Abstract
Temperature compensation is a defining feature of circadian oscillators, yet no components contributing to the phenomenon have been identified in plants. We tested 27 accessions of Arabidopsis thaliana for circadian leaf movement at a range of constant temperatures. The accessions showed varying patterns of temperature compensation, but no clear associations to the geographic origin of the accessions could be made. Quantitative trait loci (QTL) were mapped for period and amplitude of leaf movement in the Columbia by Landsberg erecta (CoL) and Cape Verde Islands by Landsberg erecta (CvL) recombinant inbred lines (RILs) at 12°, 22°, and 27°. Six CvL and three CoL QTL were located for circadian period. All of the period QTL were temperature specific, suggesting that they may be involved in temperature compensation. The flowering-time gene GIGANTEA and F-box protein ZEITLUPE were identified as strong candidates for two of the QTL on the basis of mapping in near isogenic lines (NILs) and sequence comparison. The identity of these and other candidates suggests that temperature compensation is not wholly determined by the intrinsic properties of the central clock proteins in Arabidopsis, but rather by other genes that act in trans to alter the regulation of these core proteins.
MANY biological events occur rhythmically, with frequencies ranging from fractions of a second to a matter of years. Circadian rhythms occur characteristically once per day and persist with a period close to 24 hr in the absence of daily environmental cycles. They are regulated by an endogenous clock that enables the temporal coordination of physiological and biochemical processes, allowing organisms to anticipate and respond to the predictable changes in the environment during the day-night cycle. Such anticipation is shown, for example, by the upregulated expression of the photosynthetic machinery in plants prior to dawn in preparation for the light period of the day (Harmer et al. 2000).
The proposed circadian oscillator of the higher plant Arabidopsis thaliana is based on a feedback loop involving the genes TIMING OF CAB EXPRESSION 1 (TOC1), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), and LATE ELONGATED HYPOCOTYL (LHY) (Alabadi et al. 2001). Further components of the plant circadian system, such as the phytochrome and cryptochrome photoreceptors involved in light input (Somers et al. 1998a) and the F-box protein ZEITLUPE (ZTL) involved in protein turnover, have been identified (Mas et al. 2003). No genes, however, have been implicated in the phenomenon of temperature compensation.
Temperature compensation is a defining feature of circadian rhythms and results in little change in circadian period length over a broad range of physiological temperatures (Pittendrigh 1954; Johnson et al. 2003). This allows the circadian clock to provide an accurate measure of the passage of time regardless of ambient temperature. This does not, however, mean that the clock is insensitive to temperature since oscillator period length does alter, but not at the same rate as would be expected by most biochemical reactions (Lakin-Thomas et al. 1990; Ruoff et al. 1997). Temperature steps or pulses also reset the phase of the clock in Arabidopsis (Somers et al. 1998b; McWatters et al. 2000; Salome et al. 2002), as in other organisms.
Temperature compensation has been most extensively studied in the fruit fly Drosophila melanogaster and the filamentous fungus Neurospora crassa. Studies in Drosophila suggest that factors regulating the accumulation, dimerization, and nuclear transport of the core clock components PERIOD (PER) and TIMELESS (TIM) are important to the phenomenon (Price 1997; Hamblen et al. 1998; Rothenfluh et al. 2000). This raised the possibility that temperature compensation derives directly from the intrinsic properties and interactions of core clock proteins, which function across a wide temperature range. A second suggestion, supported by data from Neurospora and Drosophila (Liu et al. 1997; Majercak et al. 1999), is that altered levels or alternative isoforms of the core proteins are important at some temperatures. This leads naturally to the hypothesis that temperature compensation may require a component, or components, distinct from the core clock proteins, which are adapted to regulate circadian period at defined temperatures by altering the regulation or function of the clock proteins. Either mechanism could achieve the necessary “antagonistic balance” of period-increasing and period-decreasing reactions in the circadian oscillator (Ruoff 1992).
The temporal coordination provided by a circadian clock is believed to impart organisms with a selective advantage, particularly when the period of the oscillator closely matches that of the external environment (Ouyang et al. 1998). For a circadian oscillator to be of adaptive significance it should provide an accurate measure of time regardless of temperature (Pittendrigh 1993). This is supported by the identification of a latitudinal cline in the length of a Thr-Gly repeat domain of the Drosophila central clock protein PER (Sawyer et al. 1997). This Thr-Gly repeat domain alters the temperature-compensation response of the flies and the observed distribution of alleles in the cline appears to correlate with the change in temperature associated with latitude. Thus, the clock of the flies oscillates with a period closer to 24 hr within their respective environments.
A large number of accessions of A. thaliana have been collected from around the world, providing a wealth of natural genetic variation for study of the genetics, ecology, and evolutionary biology of the plant (reviewed in Pigliucci 1998; Alonso-Blanco and Koornneef 2000). Given the apparent adaptive significance of temperature compensation in Drosophila (Sawyer et al. 1997), natural genetic variation may also provide a means of identifying components of the phenomenon in Arabidopsis. Quantitative trait loci (QTL) analysis has arisen as a powerful means of identifying genes contributing to polygenic traits upon the basis of such natural variation. The principle of QTL analysis lies in mapping quantitative traits to defined regions of chromosomes, allowing the identification of candidate genes. Several studies have identified circadian period QTL in Arabidopsis and mice (Hofstetter et al. 1995, 2003; Mayeda et al. 1996; Swarup et al. 1999; Shimomura et al. 2001; Salathia et al. 2002; Michael et al. 2003). We now adopt a similar approach to identify loci that contribute to temperature compensation of the Arabidopsis clock. We demonstrate extensive natural variation in the temperature-compensation response of Arabidopsis plants and utilize this variation to map QTL for the trait.
MATERIALS AND METHODS
Accessions, RILs, and NILs:
Seed for the 27 A. thaliana accessions used in leaf-movement period analysis were from the same stocks as parents of developing recombinant inbred line (RIL) populations (generously donated by Carlos Alonso-Blanco, Ian Bancroft, Maarten Koornneef, and Detlef Weigel). The Cape Verde Islands by Landsberg erecta (CvL) and the Columbia by Landsberg erecta (CoL) RILs have previously been described (Lister and Dean 1993; Alonso-Blanco et al. 1998b).
Near isogenic lines (NILs) were constructed to carry Cvi alleles around putative QTL in an isogenic Ler background through genotypic selection of progeny from crosses between RILs and Ler. NIL 42 and NIL 45 were donated by M. Koornneef and have previously been described (Alonso-Blanco et al. 1998a; Swarup et al. 1999). NILs 18, 251, and 18-32 were produced from a backcross of the line S-10 (Alonso-Blanco et al. 1998a) to Ler (F3 seed provided by C. Alonso-Blanco). NILs 26-4, 19-2, and 30-2 were produced from a backcross of CvL 125 to Ler (F3 seed provided by K. Swarup).
Plant growth conditions:
Seed were surface sterilized in 70% ethanol for 2 min, immediately followed by 8 min in 50% bleach, and then were rinsed twice in sterile distilled water. Sterile seed were stored in 0.15% agar and stratified at 4° for 4–5 days prior to sowing on Murashige-Skoog 1.5% agar medium containing 3% sucrose. Seedlings were grown for 6 days under constant light of 55–60 μmol m−2 sec−1 cool white fluorescent light and then entrained for 4 days at 21°–22° under (12 hr/12 hr) light/dark cycles of 75 μmol m−2 sec−1 cool white fluorescent light.
Measurement of leaf movement:
After 6 days growth, agar blocks carrying single Arabidopsis seedlings were transferred to 25-well square tissue culture dishes. Twelve seedlings were placed in each plate, so that they could be viewed from the side when plates were placed vertically. The seedlings were then entrained as described above. At dawn on the fourth day of entrainment, plants were transferred to Sanyo MLR350 growth chambers (Sanyo, Osaka, Japan) and imaged in front of a white background over the course of a week under constant (25 μmol m−2 sec−1) cool white fluorescent light at the indicated temperatures.
Live images of seedlings were captured using 16 Ultrak KC4300E monochromatic charge coupled device video cameras (Ultrak, Preston, UK) and transferred to a computer via a Flashbus MV Pro card (Integral Technologies, Indianapolis). A custom-built parallel port controller unit (Universal Imaging, Marlow, UK) allowed the computer program Metamorph 4.5 (Universal Imaging, West Chester, PA) to select the camera channel on a VX3 camera switcher unit via its alarm circuit (Videoswitch, Church Crookham, UK). Metamorph was programmed to capture and save images from every camera at 20-min intervals over the course of a week.
