe Y., Kawai T., Takeda K., Akira S. J. Immunol. 2003;171:4304–4310. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda A., Suzuki Y., Honda G., Muramatsu S., Matsuzaki O., Nagano Y., Doi T., Shimotohno K., Harada T., Nishida E., et al. Oncogene. 2003;22:3307–3318. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- 24.Sasai M., Oshiumi H., Matsumoto M., Inoue N., Fujita F., Nakanishi M., Seya T. J. Immunol. 2005;174:27–30. doi: 10.4049/jimmunol.174.1.27. [DOI] [PubMed] [Google Scholar]
- 25.Mizukami Y., Jo W. S., Duerr E. M., Gala M., Li J., Zhng X., Zimmer M. A., Iliopoulos O., Zukerberg L. R., Kohgo Y., et al. Nat. Med. 2005;11:992–997. doi: 10.1038/nm1294. [DOI] [PubMed] [Google Scholar]
- 26.Salcedo R., Resau J. H., Halverson D., Hudson E. A., Dambach M., Powell D., Wasserman K., Oppenheim J. J. FASEB J. 2000;14:2055–2064. doi: 10.1096/fj.99-0963com. [DOI] [PubMed] [Google Scholar]
- 27.Balkwill F. Nat. Rev. Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 28.Venetsanakos E., Mirza A., Fanton C., Romanov S. R., Tisty T., McMahon M. Exp. Cell Res. 2002;273:21–33. doi: 10.1006/excr.2001.5424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.