Abstract
For more than 80 years, the euchromatic right arm of the Drosophila fourth chromosome (101F-102F) has been one of the least genetically accessible regions of the fly genome despite the fact that many important genes reside there. To improve the mapping of genes on the fourth chromosome, we describe a strategy to generate targeted deficiencies and we describe 13 deficiencies that subdivide the 300 kb between the cytological coordinates 102A6 and 102C1 into five discrete regions plus a 200-kb region from 102C1 to 102D6. Together these deficiencies substantially improve the mapping capabilities for mutant loci on the fourth chromosome.
THE ability to manipulate the Drosophila genome has been central to the mapping and the molecular characterization of individual genes and the subsequent study of their biological function. Although the availability of the Drosophila genome sequence has accelerated molecular studies, to understand gene function, it is still necessary to determine which transcription units correspond to functional mutations. Despite significant advances over the last few years, one of the least well-characterized regions of the fly genome is the fourth chromosome. This small 5-Mb chromosome contains 1.15 Mb of euchromatic DNA on the right arm (Locke et al. 2000) and 89 annotated transcripts whose function, with few exceptions, can only be inferred from sequence comparison (Adams et al. 2000). In addition, several of these loci are likely to participate in pathways that, when mutated, lead to a number of genetic diseases in humans. Because the fourth chromosome does not recombine (Bridges 1935) gene order cannot be established by traditional meiotic mapping methods. Further, only four large deficiencies that uncover some of the genes on this chromosome [Df(4)M101-62f, Df(4)C3, Df(4)G, and T(1;4)scH] have been available so that deficiency mapping is rather low resolution. Thus, the reported locations and order of most of the mutations isolated on this chromosome are relatively crude and may not correspond to their actual positions (Sturtevant 1951; Hochman 1961, 1971, 1974).
The cytological analysis of the proximal deficiency, Df(4)M101-62f (101F-102B), as well as the molecular characterization of the loci deleted in this deficiency, suggests that it removes roughly one-third of the genes on the fourth chromosome, while the distal deficient translocations, T(1;4)scH and Df(4)G, delete genes located from approximately 102E2 onward. The deficiency Df(4)M101-4 (101F-102B or 101F-102D2) that presumably uncovered the region between these deficiencies was lost many years ago. The undeleted gap between Df(4)M101-4 and Df(4)G, (102B-102E2) led Hochman (1971) to tentatively place genes not contained within the deficiencies, between them. This gap was reduced with the isolation of Df(4)C3 (102D6-102F).
To allow a better correlation between the physical and functional maps of the genes contained in the 102B-102D6 interval, we explored the possibility of creating a series of nested terminal deficiencies (TDf) exploiting the poorly understood ability of the transposable element P to delete portions of chromosomes (Levis 1989; Ahmad and Golic 1998).
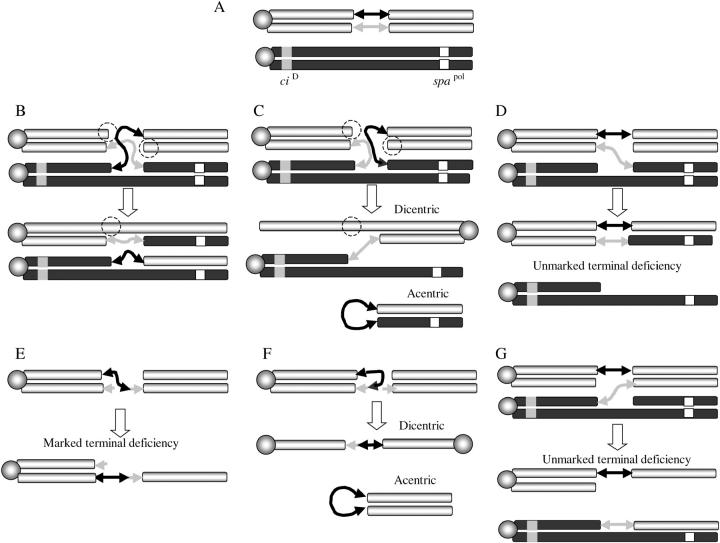
The prevailing hybrid element insertion (HEI) model (Figure 1) (Gray et al. 1996; Preston et al. 1996) proposes that terminal deficiencies can be generated during an abnormal transposition event (male recombination) in which the transposase, rather than excising the 5′ and 3′ ends of a single P element, excises a single P-element end from one chromatid and the other end from the sister chromatid to create a hybrid element (Figure 1). The creation of this hybrid element is then followed by recognition of a target site and integration of both or only one of the cleaved ends of this hybrid element into the sister chromatid or the homologous chromosome. According to the HEI model, this process involves pairing of homologous chromosomes during male mitosis and meiosis (Fung et al. 1998; Vazquez et al. 2002) and, depending on which combination of P-element ends are cleaved and the direction and chromatid that they insert into during these abnormal transpositions, these events can generate 8–13 possible products. Three of the predicted products of HEI events could be particularly useful for creating fourth chromosome deletions with marked ends. First, if the distal end of the P element inserts toward the centromere in the homolog (Figure 1C) or in the sister chromatid (Figure 1F) a dicentric chromosome will be created that can subsequently break into two unequal terminal deficiencies (Gray et al. 1996; Preston et al. 1996). Alternatively, insertions of a distal P-element end, toward the telomere in the homologous chromosome or sister chromatid, accompanied by a failure of the sister P element to either be cleaved or integrate (Figure 1, D and E) will produce two products, namely a recombinant (between either sister chromatids or homologous chromosomes) and a terminal deficiency. A similar outcome will be obtained when the proximal end of a P element is excised and the resulting genomic end fails to be repaired (Figure 1G) thus producing an unmarked terminal deficiency. Although predicted, the recovery of terminal deficiency products could not be directly verified (Gray et al. 1996; Preston et al. 1996), since large terminal deficiencies of the second chromosome are lethal. Because the fourth chromosome is viable in a single dose (Bridges 1925), it is possible to test if the terminal deficiencies predicted by HEI can indeed be recovered as a series of discrete deletions in the region between 102A6 and 102C1.
