Abstract
Multiple sclerosis (MS) and its animal model, myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis (MOG-EAE), share a complex genetic predisposition with contributions from the major histocompatibility complex class II genes and many other genes. Linkage mapping in F2 crosses between the susceptible DA rat strain and the resistant ACI or BN rat strains in various models of autoimmune neuroinflammation have repeatedly displayed suggestive linkage to a region on rat chromosome 15. A direct study of this region was undertaken in congenic strains by transferring resistant ACI alleles to the susceptible DA background. Phenotypic analysis demonstrated lower maximal and cumulative EAE scores in the DA.ACI–D15Rat6-D15Rat71 (C15), DA.ACI–D15Rat6-D15Rat48, D15Rat126-D15Rat71 (C15R3b), and DA.ACI–D15Rat23-D15rat71 (C15R4) strains compared to the parental DA rat strain. Linkage analysis was then performed in a (DA × PVG.AV1)F7 advanced intercross line, resulting in a LOD score of 4.7 for the maximal EAE score phenotype at the peak marker D15Rat71 and a confidence interval of 13 Mb, overlapping with the congenic fragment defined by the C15R3b and the C15R4 strains. Thus, a new MOG-EAE locus with the designation Eae19 is identified on rat chromosome 15. There are 32 confirmed or predicted genes in the confidence interval, including immune-responsive gene 1 and neuronal ceroid lipofuscinose gene 5. Definition of loci such as Eae19 enables the characterization of genetically regulated, evolutionary conserved disease pathways in complex neuroinflammatory diseases.
MULTIPLE sclerosis (MS) is a chronic inflammatory demyelinating disease that affects the central nervous system. Susceptibility to MS is based on interactions among several genes and influences by, largely unknown, nongenetic factors (Ebers et al. 1986, 1995; Sadovnick et al. 1993, 1996; Ebers 1996). The major histocompatibility complex (MHC) has been known to regulate MS since 1972 (Jersild et al. 1973; Ebers et al. 1986; Olerup and Hillert 1991). So far, very few individual non-MHC genes regulating MS have been identified by whole-genome scans or association studies due to the heterogeneity, polygeneity, and environmental influences in MS (Ebers et al. 1996; Haines et al. 1996, 2002; Sawcer et al. 1996; Kuokkanen et al. 1997; Chataway et al. 1998; Broadley 2001; Coraddu 2001; Akesson et al. 2002; Ban et al. 2002). Animal models of MS, such as experimental autoimmune encephalomyelitis (EAE), can circumvent these problems by minimizing the heterogeneity and controlling the environmental conditions (Lucchinetti et al. 1996; Lassmann et al. 2001). Unbiased identification of genes controlling autoimmune neuroinflammation is important, since such genes represent evolutionarily conserved disease pathways that are prime candidates for therapeutic interventions. A major problem for other approaches in MS, such as studying selected candidate regulatory molecules and cellular subsets, is to determine if the observed deviation is a cause or consequence of disease and if the pathway is involved in disease progression or protection.
EAE induced with myelin oligodendrocyte glycoprotein (MOG) in certain rat strains shares features of MS such as a relapsing/remitting disease course and a prominent demyelination (Adelmann et al. 1995; Johns et al. 1995). The formation of demyelinated lesions in MOG-EAE depends on both T cells and anti-MOG antibodies (Linington et al. 1988). MHC class II genes and multiple other genes influence this response (Weissert et al. 1998b; Dahlman et al. 1999b; Jagodic et al. 2001). An autoimmune response against MOG in MS patients suggests that MOG plays an important role also in the pathogenesis of MS (Sun et al. 1991; De Rosbo et al. 1993; Wallstrom et al. 1998). Thus, MOG-EAE is a relevant model to utilize in studies of mechanisms underlying the development of autoimmune neuroinflammation. The dark Agouti (DA) rat strain is susceptible to MOG-EAE, while the PVG.AV1 and the ACI strains are relatively resistant (Weissert et al. 1998b). The DA, PVG.AV1, and ACI rat strains all share the MHC haplotype RT1.AV1 (Hedrich 1990). This allows the establishment of intercrosses and congenic strains specifically aimed at identifying non-MHC loci regulating MOG-EAE.