The vertical position of primary leaves was measured after the experiments using Metamorph. A threshold was applied to images to identify the dark leaves against the light background. The (x, y) pixel coordinates of the central position of thresholded leaves was measured within regions set independently for each primary leaf. Data were logged into Microsoft Excel spreadsheets. A macro was custom written to plot the vertical pixel position of leaves against elapsed time (although for some traces the horizontal leaf position was used) and a window of 80 hr of data was exported for period analysis as described below. The first 24 hr of each leaf trace were excluded from the period estimation to remove any transient effects of the transfer to continuous light and, where possible, the window was started from 40 to 60 hr elapsed time for consistency. Metamorph journals to automate several steps of image capture and analysis are available at the author's website (http://www.amillar.org/downloads.html).
Genotypes were randomized among the 192 positions on the 16 leaf-movement plates to reduce the possibility of positional effects. Roughly equal numbers of each genotype were assayed per experiment.
Period data analysis:
Individual period estimates were produced from leaf-movement data by the fast Fourier transform nonlinear least-squares program (Plautz et al. 1997). Mean period estimates for each genotype were based on 10–60 leaf traces from two to four independent experiments at each temperature analyzed using REML (Patterson and Thompson 1971) in the statistical package GENSTAT 5 (Payne et al. 1993). The significance of differences between pairs of genotypes was analyzed via t-tests using the SEM estimates derived from REML. Correlations in Figure 3 are presented without multiple testing correction.
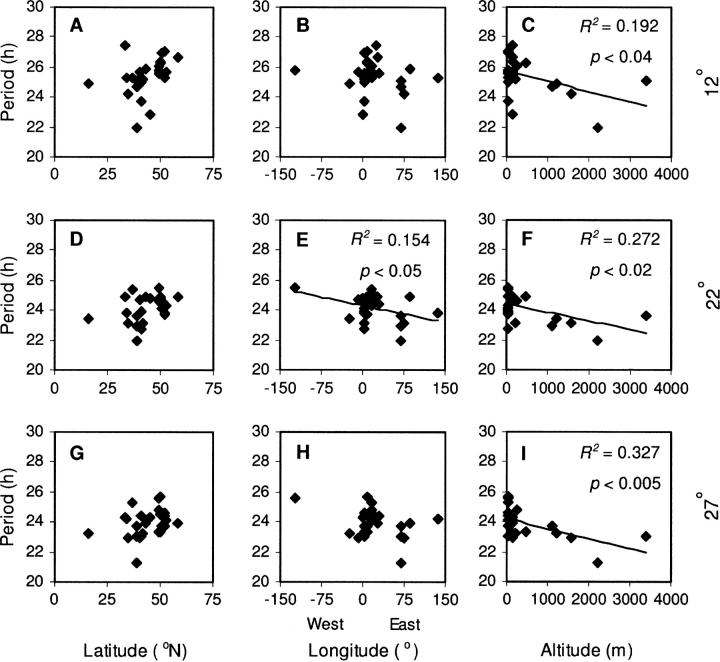
Figure 3.—
Correlations between circadian period and geographical origins of accessions. Correlation of mean circadian period to latitude (A, D, and G), longitude (B, E, and H), and altitude (C, F, and I) recorded for accession collection sites at 12° (A–C), 22° (D–F), and 27° (G–I). Correlation (R2) and probability (P) indicated for significant correlations. Data were plotted at an intermediate value for latitude, longitude, and altitude measurements recorded as ranges (see Table 1).
The variation in the circadian period of the accessions due to plate, experiment, accession, and residual variation was calculated by REML and used to estimate the proportion of total variability attributable to genetic differences among accessions at each temperature.
Q10 values were calculated for each accession from the equation Q10 = (1/τT+10)/(1/τT), where τ represents period at temperatures T and T + 10. Period values were obtained from a linear regression of accession period estimates across the three temperatures. The inverse of period was used as a shorter period corresponds with increased oscillator rate.
QTL analysis:
QTL analysis was carried out on period and leaf-movement amplitude means for the CoL and CvL RILs independently at each temperature. Genetic maps for the RILs containing markers at 5- to 15-cM intervals, as used by Swarup et al. (1999), were utilized in the QTL analysis.
Interval mapping and multiple-QTL-method (MQM) procedures of the computer program MapQTL version 4.0 (Van Ooijen and Maliepaard 1996) were used to identify putative QTL. LOD profiles displayed in Figures 5 and 6 and in Table 2 were obtained with the MQM procedure, utilizing cofactors to improve mapping accuracy. A LOD threshold of 2.7 was set for a significance level of P < 0.05 according to large-scale simulations (Van Ooijen 1999). Permutation tests in MapQTL were also used to set LOD thresholds for a significance of P < 0.05 individually for each trait on each chromosome under each temperature, and this did not alter the mapping of putative QTL.
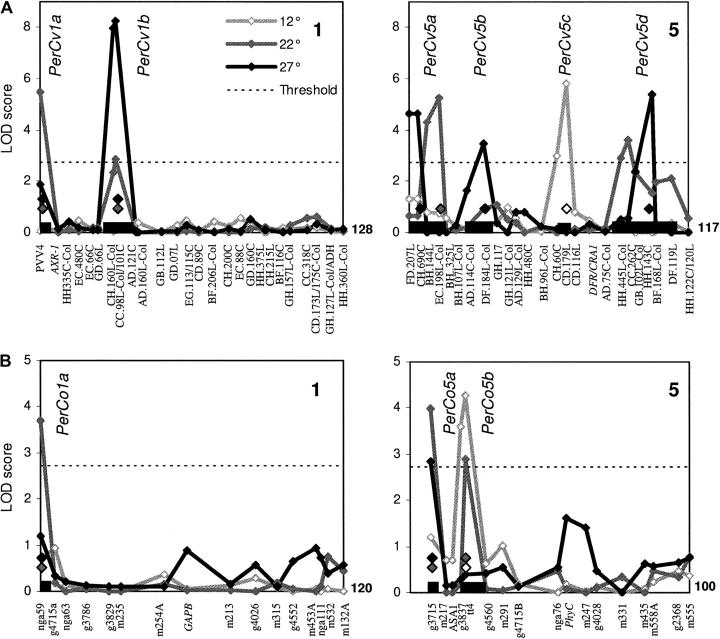
Figure 5.—
Genetic mapping of circadian period QTL in the CvL and CoL RILs. QTL likelihood (LOD) maps across Arabidopsis chromosomes 1 and 5 in the CvL (A) and CoL (B) RILs. Chromosome numbers are indicated in the upper right corner of each graph. Names on the x-axis correspond to molecular markers. QTL were mapped independently at 12° (open diamonds), 22° (shaded diamonds), and 27° (solid diamonds). Dashed line represents 2.7 LOD significance threshold of P < 0.05 (as calculated from Van Ooijen 1999). Solid boxes on x-axis span the 2-LOD support interval of mapped QTL. Putative QTL designations are indicated in italics. Selected markers used as cofactors in mapping are identified above the x-axis with diamonds shown as open (12°), shaded (22°), and solid (27°) according to temperature.
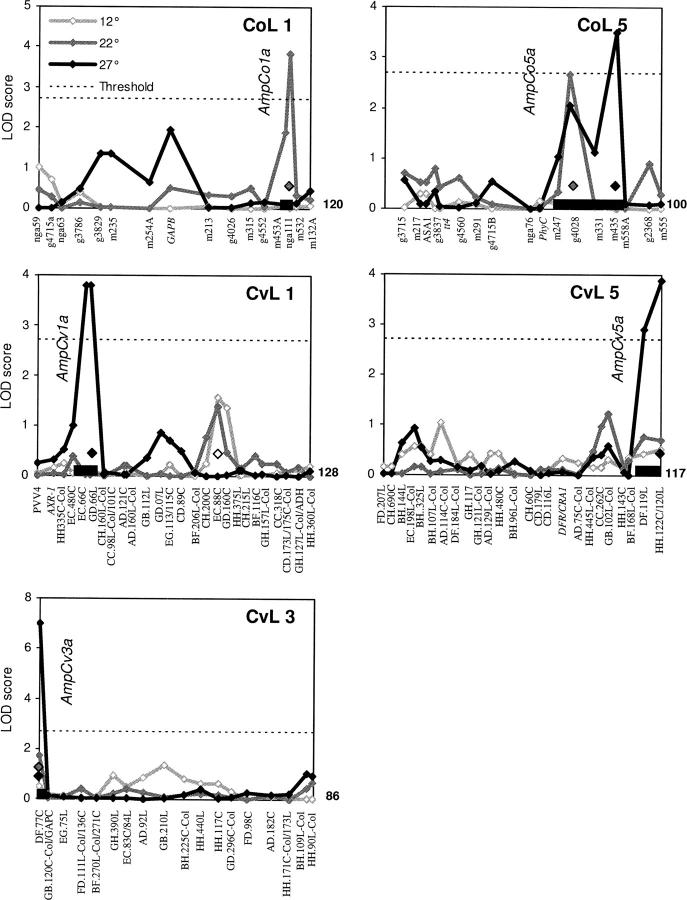
Figure 6.—
Genetic mapping of leaf-movement amplitude QTL in the CvL and CoL RILs. QTL likelihood (LOD) maps across Arabidopsis chromosomes in CvL and CoL RILs. Population and chromosome number are indicated in the upper right corner of each graph. Names on the x-axis correspond to molecular markers. QTL were mapped independently at 12° (open diamonds), 22° (shaded diamonds), and 27° (solid diamonds). Dashed line represents 2.7 LOD significance threshold of P < 0.05 (Van Ooijen 1999). Solid boxes on the x-axis span the 2-LOD support interval of mapped QTL. Putative QTL designations are indicated in italics. Selected markers used as cofactors in mapping are identified above the x-axis with diamonds shown as open (12°), shaded (22°), and solid (27°) to according to temperature.