Figure 1.—
The HEI model of aberrant transposition. The two double-headed arrows represent a single P element inserted on the fourth chromosome in a dividing cell; the sister P-element arrows are depicted in gray and black to distinguish them. Homologous chromosomes are depicted in gray and in black with sister chromatids in the same color. The centromere is represented as an shaded circle, the ciD proximal marker is represented as a shaded square, and the distal recessive marker svspa-pol is represented as an open square. (A) The pairing of two homologous chromosomes during mitosis or meiosis. (B) Two opposite ends of a replicated P element (solid and shaded arrowheads) are released by the transposase from the sister chromatids in a dividing cell leaving two breaks (dashed circles). The free P-element ends integrate at a single target in the homolog (black). (C) The integration of the distal P-element end in the homolog toward the centromere accompanied by failure of the other P end to integrate is expected to generate a dicentric. (D) The integration of the distal P-element end in the homolog toward the telomere accompanied by failure of the other P end to integrate is expected to generate an unmarked terminal deficiency. (E) The integration of the distal P-element end in the sister chromatid toward the telomere is expected to generate a marked terminal deficiency. (F) The integration of the distal P-element end in the sister chromatid toward the centromere is expected to generate a sister chromatid dicentric. (G) Finally, the excision of a proximal end and insertion in the homolog or sister chromatid would be expected to generate an unmarked terminal deficiency. In addition to these predicted configurations, the failure to integrate one of the opposite ends (shaded arrowhead) could lead to the formation of terminally marked terminal deficiencies.
We have exploited these aberrant transposition events to generate 17 terminal deficiencies in the 102A6 to 102C1 interval. Molecular analyses of 13 of the 17 deficiencies indicate that 4 of them [Df(4)A6-2b, Df(4)A6-5d, Df(4)A6-11a, and Df(4)C1-7b] result from dicentric breakage as expected by the HEI model (Figure 1, C and F), while another four deficiencies [Df(4)B2-7a, Df(4)B6-2b, Df(4)B7-6a, and Df(4)C1-10a] are equally consistent with HEI events involving dicentric breakages as depicted in Figure 1, C and F or an HEI event as shown in Figure 1E. One deficiency [Df(4)A6-23a] could have arisen either from a dicentric breakage as depicted in Figure 1, C and F or from excision events depicted in Figure 1, D and G that leave no P-element markers at the end of the chromosome. Finally, four other deficiencies [Df(4)B2-2d, Df(4)B2-5a, Df(4)B6-4a, and Df(4) C1-5b] contain distal restorations not explicitly predicted by the HEI model. The possible mechanism for these will be discussed.
MATERIALS AND METHODS
Flies and crosses:
Flies were reared at 25° on standard cornmeal molasses media. A full description of the mutations used can be found at FlyBase (http://flybase.bio.indiana.edu/). The insertion P{ry+} at 102D was previously described (Spradling and Rubin 1982). svspa-pol (102F2) is a recessive mutation that produces flies with rough eyes; svn is an allele of sv that complements svspa-pol and produces flies with reduced, absent, or malformed bristles in homozygous flies (svn/svn). svrugoso-nu (Sousa-Neves 2000) is an allele of sv that complements neither svspa-pol nor svn; T(1;4)scH is a deficient translocation marked with a wild-type copy of y+ that removes at least 11 terminal genes including sv (Sousa-Neves 2000). C(4)103 is a compound fourth chromosome derived from C(4)DRA-1 (Sousa-Neves 2000) that is marked with ci1 and eyR. All P insertions used, except P{ry+} at 102D, were single PCR mapped insertions of the vector Supor-P that carries the internal markers y+ and w+ (Roseman et al. 1995). The P elements KG02140, KG00711, KG02689, KG01667, and KG03370 as well as their orientations were mapped by the Berkeley Drosophila Genome Project at positions 102A6, 102B2, 102B6, 102B7, and 102C1, respectively.
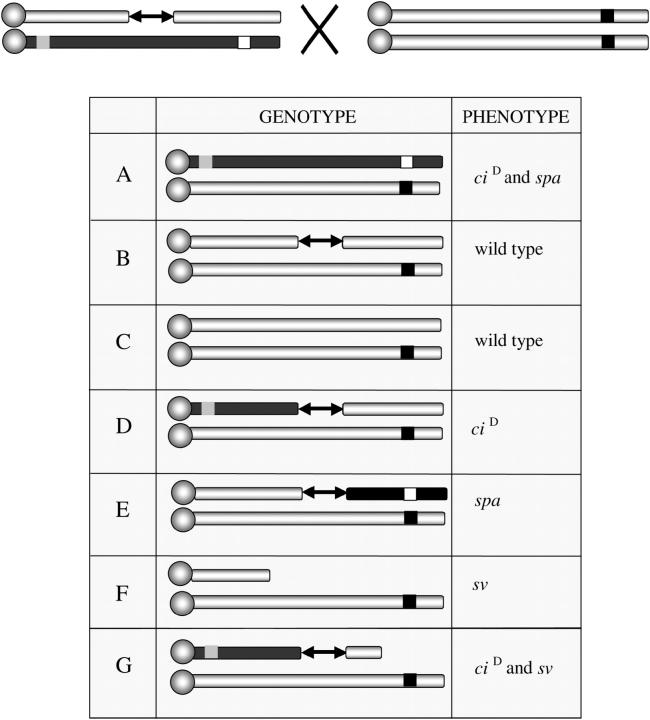
To generate interstitial duplications/deficiencies and terminal deficiencies in the pilot experiment, we made use of the unique properties of svrugoso-nu. This allele is homozygous viable and fails to complement svspa-pol and svn. Flies heterozygous for svrugoso-nu/svspa-pol have rough eyes and normal body hairs indistinguishable from svspa-pol homozygotes, while svrugoso-nu/svn animals exhibit loss of hairs similar to svn/svn homozygotes. Finally, flies heterozygous for T(1;4)scH/svrugoso-nu invariably eclose with a more severe loss of body hairs, are sterile, and die soon after eclosion. These properties allowed us to screen the F1 of heterozygous males ciD svspa-pol/ci+ P{ry+}sv+ with a source of transposase (TMSΔ2-3) and homozygous females svrugoso-nu/svrugoso-nu for recombinants that break the linkage between ciD and svspa-pol markers (ciD sv+ and ci+ svspa-pol) or carry terminal deficiencies (ciD TDf and ci+ TDf) (see Figure 2).
Figure 2.—
Scheme of the pilot experiment to measure the frequency of P-element-induced recombinants and terminal deficiencies. The scheme allows the recognition of five of the seven possible classes of progeny that may be produced. The ciD proximal marker is represented as a shaded square, the distal recessive marker svspa-pol as an open square, and svrugoso-nu as a solid square; the source of transposase is omitted. (A) Nonrecombinants of this class can be recognized by the dominant interruption of L4 (ciD) and rough eyes (svspa-pol). (B and C) Nonrecombinant with the P element as well as chromosomes with healed ends will be ci+ sv+. (D and E) Recombinants toward the centromere appear as ciD while recombinants toward the telomere appear svspa-pol. (F and G) The possible progeny classes that could be recovered from dicentric chromosome breakage between homologs.