Previous studies of MOG-EAE, whole-spinal-cord-induced EAE, and experimental autoimmune neuritis (EAN) have found a suggestive linkage (Lander and Kruglyak 1995) to a region on rat chromosome 15. In MOG-EAE and EAN, the suggestive linkage was observed in (DA × ACI)F2 rats subjected to genome scans with microsatellite markers (Dahlman et al. 1999b, 2001). In spinal-cord-induced EAE, a suggestive linkage was observed in (DA × BN)F2 rats (Dahlman et al. 1999a).
To determine if the rat chromosome 15 region indeed is important for the development of MOG-EAE, we transferred this region from the EAE-resistant ACI to the susceptible DA background with a speed congenic approach (Wakeland et al. 1997). Linkage mapping was then performed in a (DA × PVG.AV1)F7 advanced intercross line (AIL) (Darvasi and Soller 1995, 1997; Jagodic et al. 2004). An AIL is created by random intercross breeding of two inbred strains for several generations, resulting in genetically unique individuals with a mixture of founder chromosomal fragments. Theoretically, an AIL gives at least a t/2-fold reduction in the confidence interval compared to an F2 cross, given that t, where t is the number of generations, is large enough (Darvasi and Soller 1997; Xiong and Guo 1997). We combine the congenic strain and the AIL analysis to define a new MOG-EAE locus designated Eae19.
MATERIALS AND METHODS
Parental rat strains and basic conditions:
DA rats were originally obtained from the Zentralinstitut for Versuchstierzucht (Hans Hedrich, Hannover, Germany) and A × C 9935 Irish (ACI) rats were from Harlan Sprague Dawley (Indianapolis). MHC-congenic (RT1.AV1) Piebald-Viral-Glaxo (PVG) rats, PVG.AV1 (also previously referred to as PVG-RT1a), were obtained from Harlan UK (Blackthorn, UK). All the rats were locally bred in the animal facility at the Center for Molecular Medicine, Karolinska Institutet. Eight- to 15-week-old male and female rats were used in the six experiments with congenic rats. Rats were routinely tested for specific pathogens according to a health-monitoring program for rats at the National Veterinary Institute in Uppsala, Sweden. They were kept in a 12-hr light/12-hr dark cycle and housed in polystyrene cages containing aspen wood shavings, with free access to water and autoclaved standard rodent chow. The local ethical committee approved the experiments.
Breeding of the chromosome 15 congenic strains and the advanced intercross line:
Speed congenics were generated with a marker-assisted selection technique, mainly as described by Wakeland et al. (1997). An ∼25-cM fragment of ACI alleles from the D15Rat6 marker to the D15Rat71 marker was transferred to the DA rat background. Initially, (DA × ACI)F1 rats were backcrossed to DA rats. From the N2 generation, the rats were genotyped with 70 microsatellite markers outside the congenic region, with a mean distance between markers of 20 cM. One male rat from each generation, having the least amount of remaining donor (ACI) alleles in the genome, was selected for further breeding and mated with several DA female rats. In the N6 generation, all 70 background markers were fixed as DA homozygous. One further backcross was performed and heterozygous rats for the chromosome 15 region were subsequently intercrossed to produce the congenic strain DA.ACI–D15Rat6-D15Rat71 (N7F1). From the first intercross, offspring rats were genotyped with eight microsatellite markers within the congenic region to detect intraregional recombinations. We selected the full-length congenic strain DA.ACI–D15Rat6-D15Rat71 (C15) and the recombinant congenic strains DA.ACI–D15Rat6-D15rat13 (C15R1), and DA.ACI–D15Rat6-D15Rat48 (C15R3), and DA.ACI–D15Rat23-D15rat71 (C15R4) for experiments. After the fifth experiment we regenotyped the rats and found that some C15R3 rats shared a region with the C15R4 from D15Rat126 to D15Rat71, so we separated the C15R3 into DA.ACI–D15Rat6-D15Rat48 (C15R3a) and DA.ACI–D15Rat6-D15rat48, D15Rat126-D15Rat71 (C15R3b) according to the genotyping results.