TABLE 2.
QTL Summary
| 12°
|
22°
|
27°
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| QTL | Chromosome | LOD | Var | Effect | LOD | Var | Effect | LOD | Var | Effect |
| PerCv1a | 1 | — | — | — | 4.31 | 13.7 | −0.81 | — | — | — |
| PerCv1b | 1 | — | — | — | 3.69 | 11.4 | −0.73 | 8.23 | 22.3 | −1.32 |
| PerCv5a | 5 | — | — | — | 4.66 | 15.1 | 0.86 | 4.65 | 10.5 | 0.98 |
| PerCv5b | 5 | — | — | — | — | — | — | 3.48 | 7.4 | 0.75 |
| PerCv5c | 5 | 7.5 | 44.6 | −1.52 | — | — | — | — | — | — |
| PerCv5d | 5 | — | — | — | 3.09 | 7 | 0.59 | 5.41 | 12.7 | 0.99 |
| PerCo1a | 1 | — | — | — | 3.71 | 16.7 | −0.39 | — | — | — |
| PerCo5a | 5 | — | — | — | 3.99 | 18.4 | 0.49 | 3.38 | 38.4 | 0.8 |
| PerCo5b | 5 | 4.28 | 53.2 | 1.79 | 2.9 | 12 | 0.43 | — | — | — |
| AmpCv1a | 1 | — | — | — | — | — | — | 3.8 | 14.9 | −0.81 |
| AmpCv3a | 3 | — | — | — | — | — | — | 6.99 | 32.1 | 1.2 |
| AmpCv5a | 5 | — | — | — | — | — | — | 3.88 | 15.3 | 0.82 |
| AmpCo1a | 1 | — | — | — | 3.83 | 37 | −0.9 | — | — | — |
| AmpCo5a | 5 | — | — | — | — | — | — | 3.5 | 43.7 | 2.31 |
Name, chromosome, and effect of mapped QTL at 12°, 22°, and 27°. “Var” refers to the percentage of phenotypic variance explained by QTL under the particular environment. “Effect” shows the double additive effect of QTL in hours for period (Per) and in pixels for amplitude (Amp) QTL. Direction of phenotypic effect expressed in terms of the Cvi or the Col allele vs. the Ler allele of the QTL.
Genotyping:
Genotyping NILs was carried out using PCR-based markers described in The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org). Where required, new PCR-based markers were designed from publicly available single nucleotide polymorphism and INDEL data (Jander et al. 2002). Primer sequences for these markers have been submitted to TAIR.
Amplification, cloning, and sequencing of GIGANTEA:
Amplification was done by Takara ExTaq polymerase mixture in 25 μl total volume containing 2 mm MgCl2, 0.2 μm of each primer (F, 5′-cgc gga tcc ttc ttc tga att gtt gtt aca ggg ttt agc-3′; R, 5′-ggg gta ccg tta gcc aat cgc ctt cca ata ccc ttg at-3′), 50 ng of genomic DNA, and 1 unit of ExTaq polymerase. The PCR product was cloned into pBluescript SK and sequenced with T7 and gene-specific sequencing primers spaced every 400 bp. Products were amplified and sequenced from three independent plants to overcome PCR errors and the three sequences were aligned by the program ClustalW to produce a consensus sequence. Individual consensus sequences were fused with the program “blast 2 sequences” (Tatusova and Madden 1999) to reconstruct the genomic fragment.
RESULTS
Natural variation in temperature compensation:
To reveal the extent of natural variation in temperature compensation of the Arabidopsis circadian clock, we tested the effect of temperature on circadian period in plants collected from different locations around the world. Table 1 summarizes the location of collection for 27 Arabidopsis accessions from North America, Africa, Europe, and Asia. The lines were limited in range, not extending to further northern latitudes and being concentrated around Europe; however, they were selected primarily to inform future QTL mapping, because all are parents of existing or developing recombinant inbred populations.
TABLE 1.
Summary of geographic origin and circadian periods of accessions
| Collection site
|
Circadian period (hr)
|
||||||
|---|---|---|---|---|---|---|---|
| Name | Latitude (°) | Longitude (°) | Altitude (m) | 12° | 22° | 27° | Q10 |
| Ag-0 | N45 | E1–E2 | 100–200 | 22.84 (0.64) | 24.82 (0.22) | 24.31 (0.43) | 0.95 |
| An-1 | N51–N52 | E4–E5 | 1–100 | 26.98 (0.49) | 24.14 (0.26) | 23.69 (0.35) | 1.09 |
| Br-0 | N49 | E16–E17 | 200–300 | 26.08 (0.49) | 24.61 (0.27) | 24.77 (0.4) | 1.04 |
| Col-0 | N50 | E8 | 1–100 | 25.6 (0.52) | 24.94 (0.27) | 25.66 (0.46) | 1.00 |
| Ct-1 | N37–N38 | E15 | 1–100 | 25.25 (0.47) | 25.43 (0.23) | 25.31 (0.28) | 1.00 |
| Cvi-0 | N15–N17 | W23–W25 | 1200 | 24.91 (0.42) | 23.47 (0.22) | 23.21 (0.35) | 1.05 |
| Est | N58–N59 | E23–E28 | 100–200 | 26.67 (0.43) | 24.93 (0.21) | 23.95 (0.36) | 1.07 |
| Fei-0 | N40 | W8 | 100–300 | 25.72 (0.48) | 24.66 (0.21) | 22.92 (0.33) | 1.07 |
| Ga-0 | N50–N51 | E8 | 100–200 | 26.36 (0.56) | 24.8 (0.51) | 24.52 (0.45) | 1.05 |
| Gy-0 | N49 | E2 | 100 | 25.62 (0.62) | 24.6 (0.28) | 23.38 (0.39) | 1.06 |
| Kas-1 | N34–N36 | E74–E80 | 1580 | 24.2 (0.65) | 23.18 (0.21) | 22.91 (0.35) | 1.04 |
| Kin-0 | N43 | W85 | — | 25.88 (0.5) | 24.94 (0.24) | 23.94 (0.33) | 1.05 |
| Kondara | N39 | E70 | 1100 | 24.69 (0.43) | 22.92 (0.21) | 23.68 (0.39) | 1.03 |
| Ler | N53 | E15–E16 | 1–100 | 25.64 (0.5) | 24.29 (0.28) | 24.14 (0.36) | 1.04 |
| Ll-0 | N42 | E3 | 100–300 | 25.19 (0.51) | 23.11 (0.37) | 23.23 (0.36) | 1.06 |
| Mr-0 | N44–N45 | E9–E10 | 1000–1500 | 24.67 (0.62) | 22.03 (0.68) | — | 1.12a |
| Mt-0 | N33 | E23 | 100–200 | 27.46 (0.48) | 24.93 (0.28) | 24.33 (0.41) | 1.08 |
| Mz-0 | N50–N51 | E8–E9 | 4–500 | 26.31 (0.49) | 24.94 (0.24) | 23.35 (0.5) | 1.08 |
| Nok-3 | N52–N53 | E4 | 0–100 | 25.3 (0.4) | 23.8 (0.26) | 24.57 (0.41) | 1.03 |
| Sha | N39 | E70 | 3400 | 25.05 (0.47) | 23.64 (0.2) | 23.06 (0.3) | 1.06 |
| Sorbo | N39 | E70 | 2200 | 21.97 (0.49) | 21.97 (0.36) | 21.31 (0.44) | 1.02 |
| Ts-1 | N41–N42 | E3 | 1–100 | 23.77 (0.43) | 22.7 (0.27) | 23 (0.44) | 1.03 |
| Ts-5 | N41–N42 | E3 | 1–100 | 25.04 (0.44) | 23.88 (0.22) | 24.44 (0.43) | 1.02 |
| Tsu-1 | N34 | E136 | — | 25.27 (0.51) | 23.79 (0.25) | 24.22 (0.4) | 1.03 |
| Van-0 | N49–N50 | W123 | 1–100 | 25.82 (0.42) | 25.51 (0.22) | 25.54 (0.33) | 1.01 |
| Ws | N52 | E30 | — | 25.54 (0.38) | 24.44 (0.22) | 24.38 (0.33) | 1.03 |
| Wt-5 | N52–N53 | E9–E10 | 1–100 | 27.01 (0.86) | 23.68 (0.2) | 23.71 (0.34) | 1.09 |
Longitude, latitude, and altitude (where known) of the collection location for each of the 27 accessions. Leaf-movement period estimates are shown for 12°, 22°, and 27°, with standard error values shown in parentheses.
