Abstract
Misfolded proteins accumulate in many neurodegenerative diseases, including huntingtin in Huntington’s disease and α-synuclein in Parkinson’s disease. The disease-causing proteins can take various conformations and are prone to aggregate and form larger cytoplasmic or nuclear inclusions. One approach to the development of therapeutic intervention for these diseases has been to identify chemical compounds that reduce the size or number of inclusions. We have, however, identified a compound that promotes inclusion formation in cellular models of both Huntington’s disease and Parkinson’s disease. Of particular interest, this compound prevents huntingtin-mediated proteasome dysfunction and reduces α-synuclein-mediated toxicity. These results demonstrate that compounds that increase inclusion formation may actually lessen cellular pathology in both Huntington’s and Parkinson’s diseases, suggesting a therapeutic approach for neurodegenerative diseases caused by protein misfolding.
Keywords: misfolded proteins, neurodegenerative disease
The accumulation of misfolded proteins is a common feature of multiple neurodegenerative diseases, such as huntingtin (Htt) with expanded polyglutamine (polyQ) in Huntington’s disease and α-synuclein-rich Lewy bodies in Parkinson’s disease. Cells have two main defenses against such misfolded proteins: refolding by chaperones and targeted destruction by the ubiquitin-proteasome system (1, 2). The accumulation of toxic protein conformations in neurodegenerative diseases implies the insufficiency of these defense mechanisms (3). Although many of these diseases show obvious large protein inclusions, the toxicity of these inclusions has been unclear. Some studies suggest that earlier aggregation intermediates, such as oligomers, may be more toxic than larger inclusions (4–6). There is recent evidence that the inclusions may, in fact, be protective (4, 7). Screens for therapeutic compounds have generally selected for compounds that inhibit inclusion formation. We reasoned that some therapeutic compounds might not inhibit inclusion formation, yet would block downstream pathological pathways, such as proteasome dysfunction. We therefore decided to test compounds that increased inclusion formation for their effects on proteasome dysfunction and toxicity.
Results and Discussion
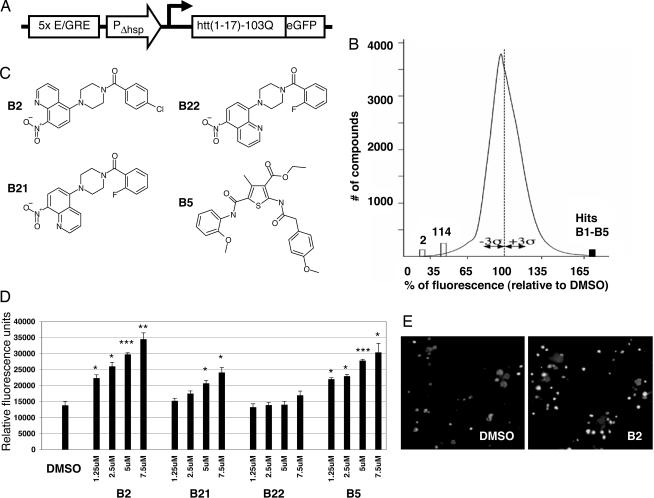
We screened a library of chemical compounds for their effects on the expression of mutant-length polyQ. We used a reporter consisting of an amino-terminal fragment of human Htt with 103 glutamines, fused to an EGFP reporter (Fig. 1A) (8). The read-out of the assay is the EGFP fluorescence level, used as a reporter of the amount of expanded polyQ. Among 37,000 compounds screened, we identified five compounds that produced an increase of ≥50% in signal, and 116 that led to a decrease in EGFP signal of ≥35% in overall fluorescent signal (i.e., at least 3 SDs from the mean) (Fig. 1B). Among the five compounds that increased reporter levels, the strongest signal was given by (5-[4-(4-chlorobenzoyl)-1-piperazinyl]-8-nitroquinoline), which we refer to subsequently as B2 (Fig. 1C). B2 increases the EGFP signal in a dose-dependent manner, ranging from a 61% increase at 1.25 μM to a 149% increase at 7.5 μM (Fig. 1D). Microscopy of cells treated with B2 confirmed the increase in reporter levels and showed an apparent increase in the size and frequency of inclusions (Fig. 1E). Three of the other four compounds that elevated the polyQ reporter were part of a single chemical scaffold, including the compound B5 (ethyl 2-([(4-methoxyphenyl)acetyl]amino)-5-([(2-methoxyphenyl)amino]carbonyl)-4-methyl-3-thiophenecarboxylate) (Fig. 1C). B5 activity also shows dose dependence, ranging from a 59% increase in signal at 1.25 μM to a 119% increase at 7.5 μM. We conclude that the compound B2, and to a lesser degree B5, increases polyQ inclusion formation in PC12 cells.