The advanced intercross line originated from the DA and the PVG.AV1 rat strains that share the RT1.AV1 MHC haplotype, thus allowing identification of non-MHC genes. One important reason for choosing the DA × PVG strain combination was to permit dense genotyping, since these strains display a high rate of polymorphic microsatellite markers: ∼60% (compared to ∼10% for the DA × ACI strain combination) according to the Whitehead Institute (http://www-genome.wi.mit.edu/rat/public/). The breeding scheme for the (DA × PVG.AV1) AIL has previously been reported (Jagodic et al. 2004). Briefly, to create the F1 generation, breeding pairs with DA female founders and PVG.AV1 female founders were established. The F2 generation was produced from seven couples each of F1 rats with DA and PVG.AV1 as female founders, respectively. The F3 generation originated from 50 breeding couples with both types of female founders. Random breeding of 50 males and females, consistently avoiding brother-sister mating, produced the subsequent generations. Three F7 litters were produced from the 50 F6 breeding couples for the MOG-EAE experiments.
Induction and clinical assessment of MOG-EAE:
The rats were anesthetized with sevoflurane and immunized intradermally in the tail base. Each rat received 200-μl inoculums containing 100 μl recombinant rat MOG (rMOG; aa 1–125) in saline emulsified in 100 μl incomplete Freund's adjuvans (Sigma-Aldrich, St. Louis). The dose of rMOG (aa 1–125) was selected upon titration in the susceptible parental DA rats. In the congenic strain experiments, the dose was 13, 20, or 65 μg/rat, depending on the batch of rMOG, and 40 μg/rat for the AIL animals. Animals were weighed and clinical signs of disease were evaluated from day 7 to day 33–40 postimmunization (p.i.). The clinical signs were scored as follows: 1, tail weakness or tail paralysis; 2, hind leg paraparesis (gait disturbance) or hemiparesis; 3, hind leg paraparalysis or hemiparalysis; 4, tetraparalysis, urinary, and/or fecal incontinence. A relapsing/remitting disease course was defined as an improvement in the disease score either from 3 or 4 to 1 or from 2, 3, or 4 to 0, which was maintained for at least 2 consecutive days and followed by an increase in the clinical score of at least 2 points that lasted for at least 2 days.
Genotyping:
A total of 152 clinically affected rats and 162 randomly selected unaffected rats in the (DA × PVG.AV1)F7 AIL were genotyped. Affected animals were selected on the basis of displaying unambiguous signs of the disease (minimum score 1 for >2 days accompanied with weight loss). Rats in the unaffected group did not display any signs of disease, including a steady increase in weight (Jagodic et al. 2004). DNA was extracted from the tail tip according to a standard protocol (Laird et al. 1991). The region analyzed in the AIL included the region defined in the full-length congenic C15 strain (Figure 1). This 25-cM (∼53 Mb) large region, extending from D15Rat6 to D15Rat71, was first genotyped with 15 microsatellite markers, and then another region from D15Rat71 to D15Rat103 (∼15 Mb) near the telomere was mapped with four additional microsatellite markers (Figure 4). The microsatellite markers were obtained from Proligo France SAS (Paris). Polymerase chain reaction (PCR) amplification was performed as previously described with [γ-33P]ATP end labeling of the forward primer (Jacob et al. 1995). The PCR products were size fractionated on 6% polyacrylamide gels and visualized by autoradiography. All genotypes were evaluated manually and double checked.