No leaf-movement period could be measured for this accession at 27°, so the Q10 value is based on 12° and 22° period estimates.
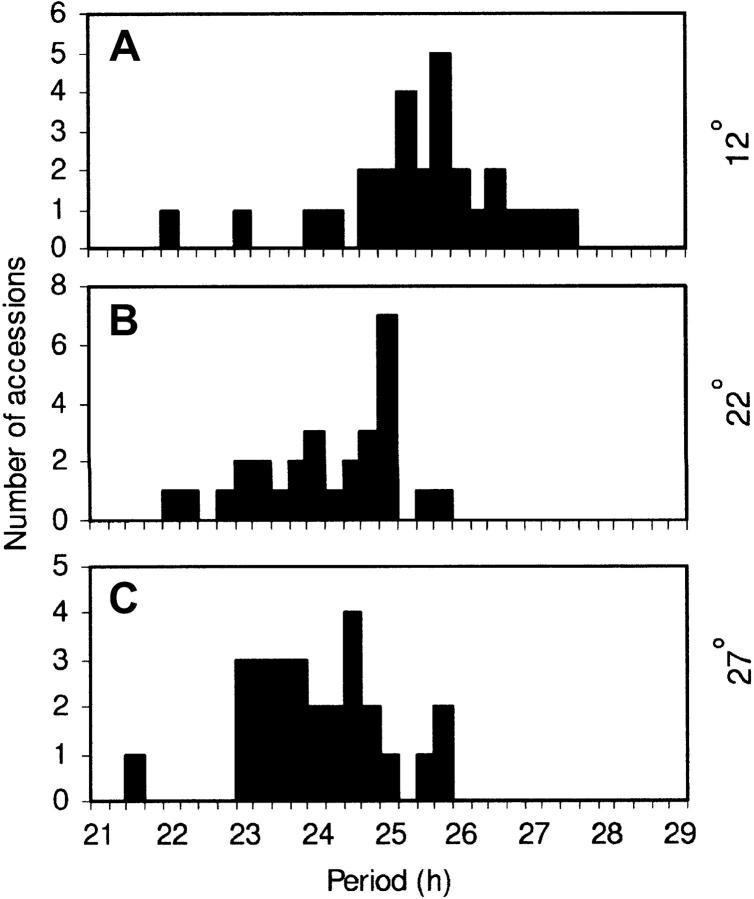
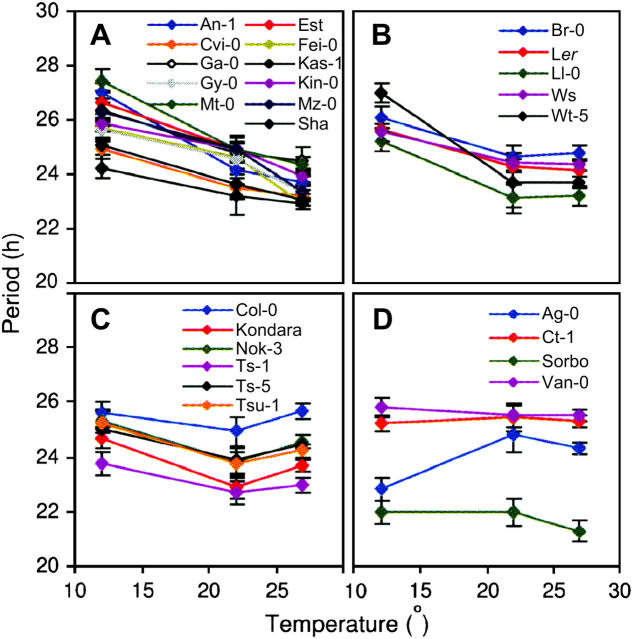
Accessions were assayed for circadian leaf-movement period at the constant temperatures of 12°, 22°, and 27°. Period estimates were returned for all accessions at each temperature, except Mr-0, which could not be successfully assayed for rhythmic leaf movement at 27°. On average, 24, 39, and 21% of the variance in plant circadian period was attributable to genetic differences among the accessions at 12°, 22°, and 27° respectively. Figure 1 shows a frequency histogram of the accession periods, which revealed a general trend of period shortening as temperature increased. The change in period shown by individual accessions across the three temperatures was relatively small, with temperature quotients (Q10) ranging from 0.95 to 1.12 (Table 1). Such values were consistent with a previously reported example for Arabidopsis (Somers et al. 1998b).
Figure 1.—
Distribution of accession periods. Frequency histograms of accession mean leaf-movement periods at 12° (A), 22° (B), and 27° (C). Periods were binned into 15-min intervals, labeled with the upper period bound of the interval.
The amplitude and direction of period change among temperatures varied greatly among accessions. Figure 2 shows the accession periods plotted against temperature and separated into panels by response. Accessions in Figure 2, A–C, all showed a decrease in period associated with an increase in temperature from 12° to 22° (Figure 2). In these lines, circadian period then continued to shorten (Figure 2A), leveled off (Figure 2B), or lengthened slightly as the temperature increased from 22° to 27° (Figure 2C). Accessions in Figure 2D showed little period change across the three temperatures, with the exception of Ag-0, which showed a short period at 12° (Figure 2D). Separating accessions on the basis of their response failed to reveal any correlations with factors of their collection sites; however, it did serve to display the extent of natural variation in the temperature-compensation response of the 27 accessions tested.
Figure 2.—
Natural variation in temperature compensation. Mean period of accessions plotted against temperature and separated by temperature-compensation response into accessions that show period shortening as temperature increases between 12° and 27° (A); accessions that show period shortening between 12° and 22°, followed by little change between 22° and 27° (B); accessions that show period shortening between 12° and 22° and period lengthening between 22° and 27° (C); and accessions that show little change in period between 12° and 27° (D). See insets for accession identities.
Figure 3 shows the circadian period of the 27 accessions plotted against the latitude, longitude, and altitude of their collection site. The effects of these factors on period were assessed by linear regression. Significant correlations were observed between period and longitude at 22° and between period and altitude at all temperatures (Figure 3, C, E, F, and I). Given the apparent effect of altitude, the effects of latitude and longitude on period were also assessed with altitude being included in the analysis as a covariate. Neither latitude nor longitude had a significant effect when the effect of altitude was thus taken into account (data not shown). Analyzing a reduced set of accessions, limited to those originating from Europe and North Africa (18 of the 27 lines), led to the loss of all significant correlations (data not shown). Similarly, the change in period between 12° and 27° revealed no significant correlation to latitude, longitude, or altitude (data not shown).
The variability in response of circadian period to temperature shown by the 27 accessions suggests that considerable natural allelic variation exists for the trait in Arabidopsis. This natural variation is clearly sufficient to justify multiple studies of temperature compensation in various accessions.
Phenotypic analysis of parental lines:
Previous studies have demonstrated the existence of natural genetic variation among the circadian systems of the Ler, Col, and Cvi accessions at a standard growth temperature (Swarup et al. 1999; Michael et al. 2003). We used the RIL populations derived from Cvi crossed with Ler (CvL) (Alonso-Blanco et al. 1998b) and Col crossed with Ler (CoL) (Lister and Dean 1993) to map circadian period QTL at different temperatures. We did this because period-altering loci displaying temperature-dependent effects could be considered likely candidates for temperature-compensation components.
Figure 4A shows the period of the Ler, Cvi, and Col accessions at a range of constant temperatures between 12° and 30°. All three accessions showed period shortening between 12° and 22° and lengthening between 22° and 30°, with Col having the longest period at all temperatures (Figure 4A). Between 18° and 27° the relationship between the accession periods remained relatively constant. Due to this and prior success in mapping circadian QTL under these conditions (Swarup et al. 1999; Michael et al. 2003), it was decided to use 22° as the intermediate assay temperature for the study. This also allowed for direct comparison of our results with the previous studies. The phenotypic difference between accessions and the unexpected period shortening in Ler and Cvi at 12° (Figure 4A) led to this temperature being selected as the low assay temperature for QTL mapping.