Fig. 1.
The compound B2 increases polyQ–EGFP reporter inclusions and overall fluorescence. (A) Schematic representation of the reporter construct used in high-throughput screen. An ecdysone-inducible promoter drives expression of a fusion of the first 17 aa of Htt plus 103 glutamines and EGFP. (B) Distribution of hits from high-throughput screen of 37,000 chemical compounds. A total of 116 compounds decreased the EGFP signal by ≥35% and 5 compounds increased the signal by ≥50% (black bar). (C) Structures of B2, its analogs B21 and B22, and B5. (D) Reporter readout from a range of doses of B2 or its analogs B21 and B22. B2 causes a dose-dependent increase in polyQ–EGFP reporter. Analog B21 is less effective than B2, and B22 shows no activity. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.005. (E) Fluorescence microscopy of cells from wells treated with either DMSO or 5 μM B2. Cells treated with B2 show an increase in polyQ–EGFP inclusion formation. (Magnification: ×10.)
We considered that B2 and B5 might act by interfering with cellular transcription, which is dysregulated in Huntington’s disease and is likely one of the major disease pathways. However, we did not observe compound-dependent histone deacetylase inhibition in cell-free and cell-based models, nor did we observe any effects on global transcription (data not shown). B2 also does not act nonspecifically on the heterodimeric ecdysone promoter; no significant increase in fluorescence was observed after identical treatment of an ecdysone-inducible 25Q-GFP/PC-12 cell line. Furthermore, B2 has no intrinsic fluorescence, ruling out the possibility of autofluorescent interference with reporter read-out. Finally, B2 and B5 are not significantly toxic at 10 μM or below, as determined by a WST-1 assay.
To identify the relevant functional groups in B2, we performed structure-activity relationship analysis. Analysis of 47 B2-related compounds revealed important chemical features of B2 that are necessary for activity. The activity profiles and structures of two analogs of B2 that are particularly revealing are shown in Fig. 1 C and D: B21 (5-[4-(2-fluorobenzoyl)-1-piperazinyl]-8-nitroquinoline) and B22 (8-[4-(2-fluorobenzoyl)-1-piperazinyl]-5-nitroquinoline). B21 differs from B2 in the addition of fluorine at position 2 on the benzene ring and the removal of the chloride at position 4 on the benzene ring. With these modifications, B21 is only half as potent as B2 and at 7.5 μM causes a 74% increase in reporter signal. B22 is identical to B21 except for the position of nitrogen in the nitroquinoline group. This modification nearly eliminates all activity; B22 does not alter levels of the polyQ reporter. An increase in signal (of 23%) is seen only at 7.5 μM. These structure-activity results suggest that B2 interacts with a molecular target in the cell with high specificity and provide a strategic approach to the discovery of this target.