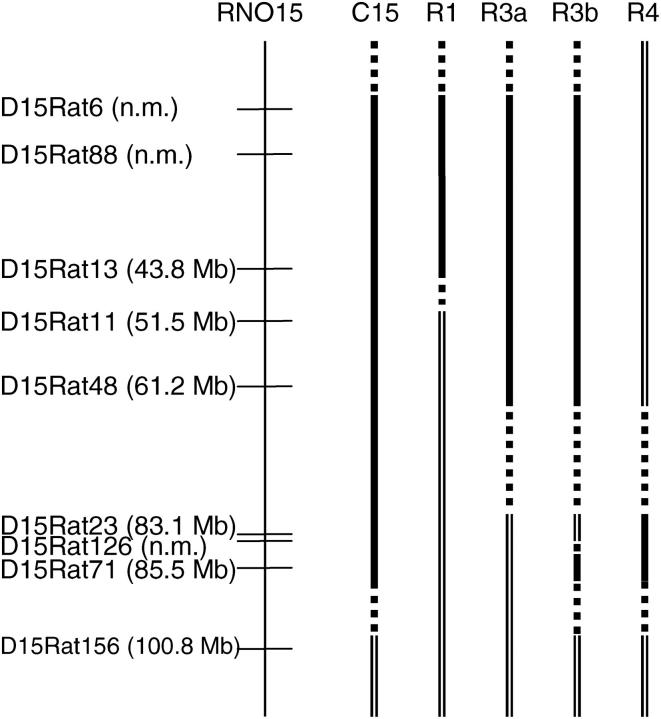
Figure 1.—
A schematic of the distal part of rat chromosome 15, aligned with the intervals defined in the congenic strains. The full-length congenic strain DA.ACI–D15Rat6-D15Rat71 (C15) and the recombinant congenic strains DA.ACI–D15rat 6-D15rat13 (C15R1), DA.ACI–D15Rat6-D15Rat48 (C15R3a), DA.ACI–D15Rat6-D15rat48, D15Rat126-D15Rat71 (C15R3b), and DA.ACI–D15Rat23-D15rat71 (C15R4) are depicted. The thin vertical line represents rat chromosome 15 along with microsatellite markers placed according to positions in megabases derived from the rat genome sequence (http://www.nsembl.org/Rattus_norvegicus/). Markers not mapped to assembly in the current Ensembl database are marked “n.m.” and positioned according to the SHRSP × BN version 7 linkage map (http://rgd.mcw.edu/). The thick black vertical lines represent different ACI rat intervals transferred to the DA rat background and the dashed lines represent the interval within which recombination has occurred. The open vertical line represents DA rat background genes.
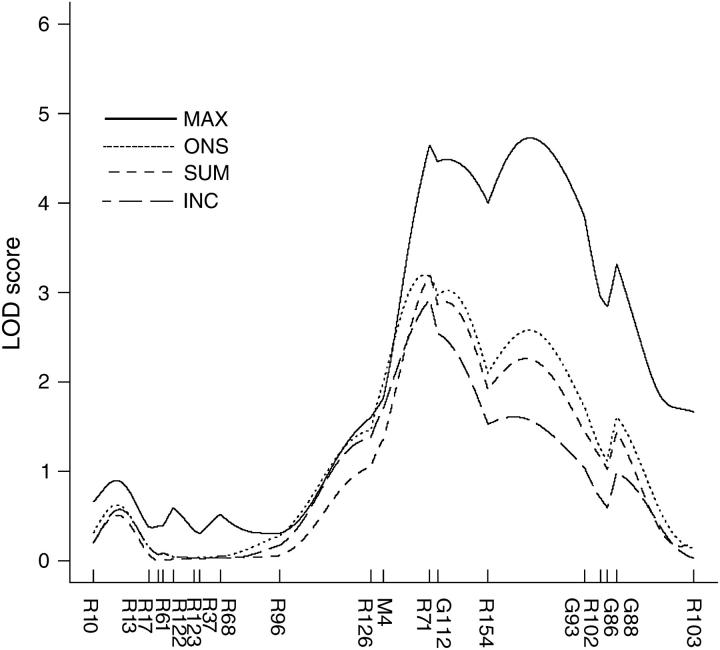
Figure 4.—
Log-likelihood plot of Eae19, identified in the (DA × PVG.AV1)F7 AIL. Eae19 displayed significant linkage to all clinical EAE phenotypes: EAE incidence and day of onset and maximum and cumulative EAE scores. The markers in the region are listed on the x-axis (R, D15Rat; M, D15Mgh; G, D15Got. Eae19 is 13 Mb and contains 32 confirmed or predicted genes according to the rat physical map retrieved from http://www.ensembl.org
Statistical analysis:
Differences in binominal traits (incidence, relapsing/remitting disease, mortality) were tested with the Fisher's exact test. Differences in the maximal score, the cumulative scores, and onset day were tested with the Wilcoxon two-sample test after normalization of the six separate experiments. Normalization was performed by subtracting the mean maximal or cumulative EAE score for the particular experiment from each individual rat's corresponding score and then the sum was divided with the standard deviation for the particular experiment. This allowed all experiments to be analyzed together, despite the variation in the severity of disease in the parental DA rat strain. The JMP 5.1 software (SAS Institute, Cary, NC) was utilized for the analysis above. Linkage analysis in the AIL was performed with the R/qtl software (Broman et al. 2003). Permutation tests in the R/qtl software were used to determine the significance levels (Churchill and Doerge 1994). The LOD levels for significant linkage generated with 5000 permutations were 2.3 for the incidence of EAE, 2.0 for the day of onset, 2.9 for the maximum EAE score, and 2.2 for the cumulative EAE score. The confidence interval was defined as a drop of 1 in the LOD score (Lander and Botstein 1989).