Figure 4.—
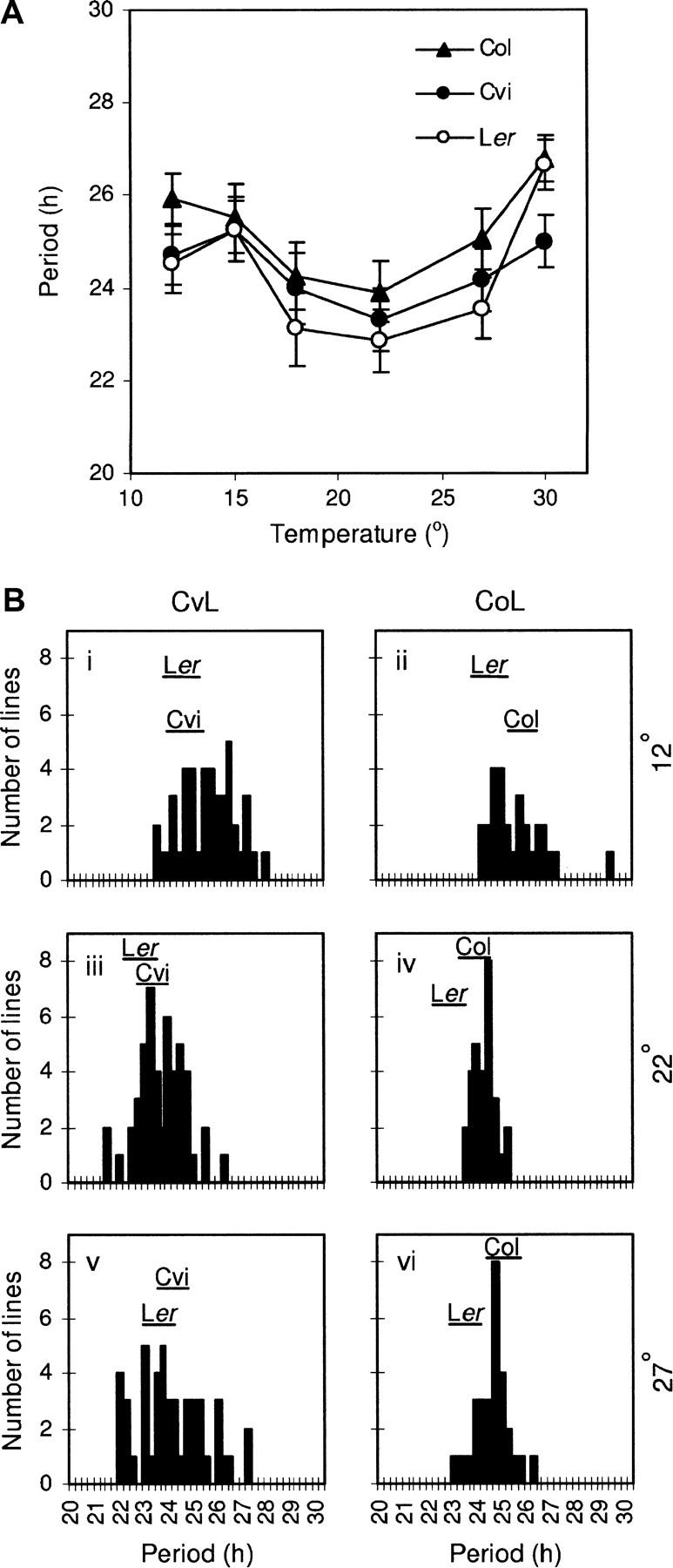
Temperature compensation in Ler, Col, Cvi, and the CvL and CoL RILs. (A) Mean leaf-movement period plotted against temperature for the Ler (open circles), Col (solid triangles), and Cvi (solid circles) accessions. Error bars show SEM for period estimates. (B) Frequency histograms of the CvL (i, iii, and v) and CoL (ii, iv, and vi) RIL mean periods at 12° (i and ii), 22° (iii and iv), and 27° (v and vi). Data are binned into 15-min intervals, labeled with the upper period bound of the interval. Horizontal lines below accession names indicate the SEM interval of parental accession periods.
Cvi's period was less affected by the higher temperatures than were the other two accessions (Figure 4A). This may be related to the adaptation of the plant to the warmer climate of the Cape Verde Islands. Although the greatest phenotypic variation was observed between the accessions at 30°, the extreme period lengthening at this temperature suggested that this might be an effect of stress, unrelated to temperature compensation. Thus, to reduce the possibility of mapping stress-related QTL, 27° was selected as the higher assay temperature.
Subsets of 30 of the CoL and 48 of the CvL RILs were assayed for leaf-movement period at 12°, 22°, and 27°. Figure 4B shows frequency histograms of the RIL periods at each temperature. Both populations showed a broad and continuous distribution of period in each environment, consistent with a polygenic trait (Figure 4B).
The CvL RILs showed transgressive variation in period in both directions from the parents at all temperatures, suggesting that despite their similar phenotypes, both parents contained multiple period-lengthening and period-shortening loci (Figure 4B, i, iii, and v). In the CoL population, however, Ler's period was consistently at the shorter end of the period range (Figure 4B, ii, iv, and vi), suggesting that Col alleles tended to have period-lengthening effects.
QTL mapping:
Circadian period QTL were mapped in the CoL and CvL populations from the RIL leaf-movement data independently at each temperature. Amplitude of leaf-movement QTL was also mapped in both populations. Due to technical reasons we could not accurately determine phase with the leaf-movement assay.
QTL were named on the basis of the trait (“Per” for period and “Amp” for amplitude), mapping population (“Cv” for CvL or “Co” for CoL), chromosome number, and an arbitrary letter to differentiate among linked loci. Figures 5 and 6 show likelihood maps of chromosomes containing putative QTL for circadian period and leaf-movement amplitude, respectively. A summary of QTL for both populations and traits can be seen in Table 2. Estimated phenotypic effects are expressed in terms of the period or amplitude phenotype associated with the Cvi or Col allele relative to the Ler allele of the locus.
Period QTL in the CvL and CoL populations:
Six putative circadian period QTL were mapped in the CvL RIL population: two onto chromosome 1 and four onto chromosome 5 (Figure 5A). All of the QTL showed some degree of temperature specificity, mapping significantly at one or two of the temperatures but never at all three (Figure 5A and Table 2).
Two QTL, PerCv1a and PerCv1b, mapped to the top of chromosome 1. In each case, Cvi alleles caused period shortening. PerCv1b was detected at 22° and 27°, whereas PerCv1a was detected at 22° and was just below the significance threshold at 27° (Figure 5A).
PerCv5a and PerCv5b mapped to the top of chromosome 5. PerCv5a showed slightly offset LOD peaks at 22° and 27°, but similar estimated allelic effects suggested that they represented the same locus (Figure 5A and Table 2). PerCv5b was mapped only at 27° (Figure 5A). PerCv5c, in the middle of chromosome 5, was the only CvL period QTL located at 12° (Figure 5A); however, it accounted for 44.6% of genetically determined variation at this temperature. The final QTL, PerCv5d, mapped to the bottom of chromosome 5 and caused period lengthening at 22° and at 27° (Figure 5A and Table 2).
The six putative CvL period QTL explained 44.6%, 47.2%, and 40.2% of the genetically determined variability in the CvL RILs at 12°, 22°, and 27°, respectively. PerCv1b, PerCv5a, and PerCv5d colocalized with the previously mapped CvL period QTL ESPRESSO (ESP), ANDANTE (AND), and RALENTANDO (RAL), respectively, estimating similar phenotypic effects (Swarup et al. 1999). The remaining three loci, PerCv1a, PerCv5b, and PerCv5c, represented novel circadian period QTL. PerCv1a, however, did colocalize with the flowering-time and hypocotyl-length QTL EARLY DAY-LENGTH INSENSITIVE (EDI) and DARK 1, as did PerCv1b with the hypocotyl-length QTL LIGHT 1 (Alonso-Blanco et al. 1998a; Borevitz et al. 2002), suggesting that the QTL for these different traits may be allelic.
Only three period QTL were mapped in the CoL RILs (Figure 5B and Table 2). As with the CvL population, all QTL were located on chromosomes 1 and 5 (Figure 5). PerCo1a mapped to the top of chromosome 1 at 22°, representing a novel CoL QTL, but colocalization and similar estimated effects to PerCv1a suggested that the QTL may be allelic (Figure 5 and Table 2).
No CoL QTL were found at the TAU1A or TAU1B locus (Michael et al. 2003), although TAU1A may colocalize with PerCv1b. PerCo5a, however, did map to the same locus as TAU5A did (Michael et al. 2003). Colocalization was also observed between PerCo5a and PerCv5a (Figure 5). The CvL and CoL QTL AND and ANOTHER ANDANTE (AAN) had colocalized in this region previously (Swarup et al. 1999). All of the QTL estimated period lengthening and similar temperature dependencies between PerCo5a and PerCv5a suggested that a common gene might cause them.
As with the CvL RILs, only a single 12° period QTL was mapped in the CoL lines. PerCo5b represented a novel CoL QTL, but colocalized with PerCv5b (Figure 5). Both QTL estimated period-lengthening effects, but with opposite temperature dependencies, indicating that they represented different polymorphisms, different loci, or possible epistatic effects. The three CoL period QTL explained a total of 53.2, 47.1, and 38.4% of the genetically determined variation at 12°, 22°, and 27°, respectively, similar to estimates from the CvL population (Table 2).