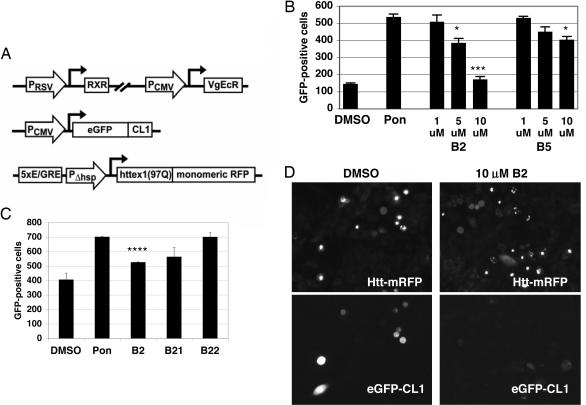
To assess the mode of action of B2, we examined pathways to toxicity in a cellular model of Huntington’s disease. It has previously been shown that expanded polyQ inhibits proteasome function in cell models and expanded polyQ is not digested by the proteasome in vitro (9–14). Proteasome impairment is highly toxic to cells and is thought to be an important pathological feature of Huntington’s disease. We developed an assay for testing the effect of compounds on mutant Htt-induced proteasome dysfunction and tested the effects of compounds B1–B5 in this assay. A CHO-K1 cell line was generated that stably expresses both a reporter of proteasome function (EGFP fused to the degradation signal peptide CL1) (15) and the first exon of Htt (with mutant 97Q) fused to monomeric red fluorescent protein (mRFP) (Fig. 2A). As expected, the induction of Htt expression produced a marked increase in the number of cells positive for proteasome reporter, reflecting reduced proteasome function. Proteasome function, as measured by EGFP–CL1, was rescued in a dose-dependent fashion by four of the five compounds. B2 exhibited the strongest rescue, with reductions of 39% at 5 μM and 93% at 10 μM (Fig. 2B). B5 also proved effective, although less potent than B2, and decreased proteasome reporter by 34% at 10 μM. As in the polyQ expression assay, the analog B21 was 20% less potent than B2, and the analog B22 was inactive (Fig. 2C). Fig. 2D shows microscopic images of the treated cells; EGFP–CL1 colocalizes with Htt inclusions in fewer cells when treated with B2. It is possible that B2 or B5 promote proteasome activity in the absence of Htt induction, but this possibility cannot be determined in the proteasome reporter because of the already low baseline levels of proteasome reporter. We conclude that B2 and B5 may prevent Htt-mediated proteasome dysfunction, while elevating overall levels of Htt.
Fig. 2.
B2 treatment prevents Htt-induced proteasome dysfunction. (A) Schematic representation of constructs stably transfected into CHO-K1 cells. CHO-K1 cells contained a heterodimeric ecdysone receptor, a CMV promoter-driven proteasome reporter (EGFP fused to the degradation peptide CL1), and an ecdysone-inducible fusion protein of the first exon of Htt (with 97Q) and mRFP. (B) Treatment of proteasome reporter cells with compounds. Ponasterone (pon) induction of mutant Htt leads to a build-up of proteasome reporter (EGFP–CL1). B2 treatment rescues the accumulation of proteasome substrate, in a dose-dependent fashion. B5 exhibits a similar, but weaker, effect. ∗, P < 0.05; ∗∗∗, P < 0.005. (C) Treatment of proteasome reporter lines with B2 vs. analogs of B2. As in the polyQ–EGFP reporter assay shown in Fig. 1C, modifications to B2 led to a decrease in activity in B21 and an elimination of activity in B22. ∗∗∗∗, P < 0.0005. (D) Fluorescence microscopy of cells from proteasome reporter line. Fewer cells exhibit build-up of EGFP–CL1 proteasome reporter when treated with B2. (Magnification: ×20.)