RESULTS
A reduced MOG-EAE severity in the C15, C15R3b, and C15R4 strains:
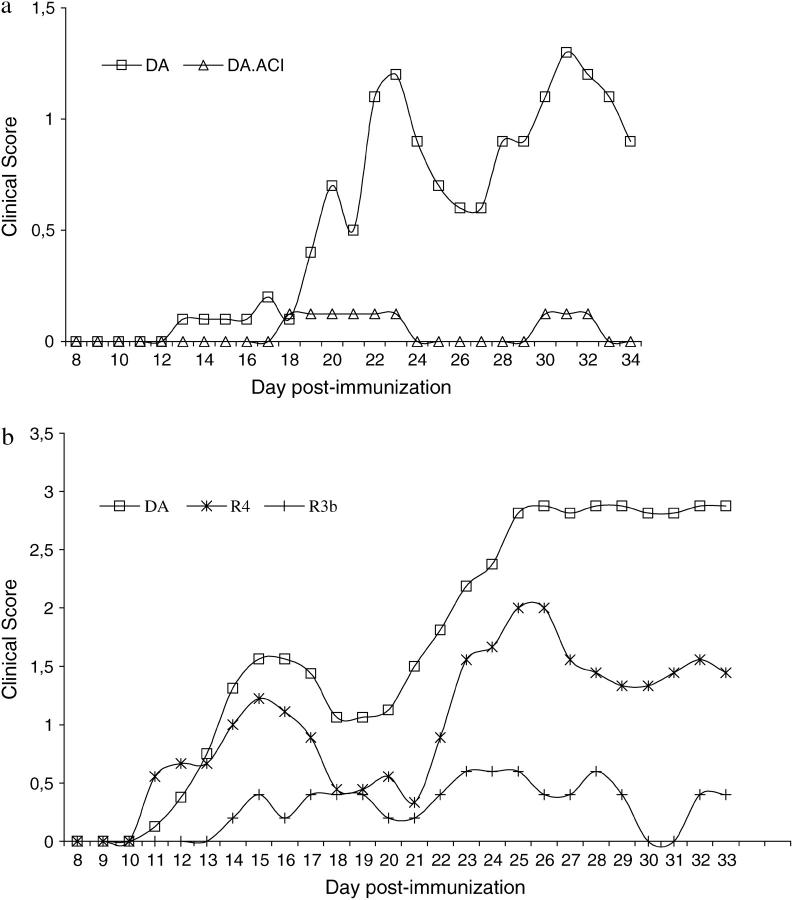
Figure 2 gives the mean maximal cumulative score and onset day of the EAE in DA rats and in congenic C15 and recombinant congenic C15R1, C15R3a, C15R3b, and C15R4 rats pooled from six separate experiments after the normalization. DA, C15R1, and C15R3a rats developed EAE with a high maximal and cumulative EAE score while the C15, C15R3b, and C15R4 rats had less severe MOG-EAE, with lower maximal and cumulative EAE scores (P < 0.05–0.001); C15R3b rats had late onset of disease compared to DA rats (P < 0.05). The disease incidence, the numbers of rats displaying a relapsing/remitting disease, or a lethal EAE were not significantly different in any of the congenic strains compared to the DA strain (data not shown). However, the power of this analysis was reduced due to the variable expression of EAE in the DA strain, as depicted in Figure 3. In experiment 3 (Figure 3a), there were only mild signs of disease in the DA rats and almost no disease signs in the C15 rats. The disease signs in the DA rats were much more severe in experiment 5, while the R3b and R4 displayed a reduced disease severity (Figure 3b). The overall disease severity was intermediate in experiment 1, mild in experiment 2, and severe in experiments 4 and 6 (data not shown).