QTL were also mapped from both populations for the change in period between the extreme temperatures. This analysis revealed one additional suggestive QTL on chromosome 2 (see supplementary data 1 at http://www.genetics.org/supplemental/), but reduced the LOD scores of existing QTL. This additional QTL, PerCv2a, colocalized with the previously mapped NON TROPPO (NOT) QTL (Swarup et al. 1999). However, NOT was at the limit of detection in our previous report and PerCv2a was only just at the significance threshold, with <1 hr difference in period change between alleles. Therefore we chose to follow up the more robust, single-temperature QTL.
Amplitude QTL in the CvL and CoL populations:
Three CvL and two CoL QTL were mapped for leaf-movement amplitude. Figure 6 shows likelihood maps for the trait in linkage groups 1, 3, and 5 of CvL and 1 and 5 of CoL. AmpCv1a, AmpCv3a, and AmpCv5a were all mapped at 27° and accounted for 62.3% of the genetically determined variability at this temperature (Figure 6 and Table 2). No significant amplitude QTL were mapped in the CvL population at 12° or 22°. Similarly, AmpCo5a was specific to 27°; however, AmpCo1a was mapped at 22° (Figure 6). The greater success in mapping amplitude QTL at 27° may be explained by morphological changes in the RILs brought about by temperatures above 27°, which has been shown to produce such phenotypes by mimicking the effects of HEAT SHOCK PROTEIN 90 (HSP90) inhibition (Queitsch et al. 2002). Given the suggested role of HSP90 as a capacitor of natural variation (Queitsch et al. 2002), 27° might provide a useful tool for Arabidopsis quantitative geneticists, allowing the QTL mapping of previously masked polymorphisms.
Failure to map an amplitude QTL to the erecta locus was surprising, since this mutation, carried by Ler, previously has been shown to reduce the amplitude of leaf-movement rhythms (Swarup et al. 1999; Michael et al. 2003). This trait, however, depends on growth rate and may have been affected by subtle differences in growth conditions between the studies.
Characterization of QTL:
PerCv1a and PerCv1b:
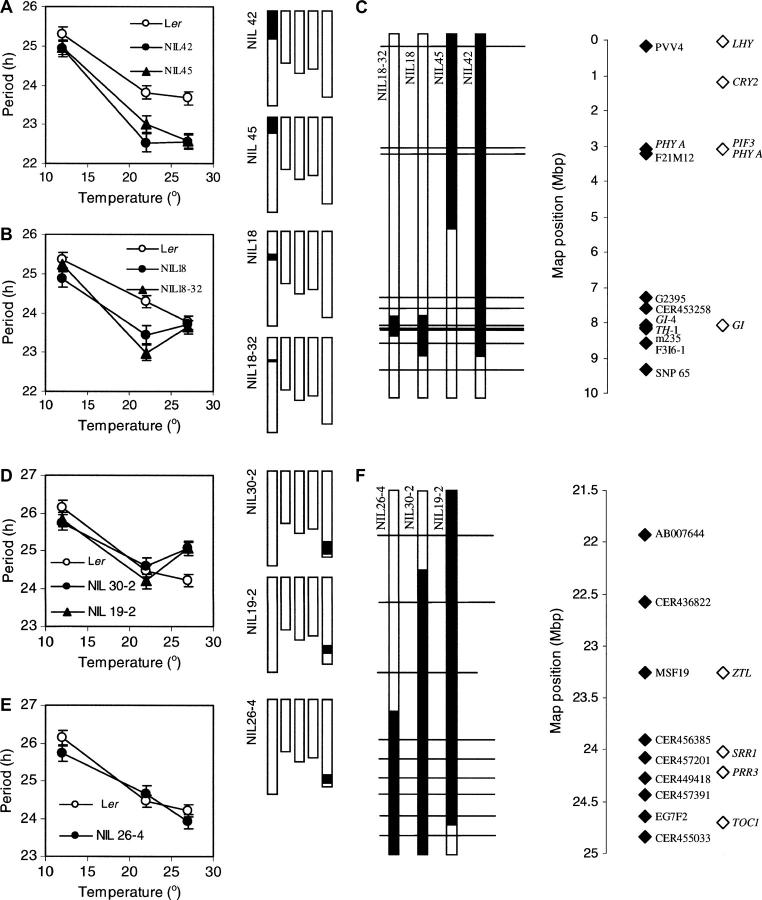
The location and effect of several of the putative CvL period QTL were confirmed using NILs. NILs were constructed by genotypic selection to contain small Cvi chromosomal regions around the putative QTL in isogenic Ler backgrounds. The circadian periods of these lines are shown in Figure 7 and Table 3. Figure 7 also shows maps summarizing the genotype of each NIL around the QTL loci.
Figure 7.—
PerCv1a, PerCv1b, and PerCv5d NILs. Mean leaf-movement period of Ler is compared to NILs for PerCv1a and PerCv1b (A and B) and for PerCv5d (D and E) at 12°, 22°, and 27°. See insets for line identification. Error bars represent SEM of period estimates. NIL genotypes across the five Arabidopsis chromosomes are shown to the right of each graph. Open regions represent the Ler genotype and solid regions represent the Cvi genotype. Detailed view of NIL genotypes around PerCv1a and PerCv1b (C) and PerCv5d (F) regions shows the position of mapping markers (solid diamonds) and candidate genes (open diamonds). Horizontal lines correspond to mapping markers and solid (Cvi) and open (Ler) regions of vertical bars represent the genotype of NILs at the markers. Breakpoints are estimated as the midpoint between mapping markers.
TABLE 3.
NIL periods
| 12°
|
22°
|
27°
|
||||
|---|---|---|---|---|---|---|
| Line | Period (hr) | SEM | Period (hr) | SEM | Period (hr) | SEM |
| Ler | 25.3 | 0.19 | 23.81 | 0.16 | 23.67 | 0.16 |
| NIL42 | 24.93 | 0.21 | 22.53*** | 0.22 | 22.56*** | 0.19 |
| NIL45 | 24.97 | 0.19 | 23.01***,a | 0.21 | 22.57*** | 0.17 |
| Ler | 25.35 | 0.25 | 24.29 | 0.27 | 23.76 | 0.37 |
| NIL18 | 24.87 | 0.34 | 23.45** | 0.25 | 23.72 | 0.26 |
| NIL18-32 | 25.23 | 0.39 | 22.99*** | 0.31 | 23.64 | 0.26 |
| Ler | 26.15 | 0.4 | 24.47 | 0.26 | 24.21 | 0.32 |
| NIL30-2 | 25.74 | 0.27 | 24.58 | 0.26 | 25.05** | 0.28 |
| NIL19-2 | 25.84 | 0.28 | 24.2 | 0.27 | 25.05* | 0.29 |
| NIL26-4 | 25.72 | 0.37 | 24.64 | 0.26 | 23.91 | 0.31 |
Asterisks indicate P-values of Student's t-test of NIL periods vs. Ler at *P < 0.05, **P < 0.02, and ***P < 0.01.
P-value < 0.1 of Student's t-test of NIL 45 vs. NIL 42.
NIL 42 and NIL 45 were used to investigate PerCv1a and PerCv1b. NIL 42 contained a Cvi introgression of ∼9 Mb at the top of chromosome 1 and NIL 45 had a similar, but slightly smaller introgression of ∼5 Mb (Figure 7C). Both NILs showed significant period shortening compared to Ler at 22° and 27° (Figure 7A), similar to the estimated effects of PerCv1a and PerCv1b at these temperatures (Table 2). This indicated that either one or both QTL mapped within the Cvi introgression of NILs 42 and 45.
The period of NIL 45 was slightly, but not significantly longer than that of NIL 42 at 22° (Figure 7A and Table 3). A previous study, however, did show a significant difference between the NILs at this temperature (Swarup et al. 1999). NILs 18 and 18-32 were produced from a backcross of NIL 42 to Ler and were selected to contain only the lower region of this NIL's introgression (Figure 7C). Both NIL 18 and NIL 18-32 showed period shortening specific to 22°, confirming that NIL 42 did contain a QTL effect independent of NIL 45 (Figure 7B and Table 3). A third NIL, 251, covering most of the unique region of NIL 42, showed the same period phenotype as NILs 18 and 18-32 did (data not shown), suggesting that a single locus responsible for this QTL effect was maintained within the <900-kb Cvi introgression of NIL 18-32.
The flowering-time gene GIGANTEA (GI) was the strongest candidate mapping within NIL 18-32 (Figure 7C). Mutant alleles of gi show circadian period effects (Park et al. 1999), and the gene was proposed as a candidate for the leaf-movement period QTL ESP (Swarup et al. 1999). PerCv1b colocalizes with ESP, so to test the possibility of GI causing the QTL, we sequenced the Cvi allele of the gene (GenBank accession AY685131) and aligned it with the Ler (GenBank accession AY682088), Col (GenBank accession AJ133786), and Wassilewskija (WS) (GenBank accession AY685132) alleles. On the basis of the At1g22770.1 gene model, five nucleotide substitutions were identified between the Ler and Cvi coding sequences (see supplementary data 2 at http://www.genetics.org/supplemental/). Translation of the coding sequences showed that only two resulted in amino acid substitutions: an isoleucine for a valine substitution at amino acid 113 in Ler and a leucine for a phenylalanine substitution at amino acid 718 in Cvi. In both cases, the substituted residues shared properties similar to the ones that they replaced.