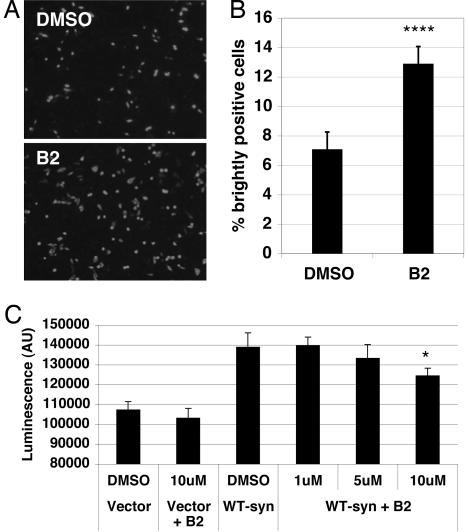
The structural specificity required for B2 action in both assays suggests that B2 targets a specific molecular target in the cell. The target of B2 could be the N-terminal portion of Htt or polyQ itself or, alternatively, a more general cellular target. To determine its specificity for Htt, B2 was tested for effects on α-synuclein inclusion formation and toxicity. To assess the effect of B2 on inclusion formation, CHO-K1 cells were transiently transfected with SynT (16), a tagged α-synuclein construct (17). Cells were processed for anti-α-synuclein immunofluorescence after 48 h. Two patterns of α-synuclein distribution were observed: (i) numerous small cytoplasmic aggregates and (ii) bright, ubiquitous immunofluorescence, with large inclusions visible through this ubiquitous fluorescence. After B2 treatment, there was a dramatic increase in the proportion of cells with ubiquitous staining and large inclusions, suggesting that B2 treatment enhances inclusion formation of α-synuclein (Fig. 3A and B). Thus, B2 can promote inclusion formation of two different neurodegenerative disease proteins, Htt and α-synuclein.
Fig. 3.
B2 treatment of cells produces more α-synuclein inclusions and lowers α-synuclein toxicity. (A) Transient transfection of CHO-K1 cells with synT construct. Cells were transfected with synT and simultaneously treated with either 10 μM B2 or an equivalent amount of DMSO for 48 h, then fixed and exposed to anti-α-synuclein antibody. Treatment with B2 nearly doubled the percentage of cells that are strongly immunofluorescent for α-synuclein; the remaining cells have numerous light cytoplasmic α-synuclein aggregates. (Magnification: ×10.) (B) Quantification of transfection shown in A. Brightly α-synuclein-positive cells and total DAPI-positive cells were counted in six random fields for each treatment. ∗∗∗, P < 0.0001. (C) α-Synuclein toxicity testing. B2 shows a dose-dependent prevention of toxicity from α-synuclein (measured by adenylate kinase release) when transiently transfected into H4 cells. ∗, P < 0.05.
To determine the effects of B2 on α-synuclein toxicity, a viability assay was performed on H4 neuroglioma cells transfected with α-synuclein. Transient overexpression of α-synuclein has been shown to be toxic to cells. This effect is physiologically relevant, as some familial Parkinson’s disease is caused by a triplication of the α-synuclein locus (18). In the presence of 10 μM B2, there was a 46% decrease in α-synuclein-mediated adenylate kinase release (a marker of toxicity) (Fig. 3C). We conclude that B2 protects against toxicity from overexpression of α-synuclein.
We transiently transfected the proteasome reporter line with the SynT construct, but failed to see significant elevation in EGFP levels; it is possible that use of a mutant form of α-synuclein would induce proteasome dysfunction in this cell line.
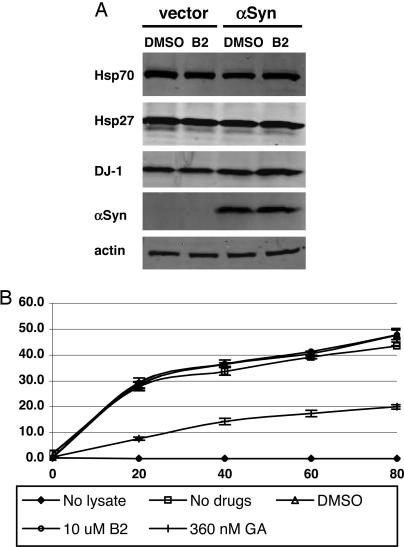
Chaperones modulate toxicity and aggregation in models of both Huntington’s disease and Parkinson’s disease (19). Increased chaperone levels could result in altered folding of Htt or α-synuclein. To determine whether B2 alters chaperone levels, we compared levels of Hsp70, Hsp27, and DJ-1 in H4 cells transfected with vector or α-synuclein and treated with carrier or 10 μM B2. B2 did not affect the levels of any of these chaperones (Fig. 4A).