To generate terminal deficiencies from the other five P insertions described above, we used a slightly different scheme. Heterozygous males ci+ sv+/ci+ P{Supor-P, y+ w+} sv+ were crossed to svspa-pol/svspa-pol and the F1 was screened for animals that exposed the recessive svspa-pol. In the F2 and F3, the deficient chromosomes were balanced over a ciD svspa-pol chromosome. To test whether these lethal chromosomes actually corresponded to large deficiencies, ciD svspa-pol/Df(4) flies were crossed to ci1 gvl1eyR svn and the non-ciD progeny were scored for the recessive markers eyR and svn. To provide a numbering system that indicates the history and origin of these deficiencies, they were named Df(4), followed by the position of the parent insertion (e.g., C1 for cytological position 102C1), the number of the male parent in which it was identified (e.g., 1, 2, 3), and the isolate number (e.g., a for the first, b for second). Thus Df(4)C1-10a derives from a parental insertion at 102C1 and was the first deficiency found in the offspring of male 10.
Primers and PCR conditions:
To define the position and integrity of P-element insertions or the presence of neighboring genomic DNA, the following procedures were followed. DNA was extracted from heterozygous deficiency lines (30–40 flies per line) using standard procedures. Each amplification reaction contained primers specifically designed for one or the other end of the P element and the corresponding neighboring genomic DNA. The DNA was amplified using 30 cycles of PCR (denaturation at 95° for 45 sec, annealing at 55° for 45 sec for KG02140, KG01667, and KG00711 or annealing at 62° for 45 sec for KG02689 and KG03370 plus an elongation at 72° for 90 sec). Amplified bands from the deficiency heterozygotes were compared to those from the parental lines by agarose gel electrophoresis. Lines that yielded a band from the proximal junction between the genomic DNA and the P element but that failed to yield a band from the distal junction of the P element and the corresponding genomic DNA were tested for dicentric origin using the distal primer of the P element and the proximal genomic primer.
The primers used were as follows:
5′ P-end specific primer—CGGTAAGCTTCGGCTATCGACGG
3′ P-end specific primer—ACGTTAAGTGGATGTCTCTTGCCG
Proximal primer for line KG03370—GCAATGGTGAGGCTCTGGTCGTTC
Distal primer for line KG03370—CAAGAGATTCGGGTGGAGTGGGC
Proximal primer for line KG02689—AGGTACGCGAATTCTAAGCAGGAC
Distal primer for line KG02689—TGTGCCGTATGTCTCCACCGTTC
Proximal primer for line KG00711—TCATCGATGCGCTTGTTTGCCG
Distal primer for line KG00711—TTTCACTTGCCTATTTCCCAATG
Proximal primer for line KG02140—CTGGCACTCAATTAAAATCTCGTC
Distal primer for line KG02140—TTAGGTACTGAAGCTGAATACTG
Proximal primer for line KG01667—TCGGCCTGAAGTCGAGTCATCG
Distal primer for line KG01667—CATCGTACTGTAATCGCCTGAG
Molecular mapping of the extent of the deficiencies:
Our terminal deficiencies were crossed to a stock bearing the ci1 mutation, which contains a Gypsy element in the upstream regulatory region of cubitus interruptus (ci). These heterozygous flies were intercrossed and single F1 embryos were picked and homogenized in 10 μl of 10 mm Tris-Cl, pH 8, 1 mm EDTA, pH 8, 200 mg/ml proteinase K solution. The tubes were incubated at 37° for 45 min and heated at 99° for 10 min. Each PCR reaction contained a pair of primers that amplify only the boundary between the Gypsy Element and ci (GE-C), a pair of primers for a region known to be present (KP), and a third pair to test for deleted regions (PD). The embryos that did not yield the expected band size using the GE-C pair and that give the expected size for the KP pair correspond to homozygous Df/Df and are analyzed for the presence or absence of the other expected PCR products using a series of PD primers as described in Podemski et al. (2004).
RESULTS
Detection of P-element-induced terminal deficiencies on the fourth chromosome and the measure of their occurrence:
To distinguish terminal deficiencies from interstitial recombinants, we conducted a pilot experiment by making use of the unique properties of the svrugoso-nu allele (Sousa-Neves 2000) that allow one to distinguish interstitial deficiency/duplication recombinants (Figure 2, D and E) from terminal deficiencies (Figure 2, F and G) among the progeny of males that are ciD svspa-pol/ci+ P{ry+}sv+ and females that are ci+ svrugoso-nu/ci+ svrugoso-nu (Figure 2). We measured the frequency of recovery of each recombinant class (Table 1). Among a total of 2162 F1 progeny from nine males (ciD spapol/ci+ P{ry+} sv+) with a source of transposase, we found six recombinants toward sv (ci+ svspa-pol, four independent), four recombinants toward ciD (ciD sv+, four independent), and 11 terminal deficiencies (3 independent).
TABLE 1.
Pilot experiment to evaluate recovery of terminal deficiencies
| Chromosomes screened |
Recombinants ciDspa+ |
Recombinants ci+spa |
ciD TDf | ci+ TDf | % of ciDspa+ | % ci+spa | % terminal deficiency ci+ |
|
|---|---|---|---|---|---|---|---|---|
| Male 1 | 286 | 0 | 0 | 0 | 6 | 0.0 | 0.0 | 2.1 |
| Male 2 | 44 | 0 | 0 | 0 | 3 | 0.0 | 0.0 | 6.8 |
| Male 3 | 351 | 0 | 0 | 0 | 0 | 0.0 | 0.0 | 0.0 |
| Male 4 | 225 | 1 | 1 | 0 | 2 | 0.4 | 0.4 | 0.9 |
| Male 5 | 252 | 0 | 0 | 0 | 0 | 0.0 | 0.0 | 0.0 |
| Male 6 | 194 | 0 | 1 | 0 | 0 | 0.0 | 0.5 | 0.0 |
| Male 7 | 284 | 2 | 1 | 0 | 0 | 0.7 | 0.4 | 0.0 |
| Male 8 | 246 | 2 | 1 | 0 | 0 | 0.8 | 0.4 | 0.0 |
| Male 9 | 105 | 1 | 0 | 0 | 0 | 1.0 | 0.0 | 0.0 |
| Totals | 2162 | 6 | 4 | 0 | 11 | 0.28 | 0.19 | 0.5 |
The offspring from nine individual males whose marker status suggested a terminal deletion are listed separately. The different classes of recombinants and the frequencies of occurrence are shown.
This result shows that terminal deficiencies are indeed produced during male recombination and are as frequent as any given recombinant class alone when normalized per number of independent events (0.18% recombinants toward ciD, 0.18% recombinants toward sv, 0.13% terminal deficiencies).