Amino acid sequences for Ler and Cvi GI protein were aligned against the Col (GenBank accession AAF00092) and WS alleles along with homologous GI protein sequences from rice, barley and wheat (GenBank accessions NP_914460, AAL08497, and AAQ11738, respectively). Both amino acid substitutions were at conserved residues in GI protein from the other Arabidopsis accessions and also rice and wheat GI (see supplementary data 2 at http://www.genetics.org/supplemental/). Leucine718 was also conserved in barley GI (see supplementary data 2 at http://www.genetics.org/supplemental/). Conservation of these amino acids is consistent with either being of functional importance to the protein; however, little is known about the domain structure of GI, so it is difficult to comment on the possible functional consequences of the substitutions.
The tight mapping of GI within NIL 18-32, together with known period effects in gi mutants (Park et al. 1999) and amino acid substitutions between the Ler and Cvi alleles of the gene, offered support for it, causing the 22° effect of PerCv1b. However, neither NIL 18 nor NIL 18-32 displayed a period phenotype at 27°, indicating that allelic variation at GI alone did not cause the QTL at this temperature.
PerCv5d:
NILs 26-4, 19-2, and 30-2 were selected to contain Cvi alleles around the PerCv5d locus on the bottom of chromosome 5, which contains many circadian-associated genes (Figures 7, D–F). Both NIL 30-2 and NIL 19-2 showed 0.8-hr period-lengthening effects specific to 27° (Figure 7D and Table 3), similar to the 1.0-hr lengthening effect estimated for the QTL at this temperature (Table 2). Neither NIL, however, showed significant period lengthening at 22°, indicating that the 22° PerCv5d effect was either overestimated or mediated by a locus outside of the NIL introgressions.
The strongest candidate genes located within both NIL 19-2's and NIL 30-2's Cvi introgressions were ZTL, SENSITIVITY TO RED LIGHT REDUCED 1 (SRR1), PSEUDO RESPONSE REGULATOR 3 (PRR3), and possibly TOC1 (Figure 7F). NIL 26-4 had a similar, but smaller, Cvi introgression to NIL 30-2 and contained the same candidate genes as the other NILs with the exception of ZTL (Figure 7F). Leaf-movement analysis of NIL 26-4 showed no significant period effect at any temperature (Figure 7E and Table 3), indicating that unlike NILs 30-2 and 19-2, NIL 26-4 did not contain the 27° QTL effect. This ruled out SRR1, PRR3, and TOC1 as candidates and offered support for ZTL being the cause of the QTL.
Previous sequence analysis identified a predicted amino acid polymorphism in the Cvi allele of ZTL (Somers et al. 2000), whereas the Ler allele shows no coding sequence polymorphisms compared to Col, C24, or WS (P. Gyula and F. Nagy, unpublished results; D. Somers and S. Kay, personal communication). The lack of a colocalizing QTL at the PerCv5d locus in the CoL population suggests that the Col and Ler alleles of the QTL are functionally comparable. Thus, the polymorphism in ZTLCvi offers support for this gene causing the QTL. Furthermore, various ztl mutants show increased period lengthening at higher temperatures (L. Kozma-Bognar, personal communication), consistent with the suggested effect of the polymorphic Cvi allele of ZTL from the NIL leaf-movement analysis.
DISCUSSION
Natural variation in temperature compensation:
All of the accessions that we tested showed temperature compensation of their circadian period, with Q10 values similar to those previously reported for Arabidopsis (Somers et al. 1998b), Drosophila (Pittendrigh 1954), and Neurospora (Lakin-Thomas et al. 1990). Sufficient variation in this response was observed to facilitate further dissection of the Arabidopsis temperature-compensation mechanisms.
No association could be made between an accession temperature-compensation response and any factors of its geographic origin. Accession period did not show any correlation with latitude, possibly due to the small sample size tested. However, it was negatively correlated with altitude at all temperatures, with the strongest tendency to shorter period in accessions from higher altitudes at 27°. This suggests that some factor related to altitude has placed a selective pressure on the Arabidopsis circadian clock, but there is insufficient information on the accession collection sites to identify this factor. Several accessions (Kondara, Sorbo, and Sha) originated from the Himalayan region at high altitude and Far Eastern longitude. This region has shown a cluster of genetic relatedness (Breyne et al. 1999), so it is possible that the period-altitude correlations and weak period-longitude correlation were partially based on this. Indeed, analysis of a reduced set of accessions, excluding these lines, led to the loss of all significant correlations.
Greater phenotypic difference was observed between Ler and Col than between Ler and Cvi (Figure 4A); however, the transgression of period in the RILs (Figure 4B) and the number of QTL mapped (Table 2) suggested that greater genetic variation existed between the Ler and Cvi parents. This is supported by the genetic relatedness of the accessions and by the previous observations of transgressive variation for flowering time in the two RIL populations (Alonso-Blanco et al. 1998a; Breyne et al. 1999). Where possible, there is clear benefit to using the available information to select more distantly related parents for QTL studies, even if they display similar phenotypes.
Identity of period QTL:
Strong candidate genes can be offered for most of the circadian period QTL mapped in this study. GI and ZTL were supported by analysis of NILs and both have polymorphisms between their Ler and Cvi alleles. The lack of a period QTL at the PerCv1b/GI locus in the CoL population suggests that the Ler and Col alleles of the QTL are functionally comparable. Thus, the substitution of leucine718 for a phenylalanine in Cvi seems the stronger candidate. However, the CoL period QTL TAU1A (Michael et al. 2003) colocalizes with PerCv1b and so the amino acid substitution in the Ler allele of GI cannot be ruled out as a possible cause of the QTL. Confirmation of either change causing PerCv1b would require substitution of the amino acid polymorphisms between the two alleles, as carried out for CRYPTOCHROME 2 (CRY2) for the flowering-time QTL EDI (El-Assal et al. 2001).
It seems likely that GI is responsible for the 22° period effect of PerCv1b and that the QTL is analogous to the previously identified period QTL ESP (Swarup et al. 1999). Hypocotyl-length phenotypes in gi mutants (Fowler et al. 1999; Huq et al. 2000) also suggest the possibility of GI causing the hypocotyl-length QTL LIGHT1 (Borevitz et al. 2002). It is clear, however, that GI alone is not responsible for the 27° effect estimated for PerCv1b and that further 22° and 27° QTL effects lie within the Cvi introgression of NIL 45. With PerCv1a mapping in this region also, it is likely that NIL 45 contains more than one period-altering locus. LHY, PHYTOCHROME A (PHYA), and PHYTOCHROME INTERACTING FACTOR 3 (PIF3) all map to this region and thus provide candidate genes for these QTL effects.
The CoL and CvL period QTL, PerCv5a and PerCo5a, colocalized at the top of chromosome 5, as observed by Swarup et al. (1999) for AND and AAN at the same locus. FLOWERING LOCUS C (FLC) was proposed as a candidate for AND, given circadian period effects observed in flc mutants and known allelic differences between the accessions (Swarup et al. 1999). FLC maps between PerCv5a and PerCv5b and so could be a candidate for either QTL. PRR7 offers an alternative candidate for PerCo5a/PerCv5a and was proposed as one for the colocalizing CoL period QTL TAU5A (Michael et al. 2003).
Temperature-compensation mechanisms in Arabidopsis:
Candidate genes for the QTL that we mapped in this study suggest that, in Arabidopsis, temperature compensation is not conferred solely by properties of the core protein components of the clock. Not all QTL mapped to such core components and NIL 26-4 showed that TOC1 did not cause a QTL effect. Of the strongest candidates identified, ZTL may directly alter the levels of the core clock component TOC1 (Mas et al. 2003) and, although the biochemical function of GI is unknown, it does affect the expression of CCA1 and LHY (Fowler et al. 1999; Park et al. 1999). Our results are consistent with a mechanism for temperature compensation that alters the abundance of clock proteins, mediated by several genes acting in trans. However, the data also indicate that different loci are important to period determination between 12° and 27°. This is consistent with the notion of “antagonistic balance” (Ruoff and Rensing 1996; Ruoff et al. 1997), because the balance is a local phenomenon at a particular temperature. Our results suggest that temperature compensation in Arabidopsis is imparted not by one balance acting at all temperatures, but by multiple balances acting at different temperatures.
Acknowledgments
We thank Carlos Alonso-Blanco for the provision of seed stocks and for helpful discussions. We are also very grateful to Maarten Koornneef, Detlef Weigel, Ian Bancroft, and Kamal Swarup for provision of seed stocks and crosses; to Jo Putterill, David Somers, and Steve Kay for sequence information; and to Nigel Murphy (Universal Imaging, Marlow, UK) for technical assistance. K.D.E. was funded by a Biotechnology and Biological Sciences Research Council postgraduate studentship. Sequencing work carried out in Hungary was supported by Hungarian Scientific Research Fund T-046710 and by Howard Hughes Medical Institute International Scholarship grants to F.N.