Fig. 4.
B2 does not alter chaperone levels or activity. (A) Western blot for levels of chaperones in H4 cells transfected with vector or α-synuclein and treated with B2 or DMSO. Hsp70, Hsp27, and DJ-1 are not significantly altered by B2 treatment. (B) Refolding of heat-denatured firefly luciferase protein in rabbit reticulocyte lysate, in the presence of DMSO, B2, or Hsp90 inhibitor geldanamycin (GA). GA decreases the rate of refolding by >50% in this assay, whereas DMSO and B2 have no effect. Values are the mean of two assays, and error bars show SD.
We reasoned that even if B2 was not altering levels of chaperones it might alter their activity. We compared the effects of B2 and the known Hsp90 inhibitor geldanamycin (20) on chaperone-mediated refolding of heat-denatured luciferase in chaperone-rich rabbit reticulocyte lysate. As in prior reports (21), geldanamycin decreased the refolding activity by >50% relative to controls, demonstrating that the refolding rate in this assay can be altered by the addition of chaperone modulators. In contrast, DMSO and B2 had no effect (Fig. 4B). We conclude that B2 does not act directly on chaperone activity, but may otherwise alter protein quality-control mechanisms.
In summary, we have identified a compound that lowers pathological consequences of Htt and α-synuclein expression, while increasing inclusion formation. B2’s efficacy of action was dramatically influenced by the alteration of single atoms in the chemical structure of the compound, suggesting that there is a very specific chemical interaction between the compound and its target within the cell. The effectiveness of compound B2 in models of both Huntington’s disease and Parkinson’s disease is highly unusual. The target of B2 is still unknown, but it does not appear to act by altering the levels or activity of chaperones. B2 likely acts by altering some aspect of protein quality control that has a common impact on the misfolded protein underlying pathology in both Huntington’s disease and Parkinson’s disease. Alternatively, B2 might affect a protein conformation common to Htt and α-synuclein (e.g., oligomeric). Our observations are consistent with previous studies that suggest that the formation of inclusions may be beneficial to compromised cells in Huntington’s disease and Parkinson’s disease; however, the reversal of toxicity or proteasome dysfunction that we observe may be caused by lowered levels of another conformation of Htt or α-synuclein, such as oligomers. Developing more potent analogs of B2 may prove therapeutic for Huntington’s disease and Parkinson’s disease and other neurodegenerative diseases caused by protein misfolding.
Materials and Methods
Compounds.
The compound collection included 30,000 compounds (Diverse and CNS sets) from Chembridge (San Diego), 5,000 compounds from Maybridge (Cornwall, U.K.), and 2,000 highly purified natural products from TimTec (Newark, DE).
High-Throughput PolyQ Expression Screen.
To screen for potential modifiers of polyQ levels, the cell line 14A2.6 was used (8). This previously published cell line is stably transfected with both the heterodimeric ecdysone receptor and an ecdysone-inducible fusion of the N-terminal 17 aa of Htt to 97 glutamines and EGFP. Briefly, cells were plated in black, clear-bottom 96-well plates at a density of 3.5 × 104 cells per well followed by addition of compounds (at 10–25 μM) in triplicate plates on an EP3 liquid handling robot (PerkinElmer). After a 72-h incubation, cells were lysed in RIPA buffer [50 mM Tris, pH 8/150 mM NaCl/25 mM EGTA/1% (vol/vol) Nonidet P-40/0.5% (wt/vol) Na deoxycholate/0.1% (wt/vol) SDS]. Total GFP was quantitated on a Wallac Victor2V multilabel plate reader (PerkinElmer). Although we cannot rule out that the aggregation of GFP increases its intrinsic fluorescence, we believe that the increase in total GFP signal represents a real increase in absolute GFP reporter levels. For all five compounds (B1–B5), microscopic analysis was used to confirm the presence of more GFP inclusions.