Recovery of terminal deficiencies along the fourth chromosome:
On the basis of the results of our pilot experiment, we designed a large-scale experiment in which rather than identifying the recombinants on the basis of the exchange of flanking markers, we screened directly for animals mutant for svspa-pol in descendants of P{w+y+}/+ males and svspa-pol/svspa-pol females. In the absence of transposase, this cross is expected to yield an F1 of only sv+ flies (P{w+y+} +/svspa-pol). However, in the presence of transposase, sv F1 animals that appear can correspond to flies with a single fourth chromosome (haplo-4), to fortuitous integrations in the sv gene, or to terminal deficiencies. Haplo-4 flies are readily distinguished from the other two classes because they are invariably Minute (a slight reduction in the body size, thinning of the bristles, reduced viability, and late emergence) due to the presence of a single copy of the Minute gene RpSA3(101F1) (van Beest et al. 1998). The other two classes (new mutation of sv vs. terminal deficiency) can be distinguished by the fact that svspa-pol/Df heterozygous exhibit a more severe svspa-pol phenotype than do svspa-pol/svspa-pol homozygous flies and the fact that terminal deficiencies uncover all recessive mutant loci distal to the insertion point, while insertions in the sv gene exhibit only sv.
We screened a total of 27,899 chromosomes from 5 different fourth chromosomes with single P-element insertions that were molecularly mapped by the Berkeley Drosophila Genome Project at 102A6, 102B2, 102B6, 102B7, and 102C1 (Figure 5). This screening led to the recovery of 52 deficiencies (svspa-pol/Df): 13 (0.66%) from the insertion at 102C1 (5 independent, 0.25%), 3 (0.16%) from the insertion at 102B7 (2 independent, 0.11%), 18 (0.29%) from the insertion at 102B6 (5 independent, 0.08%), 13 (0.21%) from the insertion at 102B2 (4 independent, 0.06%), and finally 5 (0.06%) from the insertion at 102A6 (2 independent, 0.04%). These results are comparable to the result obtained in the pilot experiment since terminal deficiencies were recovered at frequencies that range between 0.04% (102A6) and 0.25% (102C1) of the total number of chromosomes screened. However, there was a notable progressive decrease in the recovery of independent terminal deficiencies as the size of the region deleted increased. This distribution is in agreement with a previous observation that less than two copies of the fourth chromosome reduce the viability of the carriers (Ahmad and Golic 1998).
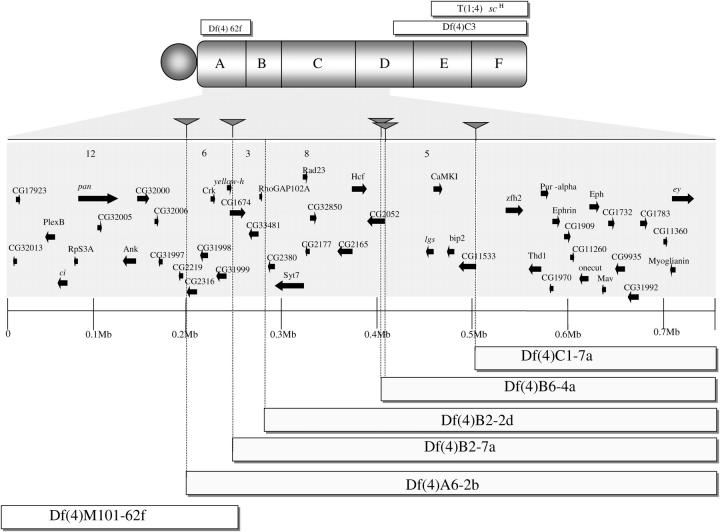
Figure 5.—
The right arm of the fourth chromosome and its subdivisions 102A–F. The position of three previously existing deficiencies, Df(4)M101-62f, T(1;4)scH, and Df(4)C3 is indicated. Below the fourth chromosome is a blowup of the region 102A6-102C4. Solid triangles indicate the positions of the P-element insertions used to generate the deficiencies and arrows indicate the coding sequences within the region. Df(4)A6-2b is the largest deficiency on the fourth chromosome isolated to date and the combination of this deficiency with Df(4)M101-62f uncovers virtually the entire right arm of the fourth chromosome.
Of the 17 males recovered, 15 were fertile, 1 died before mating, and 1 was sterile. In contrast, 18 females were recovered, 1 was fertile, 16 were sterile, and 1 died before mating.
To provide further evidence that these deficiencies are actually large terminal deficiencies as opposed to insertions in the sv gene, we tested each isolate against the multiple marked chromosome ci1(101F) gvl1 eyR(102D4-D6) svn (102F2). If the svspa-pol animals bear large deficiencies with breakpoints between 102A6 and 102C1, all these deficiencies should at least uncover the ey and sv markers present in the ci gvl ey sv chromosome. Indeed all deficiencies tested (a total of 13 independent) uncovered the two recessive markers eyR and svn, but all complemented ci1 (101F) and gvl1.
Mapping the deficiency breakpoints and P elements:
Of the 13 independent deficiencies that could be established as stable stocks, 9 of 13 of the terminal deficiencies (70%) retained the P element as indicated by the retention of the y+ or w+ markers carried by the P element. Specifically, 5 carried both internal markers (w+ and y+), 3 carried only y+, 1 carried only w+, and only 4 did not express any of the internal markers of the P element (w− y−).
To physically confirm the terminal deficiencies and determine whether these deficiencies retained P elements in the same position as the parental insertion, we performed a series of DNA analyses on the 10 of the original 13 lines that survived. Initially, DNA was amplified from the boundary between the P element and the flanking genomic DNA using primers for both the proximal and the distal ends of the P-element insertion and the respective proximal and distal flanking DNA. Amplification of the expected PCR product indicates the presence of the P element in the original location while failure to amplify indicates that the P element is either deleted or no longer in the same location. Control PCR reactions using the parent insert confirm the insert location and amplification potential of all of the primers used (Figure 3).
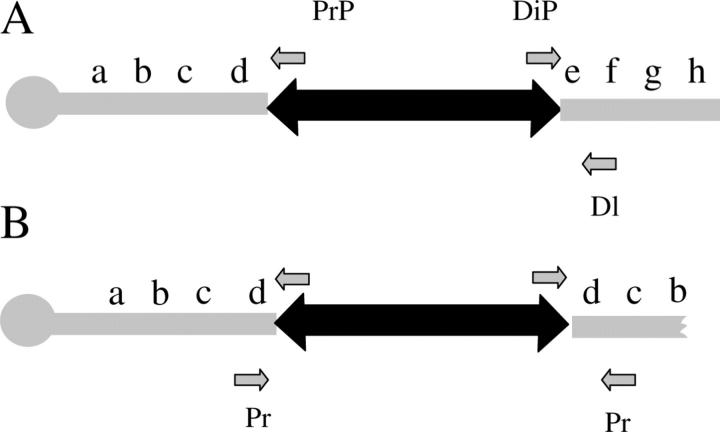
Figure 3.—
Primers used to distinguish between different classes of terminal deficiency structure. (A) A P element is depicted (double-headed arrow), with the proximal P inverted repeat (PrP) and distal inverted repeat (DiP) primers, as well as the respective proximal (Pr) and distal (Dl) genomic primers used to verify the position of the P element in the terminal deficiencies. The sequence of the chromosome, increasing in cytology, is arbitrarily represented as a b c d e f g h. (B) Terminal deficiencies that derive from dicentric breakage are expected to generate a reverse duplication of the proximal genomic DNA (d c b a) after the distal end of the P element. Only events deriving from breakage of a dicentric produce PCR products when the DiP and Pr primer pair is used.