References
- Alabadi, D., T. Oyama, M. J. Yanovsky, F. G. Harmon, P. Mas et al., 2001. Reciprocal regulation between TOC1 and LHY/CCA1. Science 293: 880–883. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco, C., and M. Koornneef, 2000. Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci. 5: 22–29. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco, C., S. E. D. ElAssal, G. Coupland and M. Koornneef, 1998. a Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco, C., A. J. M. Peeters, M. Koornneef, C. Lister, C. Dean et al., 1998. b Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14: 259–271. [DOI] [PubMed] [Google Scholar]
- Borevitz, J. O., J. N. Maloof, J. Lutes, T. Dabi, J. L. Redfern et al., 2002. Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics 160: 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne, P., D. Rombaut, A. Van Gysel, M. Van Montagu and T. Gerats, 1999. AFLP analysis of genetic diversity within and between Arabidopsis thaliana ecotypes. Mol. Gen. Genet. 261: 627–634. [DOI] [PubMed] [Google Scholar]
- El-Assal, S. E., C. Alonso-Blanco, A. J. M. Peeters, V. Raz and M. Koornneef, 2001. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 29: 435–440. [DOI] [PubMed] [Google Scholar]
- Fowler, S., K. Lee, H. Onouchi, A. Samach, K. Richardson et al., 1999. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18: 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblen, M. J., N. E. White, P. T. J. Emery, K. Kaiser and J. C. Hall, 1998. Molecular and behavioral analysis of four period mutants in Drosophila melanogaster encompassing extreme short, novel long, and unorthodox arrhythmic types. Genetics 149: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S. L., J. B. Hogenesch, M. Straume, H. S. Chang, B. Han et al., 2000. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113. [DOI] [PubMed] [Google Scholar]
- Hofstetter, J. R., A. R. Mayeda, B. Possidente and J. I. Nurnberger, 1995. Quantitative trait loci (QTL) for circadian rhythms of locomotor activity in mice. Behav. Genet. 25: 545–556. [DOI] [PubMed] [Google Scholar]
- Hofstetter, J. R., J. A. Trofatter, K. L. Kernek, J. I. Nurnberger and A. R. Mayeda, 2003. New quantitative trait loci for the genetic variance in circadian period of locomoter activity between inbred strains of mice. J. Biol. Rhythms 18: 450–462. [DOI] [PubMed] [Google Scholar]
- Huq, E., J. M. Tepperman and P. H. Quail, 2000. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 97: 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander, G., S. R. Norris, S. D. Rounsley, D. F. Bush, I. M. Levin et al., 2002. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C. H., J. Elliott, R. Foster, K.-I. Honma and R. Kronauer, 2003 Fundamental properties of circadian rhythms, pp. 67–105 in Chronobiology: Biological Timekeeping, edited by J. Dunlap, J. J. Loros and P. J. DeCoursey. Sinauer Associates, Sunderland, MA.
- Lakin-Thomas, P. L., G. G. Cot and S. Brody, 1990. Circadian rhythms in Neurospora crassa: biochemistry and genetics. CRC Crit. Rev. Microbiol. 17: 365–416. [DOI] [PubMed] [Google Scholar]
- Lister, C., and C. Dean, 1993. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4: 745–750. [DOI] [PubMed] [Google Scholar]
- Liu, Y., N. Y. Garceau, J. J. Loros and J. C. Dunlap, 1997. Thermally regulated translational control of FRQ mediates aspects of temperature responses in the Neurospora circadian clock. Cell 89: 477–486. [DOI] [PubMed] [Google Scholar]
- Majercak, J., D. Sidote, P. E. Hardin and I. Edery, 1999. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24: 219–230. [DOI] [PubMed] [Google Scholar]
- Mas, P., W. Kim, D. Somers and S. Kay, 2003. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570. [DOI] [PubMed] [Google Scholar]
- Mayeda, A. R., J. R. Hofstetter, J. K. Belknap and J. I. Nurnberger, 1996. Hypothetical quantitative trait loci (QTL) for circadian period of locomotor activity in CxB recombinant inbred strains of mice. Behav. Genet. 26: 505–511. [DOI] [PubMed] [Google Scholar]
- McWatters, H. G., R. M. Bastow, A. Hall and A. J. Millar, 2000. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720. [DOI] [PubMed] [Google Scholar]
- Michael, T. P., P. A. Salome, H. J. Yu, T. R. Spencer, E. L. Sharp et al., 2003. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053. [DOI] [PubMed] [Google Scholar]
- Ouyang, Y., C. R. Andersson, T. Kondo, S. S. Golden and C. H. Johnson, 1998. Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. USA 95: 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D. H., D. E. Somers, Y. S. Kim, Y. H. Choy, H. K. Lim et al., 1999. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582. [DOI] [PubMed] [Google Scholar]
- Patterson, H. D., and R. Thompson, 1971. Recovery of inter-block information when block sizes are unequal. Biometrika 58: 545–554. [Google Scholar]
- Payne, R. W., P. Lane, P. Digby, S. Harding, P. Leech et al., 1993 Genstat 5 Release 3 Reference Manual. Oxford University Press, Oxford.
- Pigliucci, M., 1998. Ecological and evolutionary genetics of Arabidopsis. Trends Plant Sci. 3: 485–489. [Google Scholar]
- Pittendrigh, C. S., 1954. On temperature independence in the clock system controlling emergence time in Drosophila. Proc. Natl. Acad. Sci. USA 40: 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh, C. S., 1993. Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55: 17–54. [DOI] [PubMed] [Google Scholar]
- Plautz, J. D., M. Straume, R. Stanewsky, C. F. Jamison, C. Brandes et al., 1997. Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms 12: 204–217. [DOI] [PubMed] [Google Scholar]
- Price, J. L., 1997. Insights into the molecular mechanisms of temperature compensation from the Drosophila PERIOD and TIMELESS mutants. Chronobiol. Int. 14: 455–468. [DOI] [PubMed] [Google Scholar]
- Queitsch, C., T. A. Sangster and S. Lindquist, 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624. [DOI] [PubMed] [Google Scholar]
- Rothenfluh, A., M. Abodeely, J. L. Price and M. W. Young, 2000. Isolation and analysis of six timeless alleles that cause short- or long-period circadian rhythms in Drosophila. Genetics 156: 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoff, P., 1992. Introducing temperature-compensation in any reaction kinetic oscillator model. J. Interdiscipl. Cycle Res. 23: 92–99. [Google Scholar]
- Ruoff, P., and L. Rensing, 1996. The temperature-compensated Goodwin model simulates many circadian clock properties. J. Theor. Biol. 179: 275–285. [Google Scholar]
- Ruoff, P., L. Rensing, R. Kommedal and S. Mohsenzadeh, 1997. Modeling temperature compensation in chemical and biological oscillators. Chronobiol. Int. 14: 499–510. [DOI] [PubMed] [Google Scholar]
- Salathia, N., K. Edwards and A. J. Millar, 2002. QTL for timing: a natural diversity of clock genes. Trends Genet. 18: 115–118. [DOI] [PubMed] [Google Scholar]
- Salome, P. A., T. P. Michael, E. V. Kearns, A. G. Fett-Neto, R. A. Sharrock et al., 2002. The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol. 129: 1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, L. A., J. M. Hennessy, A. A. Peixoto, E. Rosato, H. Parkinson et al., 1997. Natural variation in a Drosophila clock gene and temperature compensation. Science 278: 2117–2120. [DOI] [PubMed] [Google Scholar]
- Shimomura, K., S. S. Low-Zeddies, D. P. King, T. D. L. Steeves, A. Whiteley et al., 2001. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 11: 959–980. [DOI] [PubMed] [Google Scholar]
- Somers, D. E., P. F. Devlin and S. A. Kay, 1998. a Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490. [DOI] [PubMed] [Google Scholar]
- Somers, D. E., A. A. R. Webb, M. Pearson and S. A. Kay, 1998. b The short-period mutant, toc1–1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125: 485–494. [DOI] [PubMed] [Google Scholar]
- Somers, D. E., T. F. Schultz, M. Milnamow and S. A. Kay, 2000. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329. [DOI] [PubMed] [Google Scholar]
- Swarup, K., C. Alonso-Blanco, J. R. Lynn, S. D. Michaels, R. M. Amasino et al., 1999. Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 20: 67–77. [DOI] [PubMed] [Google Scholar]
- Tatusova, T. A., and T. L. Madden, 1999. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174: 247–250. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J. W., 1999. LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83: 613–624. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J. W., and C. Maliepaard, 1996 MapQTL Version 3.0: Software for the Calculation of QTL Positions on Genetic Maps. CPRO-DLO, Wageningen, The Netherlands.