Htt Proteasome Dysfunction Assay.
EcR-CHO cells were purchased from Invitrogen. This line (no longer commercially available) consisted of CHO-K1 cells stably transfected for a heterodimeric ecdysone receptor. Proteasome reporter lines were made by stable transfection with a fusion of EGFP with the CL1 degron (ACKNWFSSLSHFVIHL) in pcDNA3.1 (CL1 degron DNA: GCCTGCAAGAACTGGTTCAGCAGCCTGAGCCACTTCGTGATCCACCTG). Stable lines were chosen for response to 24-h treatment with the proteasome inhibitor epoxomicin (Calbiochem), as measured by a Victor2 plate reader. The strongest line was then stably transfected with a fusion between the first exon of Htt and mRFP (a gift from Roger Tsien, University of California, San Diego), subcloned into the pIND vector, which creates an ecdysone-inducible expression construct. Cells were maintained in zeocin, G418, and hygromycin to maintain the three constructs; however, these antibiotics were not present during Htt induction with ponasterone.
Htt-mediated proteasome dysfunction cells, described above, were grown overnight in 96-well TC plates (Wallac), then treated for 48 h with DMSO (negative control) or 25 μM ponasterone A (to induce Htt-97Q-mRFP) and either test compounds or vehicle (DMSO). Compounds were tested at 1, 5, and 10 μM in duplicate wells. Final DMSO concentration was 0.35% in all wells. Total EGFP-positive cells (without regard to EGFP levels) were counted for each well. Statistical significance was determined by using t test.
α-Synuclein Cell Counts.
One day before transfection, CHO-K1 cells were plated at 50,000 cells per well in 4-well chamber slides (LabTek II, Nunc). Cells were transfected with wild-type α-synuclein, synT, or equimolar amounts of synT and synphilin by using Fugene (Roche Diagnostics). B2 dissolved in DMSO, or an equivalent amount of DMSO, was added to the wells at the time of transfection. After 48 h, cells were fixed for 10 min with 4% formaldehyde in PBS and rinsed in PBS. The samples were blocked with 10% goat serum in PBS and incubated for 1 h at room temperature with mouse anti-α-synuclein (1:100; 610786,BD Biosciences) in 10% goat serum/0.1% Tween/PBS and 30 min with Alexa 594 donkey anti-mouse (1:200; A-21203, Molecular Probes). Washes were done with PBS and 0.2% Tween. Slides were mounted by using Vectashield with DAPI (Vector Laboratories) and photographed by using openlab software (Improvision, Lexington, MA) on a Zeiss Axioplan II microscope. Six random fields were photographed per condition. Total nuclei and brightly α-synuclein-positive cells were counted (blinded to treatment identity) by using openlab. Statistical significance was determined by using χ2.
Plasmid Construction.
The constructs for human wild-type untagged α-synuclein and its C-terminal-tagged version (referred to as synT) have been described (16, 22).
Cell Culture and Transfection.
Human H4 neuroglioma cells (HTB-148, ATCC) were maintained in OPTI-MEM (Life Technologies, Grand Island, NY) supplemented with 10% FBS. H4 cells were passaged 24 h before transfection and plated in 24-well plates. Cells were transfected by using Superfect (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Compounds or DMSO were added after the transfection procedure was concluded. For the immunoblotting experiments, cells were plated in 60-mm dishes 24 h before transfection. Transfections were performed as described above.
α-Synuclein Toxicity Assay.