We characterized 10 deficiencies of independent origin derived from five different insertions. Eight of these yielded at least one product (5 with only a single product and 3 with two products), indicating that 80% of the deletions retain an insert at the original position (Table 2). In all cases where PCR detected only one end of the P element, the missing end was the distal end. Two of 10 deficiencies analyzed [Df(4)A6-23a and Df(4)B2-2d] did not yield any product from either end (Table 2). These are consistent with the possibility that the original P element jumped before generating the terminal deletion or the possibility that these deficiencies derive from a dicentric breakage.
TABLE 2.
Structure of terminal deficiencies recovered as hemizygotes
| Parental line | Deficiency | Proximal flanking |
Distal flanking |
Markers | Notes |
|---|---|---|---|---|---|
| KG02140 | 5′ P | 3′ P | w+ y+ | ||
| A6-23a | Lost | Lost | w− y− | ||
| KG00711 | 3′ P | 5′ P | y+ w+ | ||
| B2-2d | Lost | Lost | y+ w+ | +∼52 kb | |
| B2-5a | Retained | Retained | y+ w− | +3–6 kb | |
| B2-7a | Retained | Lost | y+ w− | ||
| KG02689 | 5′ P | 3′ P | w+ y+ | ||
| B6-2a | Retained | Lost | w+ y+ | ||
| B6-2b | Retained | Lost | w+ y− | ||
| B6-2d | Retained | Lost | w+ y− | ||
| B6-2e | Retained | Lost | w+ y− | ||
| B6-2j | Retained | Lost | w+ y− | ||
| B6-4a | Retained | Retained | w+ y+ | +3–6 kb | |
| KG01667 | 3′ P | 5′ P | y+ w+ | ||
| B7-6a | Retained | Lost | y+ w+ | ||
| KG03370 | 3′P | 5′ P | y+ w+ | ||
| C1-5a | Retained | Lost | y+ w+ | ||
| C1-5b | Retained | Retained | y+ w+ | +>6 kb | |
| C1-7a | Retained | Lost | y− w+ | ||
| C1-7b | Retained | Lost | y− w+ | Sister dicentric | |
| C1-7d | Retained | Lost | y− w+ | Sister dicentric | |
| C1-10a | Retained | Lost | y+ w+ |
The first column indicates the name of the original parental insertion; the second indicates deficiencies isolated, with the capital letter and number referring to the cytological location of the parent P element (described in materials and methods) and the hyphen and number followed by a letter indicating all the progeny from a single male. Since these could originate from a single cell that underwent mitosis they are considered sibling deficiencies and not independent events (e.g., C1-7a, b, d). The third and fourth columns indicate the position of the proximal and distal P-element ends of each insertion and whether these yielded proximal and distal fragments by regular PCR. The fifth column indicates the P-element markers and the orientation carried by each parental insertion and its derivatives. All of the deficiencies were isolated over a marked chromosome. An improved scheme shown in Table 3 employed recovery over a compound fourth chromosome and yielded better survival of large deficiencies.
To test whether any deficiencies that did not yield a distal PCR fragment might derive from the breakage of a dicentric chromosome, we amplified DNA using the distal primers of the P element and the proximal genomic primers. Only the sibling pair of deficiencies, Df(4)C1-7b and Df(4)C1-7d, produced products from these reactions. Sequencing of the amplified fragment showed that the distal end of the P element had inserted into the y+ in the direction of the centromere on the sister chromatid. The fact that these two deficiencies produce fragments from both the proximal and the distal P-element ends with the same proximal genomic primer indicates that they were generated by breakage of a dicentric chromosome involving sister chromatids (Table 2).
To further confirm the physical nature of the terminal deficiencies, two examples [Df(4)B2-2d and Df(4)C1-10a] were mapped by testing individual homozygous Df embryos for the presence or absence of a set of PCR markers across the fourth chromosome. By using PCR primers for various loci along chromosome four (Podemski et al. 2004), we were able to physically confirm the deleted regions and locate the breakpoints. The starting P element for the deficiencies at 102C1 is located between CG11533 and CG1449 and by PCR analysis, Df(4)C1-10a, y+ w+ is a terminal deletion with the breakpoint between CG11533 and CG1449 (zfh-2). The first coding sequence is present on the chromosome and the second is absent. All the loci distal to CG1449, which span ∼700 kb, are absent. PCR analysis of the insertion site indicated that the proximal end of the P element was still in its original location but that the sequences flanking the distal end of the P element were deleted.
The starting P element for the 102B2 deficiencies is located in the proximal end of CG1674. However, upon PCR analysis, it is apparent that Df(4)B2-2d, y+ w+ is a terminal deletion with the breakpoint between CG1748 (RhoGAP102A) and CG2380, roughly 35 kb distal to the original P insertion. All of the loci distal to and including CG2380, a region of ∼912 kb, are absent. In this deficiency, the P element has locally hopped and a new P insert is now located 17 kb proximal to the original insertion site so that the end point of the deficiency is located roughly 52 kb distal from the new insertion point.
Four independent terminal deficiencies (B2-2d, B2-5a, B6-4a, and C1-5b) that genetically delete distal markers, nevertheless, contained sequences that extend beyond the distal end of the P element. To better understand the structure of these deficiencies, we performed inverse PCR of the distal junction of the P element and the genomic DNA in Df(4)B6-4AT, which yielded a single band, indicating the presence of a single distal P-element sequence in this chromosome. The sequence obtained from this product is collinear with the distal genomic DNA of the parental insertion indicating the presence of a distal extension beyond the P location. To evaluate the size of the distal extensions in Df(4)B2-5a, Df(4)B6-4a, and Df(4)C1-5b (Table 2), we used primers anchored in the P-element end and in the surrounding genomic DNA at 3 and 6 kb away distal from the P element. Df(4)B2-5a and Df(4)B6-4a yield a fragment of 3 kb but both failed to produce the 6-kb fragment, suggesting that these terminal deficiencies end 3 to 6 kb distal to the original insertion point. Df(4)C1-5b yielded PCR fragments of 3 and 6 kb, showing that the distal restoration extends at least 6 kb. However, the largest distal restoration seems to be the one found in Df(4)B2-2d. In Df(4)B2-2d, we found a single inverse PCR band whose sequence maps 17 kb proximal to the original insertion while the breakpoint of this deficiency is 35 kb distal to the original insertion and 52 kb from the current P insertion.
Improving the deficiency recovery:
Since in the previous experiments we obtained only two terminal deficiencies from the insertion at 102A6, and only one could be established as a stock, it would be desirable to improve the number of marked deficiencies that start in this region. To improve deficiency recovery in this region, we used a scheme in which the males with the transposase, heterozygous for the P element at 102A6 and a wild-type fourth chromosome (P{w+y+}/+) were crossed to an attached fourth chromosome marked proximally with ci1 and distally with eyR [C(4)103 ci1 eyR].
Although this scheme does not increase the frequency of events, it should allow a better propagation of the deleted chromosomes and thus the number of individual stocks with terminal deficiencies. This experiment should also provide a chance to obtain products that the previous scheme might have missed.
In this experiment, we screened 3433 chromosomes and identified 13 putative terminal deficiencies from eight independent males (0.23%) (ci+ ey−). One independent line did not yield progeny and the remaining 7 were retrieved over C(4)103 ci1 eyR. Three of the 7 independent putative terminal deficiencies carried both markers of the P element, 2 did not carry any markers [Df(4)A6-7a and Df(4)A6-9a], and one male produced progeny carrying different P-element markers, namely one carried only the proximal marker y+ [Df(4)A6-2b] while its sibling was y− w− [Df(4)A6-2a]. Two independent terminal deficiencies that originally carried both P-element markers [Df(4)A6-4b, y+ w+ and the sibling pair Df(4)A6-5a, y+ w+/Df(4)A6-5b, y+ w+] gave rise to progeny that lost one P-element marker and in each case the marker retained was the originally distal marker [i.e., Df(4)A6-4b1 y+ w− and Df(4)A6-5a1, y+ w−/Df(4)A6-5b1, y+ w−] (Table 3). These results suggest that these deficiencies may derive from dicentric chromosomes.
TABLE 3.
Terminal deficiencies recovered over a compound fourth chromosome
| Parental line | Deficiency | Proximal | Distal | Markers | Notes |
|---|---|---|---|---|---|
| KG02140 | 5′ P | 3′ P | w+ y+ | ||
| A6-2a | w− y− | ||||
| A6-2b | Lost | Retained | w− y+ | P markers inverted | |
| A6-4a | w+ y+ | ||||
| A6-4b | w+ y+ | 4b1 w− y+ | |||
| A6-5a | w+ y+ | 5a1, w− y+− | |||
| A6-5b | w+ y+ | 5b1, w− y+ | |||
| A6-5d | Lost | Retained | w+ y+ | P markers inverted | |
| A6-7a | w− y− | ||||
| A6-7b | w− y− | ||||
| A6-9a | w− y− | ||||
| A6-10a | w− y+ | ||||
| A6-11a | Lost | Retained | w+ y+ | P markers inverted |
See Table 2 legend. These deficiencies were recovered using the improved method of recovering over a compound fourth chromosome. The status of the terminal P-element markers were evaluated for all lines. The arrangement of the P element vis à vis the flanking genomic DNA was evaluated by PCR for A6-2b, A6-5d, and A6-11a and found to be inverted, suggesting the involvement of an intermediate dicentric chromosome.
We tested all 12 of these chromosomes to determine whether they uncover svspa-pol, a recessive visible that is distal to ey in the compound C(4)RM svspa-pol chromosome. In all cases, they uncovered svspa-pol indicating the presence of large terminal deficiencies.
Three of the independent deficiencies [Df(4)A6-2b, Df(4)A6-5d, and Df(4)A6-11a] were molecularly mapped. None of these yielded products with the primers that identify the junction of the genomic DNA and proximal end of the P element (Pr-PrP) but they did yield products with the proximal genomic primer (Pr) and the primer anchored in the distal end of the P element. Thus, in these three cases the distal end of the P element inserted into either the homolog or the sister chromatid in the direction of the centromere to create a dicentric chromosome.
Mapping lethals using nested terminal deficiencies:
To demonstrate the utility of these deficiencies for mapping purposes, we mapped the recessive lethal gene in the P{102A8} chromosome bearing a P insertion at 102A8. To map the mutant gene, we tested whether the P{102A8} chromosome was lethal or viable over Df(4)M101-62f, Df(4)B2-2d, Df(4)B7-6a, and Df(4)C1-7a. If the lethality is caused by the insertion, only Df(4)M101-62f should be lethal in heterozygous flies, i.e., Df(4)M101-62f/P{102A8}. Since Df(4)M101-62f/P{102A8} animals are viable, the lethality of the chromosome that bears P{102A8} can not be ascribed to the insertion.
We then tested this chromosome over Df(4)B2-2d(102B2), Df(4)B7-6a(102B7), and Df(4)C1-7a(102C1). The P{102A8} chromosome is lethal over Df(4)B2-2d(102B2) and Df(4)B7-6a(102B7) but not over Df(4)C1-7a(102C1). Thus, the lethal gene, here named l(4)B6-C1, corresponds to one of the four loci within the 111 kb between Df(4)B7-6a and Df(4)C1-7a (lgs, CamK1, bip 2, and CK1).
DISCUSSION
P-element-generated terminal deficiencies are consistent with the HEI model:
The HEI model predicts two types of insertions that could be useful for generating large marked terminal deficiencies from fixed points. The first is the insertion of the distal end of a P element toward the centromere in the sister chromatid or homolog to create a dicentric chromosome. The second type of HEI event involves insertion events in which only one cleaved end of the hybrid element inserts in the sister chromatid toward the telomere (Figure 1E). The first type of insertion creates a dicentric chromosome that subsequently breaks (dicentric break) to produce two unequal terminal deficiencies. The second type of insertion is expected to produce an unequal recombination event yielding one recombinant chromosome and one terminal deficiency. If the distal end of the element in one chromatid is inserted before the element on its sister, we should expect a terminal deficiency without P-element sequences. If the insertion of the distal end of the element is within the sister element, we should expect a deficiency with a P element truncated distally. Finally, if the insertion is after the distal end of the P element in the sister element, we should expect the presence of a complete P element. In addition to the two predicted configurations of HEI described above, the excision of a distal end followed by a failed integration could in principle lead to a terminally marked terminal deficiency (Figure 1).
The best evidence that some of these deficiencies were generated after an HEI event comes from the sequencing of the DNA that abuts the distal end of Df(4)C1-7b and Df(4)C1-7d. In both cases, while the proximal P-element border is intact, the distal P-element end is followed by a reverse duplication of the proximal marker yellow, which is flanked by a reverse duplication of the genomic DNA that flanks the proximal end of the P element. The structure of the P insertion in these sibling deficiencies indicates that these terminal deficiencies derive from a bona fide dicentric breakage between sister chromatids.