Toxicity was analyzed 24 h after transfection by measuring the release of adenylate kinase from damaged cells into the culture medium by using the ToxiLight kit (Cambrex, East Rutherford, NJ) according to the manufacturer’s protocol. Statistical significance was determined by t test.
SDS/PAGE and Immunoblotting.
Twenty four hours after transfection, H4 cells were washed with cold PBS, harvested by scraping in cold lysis buffer without detergents [50 mM Tris·HCl, pH 7.4/175 mM NaCl/5 mM EDTA, pH 8.0/protease inhibitor mixture (Roche Applied Sciences)], and sonicated for 10 s. Protein concentration was estimated by using the BCA method, and the appropriate volume of each sample was diluted in 4× SDS sample buffer. Lysates were subjected to SDS/PAGE by using 10–20% Tris–Glycine gels (NOVEX, San Diego) for Western blot analysis. Protein was transferred to Immobilon-P membrane (Millipore) and blocked in blocking buffer (LI-COR, Lincoln, NE) for 1 h before the addition of the primary antibody [anti-Hsp70 (Stressgen Biotechnologies, Victoria, CA), anti-Hsp27 (Stressgen Biotechnologies), anti-DJ-1 (Chemicon), anti-α-synuclein (syn1, BD Biosciences, Franklin Lakes, NJ) or anti-actin (Sigma)] at room temperature for 1–2 h or overnight at 4°C. The blots were washed three times in Tris-buffered saline with 0.2% Tween (TBS-T, pH 7.4) and incubated at room temperature for 1 h in fluorescent labeled secondary antibodies [IRDye 800 anti-rabbit or anti-mouse (Rockland Immunochemicals), 1:3,000 or Alexa-680 anti-rabbit or anti-mouse (Molecular Probes), 1:3,000]. After three washes in TBS-T, immunoblots were processed and quantified by using the Odyssey infrared imaging system (LI-COR).
Rabbit Reticulocyte Assay.
Luciferase refolding assays were done as described (23) with some modifications. Briefly, Quantilum recombinant firefly luciferase (Promega) was diluted to 100 nM in stability buffer (25 mM Tricine·KOH, pH 7.8/8 mM MgSO4/0.1 mM EDTA/10% glycerol/0.25% Triton X-100/10 mg/ml BSA). The luciferase was denatured at 40°C for 15 min, followed by incubation for 10 min on ice and 5-fold dilution into Tris buffer (TB; 10 mM Tris·HCl, pH 7.5/3 mM MgCl2/50 mM KCl/2 mM DTT/2 mM ATP/10 mM phosphocreatine/35 units/ml creatine kinase). The denatured luciferase was then added at a final concentration of 10 nM to reactions containing TB, 10% untreated rabbit reticulocyte lysate (Promega), and DMSO, B2, or geldanamycin (Calbiochem) in a final volume of 100 μl. All conditions were done in duplicate. Reactions were incubated at 25°C. At time points from 0 to 80 min, 5 μl of renaturation sample was added to 120 μl of Luciferase Assay Reagent (Promega). Light production was measured for 10 s in an Optocomp I luminometer (MGM Instruments, Hamden, CT). Renaturation activity was expressed as a percentage of the total signal from native luciferase incubated in reticulocyte lysate, as above.
Acknowledgments
We thank Roger Tsien for the mRFP plasmid and J. Michael Andresen, Alain Charest, Jill Crittenden, John Doench, and Erik Wilker for valuable discussions. R.A.B. is supported by a postdoctoral fellowship from the Hereditary Disease Foundation. T.F.O. is supported by a Massachusetts Biotechnology Research Council Tosteson Award Fellowship. T.F.O., P.J.M, and B.T.H. were supported by National Institutes of Health Grant 5P50NS38372A-06. P.J.M. is also supported by the American Parkinson’s Disease Association and a Claflin Distinguished Scholars Award. A.B.Y. and A.G.K. received gift support from Discovery of Novel Huntington’s Disease Therapeutics, MassGeneral Institute for Neurodegenerative Disease.
Abbreviations
- Htt
huntingtin
- polyQ
polyglutamine
- B2
(5-[4-(4-chlorobenzoyl)-1-piperazinyl]-8-nitroquinoline)
- B5
(ethyl 2-([(4-methoxyphenyl)acetyl]amino)-5-([(2-methoxyphenyl)amino]carbonyl)-4-methyl-3-thiophenecarboxylate)
- B21
(5-[4-(2-fluorobenzoyl)-1-piperazinyl]-8-nitroquinoline)
- B22
(8-[4-(2-fluorobenzoyl)-1-piperazinyl]-5-nitroquinoline)
- mRFP
monomeric red fluorescent protein.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Frydman J. Annu. Rev. Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 2.Glickman M. H., Ciechanover A. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 3.Ross C. A., Pickart C. M. Trends Cell Biol. 2004;14:703–711. doi: 10.1016/j.tcb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 5.Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C. M., Stefani M. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 6.Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 7.Taylor J. P., Tanaka F., Robitschek J., Sandoval C. M., Taye A., Markovic-Plese S., Fischbeck K. H. Hum. Mol. Genet. 2003;12:749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- 8.Apostol B. L., Kazantsev A., Raffioni S., Illes K., Pallos J., Bodai L., Slepko N., Bear J. E., Gertler F. B., Hersch S., et al. Proc. Natl. Acad. Sci. USA. 2003;100:5950–5955. doi: 10.1073/pnas.2628045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bence N. F., Sampat R. M., Kopito R. R. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 10.Bennett E. J., Bence N. F., Jayakumar R., Kopito R. R. Mol. Cell. 2005;17:351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 11.Holmberg C. I., Staniszewski K. E., Mensah K. N., Matouschek A., Morimoto R. I. EMBO J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jana N. R., Zemskov E. A., Wang G., Nukina N. Hum. Mol. Genet. 2001;10:1049–1059. doi: 10.1093/hmg/10.10.1049. [DOI] [PubMed] [Google Scholar]
- 13.Venkatraman P., Wetzel R., Tanaka M., Nukina N., Goldberg A. L. Mol. Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- 14.Verhoef L. G., Lindsten K., Masucci M. G., Dantuma N. P. Hum. Mol. Genet. 2002;11:2689–2700. doi: 10.1093/hmg/11.22.2689. [DOI] [PubMed] [Google Scholar]
- 15.Gilon T., Chomsky O., Kulka R. G. EMBO J. 1998;17:2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLean P. J., Kawamata H., Hyman B. T. Neuroscience. 2001;104:901–912. doi: 10.1016/s0306-4522(01)00113-0. [DOI] [PubMed] [Google Scholar]
- 17.Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 18.Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., et al. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 19.Klucken J., Shin Y., Masliah E., Hyman B. T., McLean P. J. J. Biol. Chem. 2004;279:25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 20.Whitesell L., Mimnaugh E. G., De Costa B., Myers C. E., Neckers L. M. Proc. Natl. Acad. Sci. USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thulasiraman V., Matts R. L. Biochemistry. 1996;35:13443–13450. doi: 10.1021/bi9615396. [DOI] [PubMed] [Google Scholar]
- 22.McLean P. J., Kawamata H., Shariff S., Hewett J., Sharma N., Ueda K., Breakefield X. O., Hyman B. T. J. Neurochem. 2002;83:846–854. doi: 10.1046/j.1471-4159.2002.01190.x. [DOI] [PubMed] [Google Scholar]
- 23.Schumacher R. J., Hansen W. J., Freeman B. C., Alnemri E., Litwack G., Toft D. O. Biochemistry. 1996;35:14889–14898. doi: 10.1021/bi961825h. [DOI] [PubMed] [Google Scholar]