In most cases (6/13 independent) (B2-7a, B6-2b, B7-6a, C1-5a, C1-7a, and C1-10a), we obtained PCR products of the proximal P-element end but none from the distal end (Table 2). This correlation between deleted or truncated ends and the lack of the distal markers can be easily understood as being the result of either a failed insertion (Figure 1B) or the insertion of a single distal end of the P element toward the telomere that creates an unequal recombinant (Figure 1E). However, clearly dicentric bridges can form and some of these can exhibit breaks at or within the P element perhaps due to cleavage followed by failed integration (e.g., Df's A6-2b, -5d, and -11a)(Figure 1, C and F).
Distal restorations:
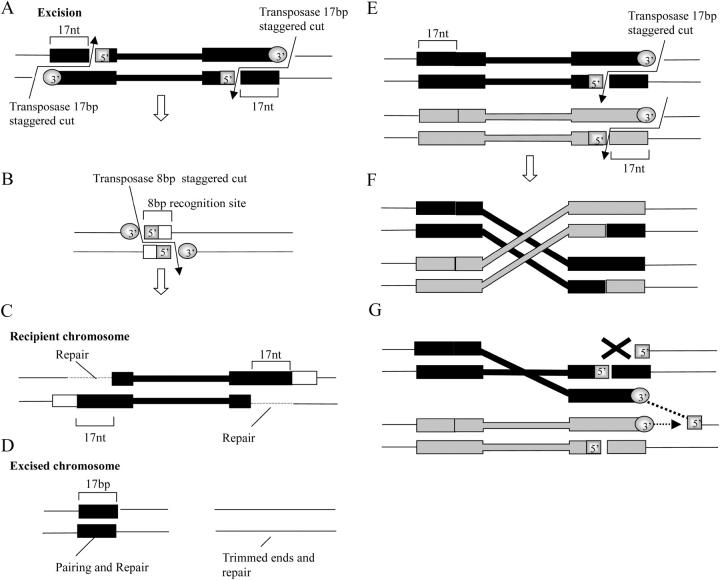
Several of the deficiencies have sequences that extend a variable distance past the P element before the deletion starts. To test whether distal restorations that extend from 3 to 50 kb beyond the parental P insertion could be due to the activity of two P elements, one that remains in the original location and another that creates a double-strand break further away, we amplified and sequenced the original insertion point and performed inverse PCR with DNA from Df(4)B2-2d to specifically identify the 17-bp footprint of the P-element inverted repeat anywhere in the genome. Sequencing of the original insertion site revealed no P-element sequences and inverse PCR identified a single truncated end 17 kb proximal to the original insertion site. Thus, although not impossible, it is unlikely that these restorations may have resulted from the activity of two P elements. However, a modification of the basic mechanism of P-element transposition provides a plausible explanation for these restorations. Normal transposition involves a staggered cut at the end of the P-element inverted repeat (Beall and Rio 1997) (Figure 4A) that leaves a 17-nt footprint with sequences from the very end of the P element in the cleaved chromosome and a P-element end with a 17-nt single stranded overhang (Figure 4A). This is followed by a staggered 8-nt cut in the target chromosome (Figure 4B). The 3′ end of the P element is then ligated to the 5′ of the recipient chromosome and the remaining 3′ end is used as the primer to repair the 17 nt of single-strand DNA of the P-element inverted repeat (Figure 4C).
Figure 4.—
Possible mechanism for the generation of distally restored terminal deficiencies. A–D depict the steps of normal transposition events and E–G depict the possible mechanism that results in distal restorations. The diagrams depict double strands of DNA. The P-element ends are represented as black or as gray boxes and thin and thick lines indicate the internal P-element sequences or genomic DNA, respectively. (A) Excision begins with two staggered 17-nt cuts at each end of the P element. (B) Recognition of a target results in a staggered 8-nt cut in the recipient site or chromosome. (C) The long 3′ overhanging end of the excised element is ligated to the exposed 5′ genomic and the 17-nt single-stranded gap is repaired from the genomic 3′. (D) The excised chromosome is either left with a footprint of the P element at the original location or trimmed by nucleases. (E) If only one cut is made at the distal end in the two sister chromatids, there are two possible outcomes. (F) In one, the double-stranded end of each element can be reciprocally ligated to its sister, resulting in an exchange event. In the other (G), the ligation of a single-strand 3′ end to the 5′ genomic (dashed line) exposes a genomic 3′ that could serve as a source of break-induced repair initiation (dashed arrow). Repair in this configuration would displace the transligated single-strand DNA and would be stopped either by a single-stranded nick or by breakage during sister chromatid segregation to the daughter cells, resulting in a distal restoration.
Transposition events in which a distal P-element end is released from both sister chromatids (Figure 4E) can lead to several outcomes. One is repair. A second is sister chromatid exchange, i.e., when the 17-nt staggered excision of one chromatid “integrates” into the end of the P element excised from its sister (Figure 4F). However, aberrant events that occur at a low frequency could lead to partial distal restorations. For example, if only the 3′ end of the 17 nt of one chromatid is ligated to the 5′ of its sister (Figure 4G), the 3′ end that remains exposed can serve as a primer for the initiation of a repair event that displaces the transligated strand in a distal direction (Figure 4G). Such a mechanism would be very similar to the break-induced repair events observed in yeast (Kraus et al. 2001). In this case, a unidirectional conversion tract would be extended until it is eventually aborted as the chromatids are assorted into the daughter cells and the single-stranded gap holding the sister chromatids together is either broken or degraded (Figure 4G). Formally, the model above does not demand homology searches between single-stranded and DNA duplexes (Tsubouchi and Roeder 2003) or a single-strand invasion (Petukhova et al. 2003) although one or more of these mechanisms could be involved.
This hypothesis explicitly predicts that terminal deficiencies with distal restorations should not contain a new 8-bp duplication at the end of the P element if the “integration” in the sister chromatid occurs exactly at the excision point and does not necessarily require an 8-bp staggered cut. Our data support this prediction since the sequences obtained from the boundary of the P element and genomic DNA of Df(4)B2-5a, Df(4)B6-4a, and Df(4)C1-5b have exactly the same 8-bp duplication found in the each parental line.
Stability and segregation of terminal deficiencies:
The terminal deficiencies described here have remained stable as evaluated by the presence of their terminal markers for over 30 generations. This suggests that either these chromosomes are rapidly protected against degradation or the degradation of the ends of these particular chromosomes is a slow process. In addition, the segregation properties of these chromosomes exhibit changes over generations. With increasing generations, there is a tendency for the larger deficiencies to accumulate a second copy of the marked balancing chromosome yielding a partial triplo-4 stock (Podemski et al. 2004). In most of the cases, we balanced these chromosomes in two ways: one using a regular ciD spapol chromosome and the other using compound fourth chromosomes. Animals with deleted chromosomes balanced with ciD spapol progressively became more viable, this correlates with the accumulation of a second copy of the ciD spapol chromosome and these stocks show a markedly reduced transmission of the deficiency alone (i.e., only ∼1/26 offspring inherit just the deficiency chromosome without the ciD spapol). Conversely chromosomes balanced over compounds transmit the deficiency chromosome more faithfully possibly because the covering compound fourth segregates as a single chromosome. For this reason, it is strongly advisable to maintain the larger deficiencies over dominantly marked compound chromosomes and to use appropriate markers on the lethal stocks to be mapped.
Mapping the 102B2-102C1 region using terminal deficiencies:
The deficiencies described here are useful for the fine structure mapping of genes on the fourth chromosome. To facilitate generating deficiencies whose breakpoints can be readily identified at the molecular level, we used P inserts whose integration points and orientation are known. In principle, knowing the locations of these P elements provides a fast means of localizing potential breakpoints and thus individual coding sequences in a defined interval. On the other hand, the fact that we have observed a significant number of distal restorations that produce deficiencies with somewhat different termini provides a means of isolating nested sets of deletions that cluster near a P insertion that can be valuable reagents for fine structure mapping on the fourth chromosome. For instance, the breakpoint of Df(4)A6-2b is ∼50 kb of Df(4)B2-7a and there are six genes in this interval (CG2316, CG31998, Crk, CG31999, yellow-h, and CG1674) (Figure 5). The breakpoint of Df(4)B2-2d is approximately 35 kb distal from the two deficiencies that occur at the original P-insert site at 102B2 [Df(4)B2-5a and Df(4)B2-7a]. Together, these define a three-gene interval (CG1674, CG33481, and RhoGAP102A). Df(4)B2-2d and the deficiencies with breakpoints at 102B6 [Df(4)B6-4a, Df(4)B6-2b, Df(4)B6-2d, Df(4)B6-2e, and Df(4)B6-2j] define a 112-kb region that contains eight genes (CG2380, Syt7, Rad 23, CG2177, CG32650, CG2165, Hcf, and CG2052). The deficiencies at 102B6 are only 2 kb from the deficiency break at 102B7 and only one gene is deleted (CG2052). Finally, the deficiency with a breakpoint at 102B7 [Df(4)B7-6a] is 111 kb from the breakpoints of the deficiencies at 102C1 [Df(4)C1-5a, Df(4)C1-7a, Df(4)C1-7b, Df(4)C1-7d, and Df(4)C1-10a] and there are five genes in the interval (CG2052, lgs, CamKI, bip2, and CG11533).
These deficiencies subdivide the 300-kb segment between 102A6 and 102C1 into four discrete regions plus an extra segment of ∼200 kb between 102C1 and Df(4)C3 at 102D6. Together these deficiencies greatly expand our ability to correlate mutations with computationally predicted transcripts.
Acknowledgments
The authors thank Kathy Matthews and Kevin Cook and the National Drosophila Stock Center in Bloomington, Indiana for the stocks used in this work; Dan Lindsley for his criticism and suggestions; and members of the Marsh Laboratory for helpful discussions. This work was supported by National Institutes of Health awards HD36081 and HD36049 to J.L.M.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Ahmad, K., and K. G. Golic, 1998. The transmission of fragmented chromosomes in Drosophila melanogaster. Genetics 148: 775–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall, E. L., and D. C. Rio, 1997. Drosophila P-element transposase is a novel site-specific endonuclease. Genes Dev. 116: 2137–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, C. B., 1925. Elimination of chromosomes due to a mutant (Minute-N) in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 11: 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, C. B., 1935. The mutants and linkage data of chromosome four of Drosophila melanogaster. Biol. Zh. 4: 401–420. [Google Scholar]
- Fung, J. C., W. F. Marshall, A. Dernburg, D. A. Agard and J. W. Sedat, 1998. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J. Cell Biol. 141: 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, Y. H., M. M. Tanaka and J. A. Sved, 1996. P-element-induced recombination in Drosophila melanogaster: hybrid element insertion. Genetics 144: 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman, B., 1961. On fourth chromosome lethals from a natural population of Drosophila melanogaster. Am. Nat. 95: 375–382. [Google Scholar]
- Hochman, B., 1971. Analysis of chromosome 4 in Drosophila melanogaster. II. Ethyl methanesulfonate induced lethals. Genetics 67: 235–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman, B., 1974. Analysis of a whole chromosome in Drosophila. Cold Spring Harbor Symp. Quant. Biol. 38: 581–589. [DOI] [PubMed] [Google Scholar]
- Kraus, E., W. Y. Leung and J. E. Haber, 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98: 8255–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis, R. W., 1989. Viable deletions of a telomere from a Drosophila chromosome. Cell 58: 791–801. [DOI] [PubMed] [Google Scholar]
- Locke, J., L. Podemski, N. Aippersbach, H. Kemp and R. Hodgetts, 2000. A physical map of the polytenized region (101EF–102F) of chromosome 4 in Drosophila melanogaster. Genetics 155: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova, G. V., P. J. Romanienko and R. D. Camerini-Otero, 2003. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell 5: 927–936. [DOI] [PubMed] [Google Scholar]
- Podemski, L., R. Sousa-Neves, J. L. Marsh and J. Locke, 2004. Molecular mapping of deletion breakpoints on chromosome 4 of Drosophila melanogaster. Chromosoma 112: 381–388. [DOI] [PubMed] [Google Scholar]
- Preston, C. R., J. A. Sved and W. R. Engels, 1996. Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics 144: 1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman, R. R., E. A. Johnson, C. K. Rodesch, M. Bjerke, R. N. Nagoshi et al., 1995. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Neves, R., 2000. Synthesis of a double fourth chromosome marked with y+. Dros. Inf. Serv. 83: 8–10. [Google Scholar]
- Spradling, A. C., and G. M. Rubin, 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347. [DOI] [PubMed] [Google Scholar]
- Sturtevant, A. H., 1951. A map of the fourth chromosome of Drosophila melanogaster, based on crossing over in triploid females. Proc. Natl. Acad. Sci. USA 37: 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi, H., and G. S. Roeder, 2003. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell 5: 915–925. [DOI] [PubMed] [Google Scholar]
- van Beest, M., M. Mortin and H. Clevers, 1998. Drosophila RpS3a, a novel Minute gene situated between the segment polarity genescubitus interruptus and dTCF. Nucleic Acids Res. 26: 4471–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, J., A. S. Belmont and J. W. Sedat, 2002. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12: 1473–1483. [DOI] [PubMed] [Google Scholar]