Figure 2.—
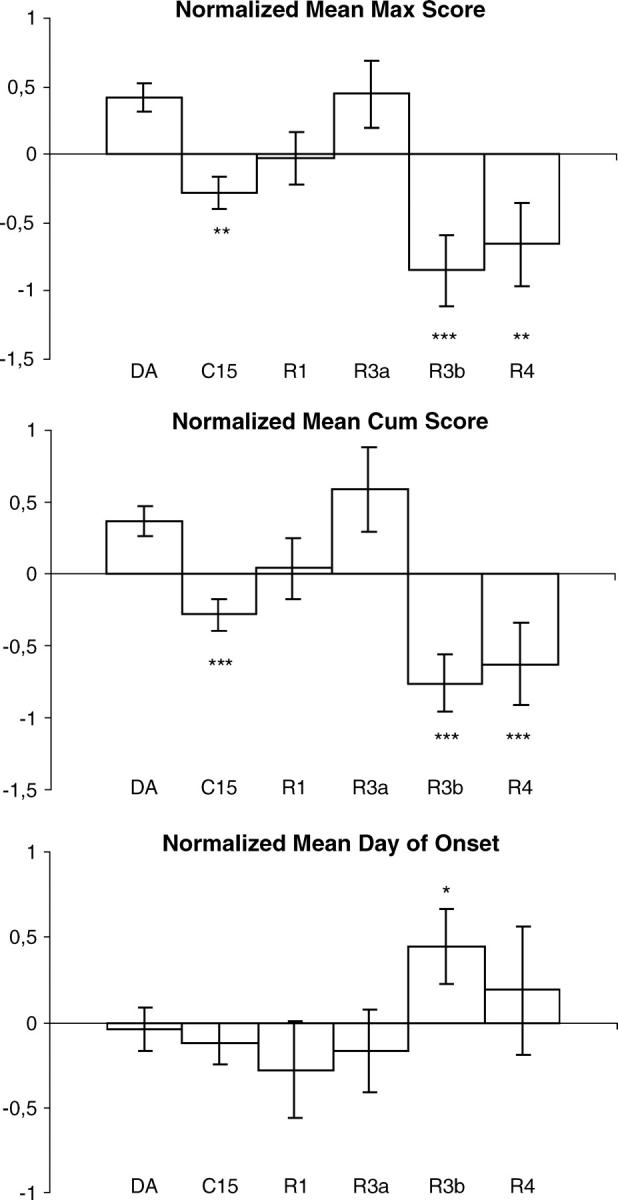
Combined analysis of the clinical MOG-EAE phenotypes in six separate experiments encompassing the DA (n = 72), C15 (n = 49), C15R1 (n = 23), C15R3a (n = 9), C15R3b (n = 14), and C15R4 (n = 17) strains. Maximum EAE score, cumulative EAE score, and onset day were tested with the Wilcoxon two-sample test after normalization; *P < 0.05, **P < 0.01, and ***P < 0.001. Pairwise comparisons were made with the congenic strains and the DA strain. Mean values and SEM for disease onset day after immunization were calculated only for affected rats.
Figure 3.—
The clinical course of rMOG (aa 1–125)-induced EAE in selected strains and experiments. (a) Experiment 3: a mild disease course in DA rats (n = 11) and almost complete protection in C15 rats (n = 8). (b) Experiment 5: a severe disease in DA rats (n = 16) and reduced disease severity in C15R3b (n = 5) and C15R4 (n = 9) rats.
Eae19 delineated by linkage mapping in a (DA × PVG.AV1) AIL:
A total of 1068 MOG-immunized (DA × PVG.AV1)F7 rats were monitored 31 days p.i. for clinical signs of EAE. Unambiguous signs of EAE were recorded in 14.8% (158/1068) of the rats. A detailed account of the EAE disease outcome in the F7(DA × PVG.AV1) AIL rats has been published (Jagodic et al. 2004). All available EAE-affected rats were selected for genotyping (n = 152). Randomly selected healthy rats, displaying no signs of EAE and no weight loss, were genotyped in parallel (n = 162). The DA × PVG.AV1 strain combination provided a substantially higher degree of polymorphic microsatellite markers than the DA × ACI combination did, as expected. Linkage analysis resolved the C15 region into a significant locus, named Eae19 (http://ratmap.org/), displaying linkage to several EAE phenotypes. Interestingly, the disease incidence and the day of onset was linked to Eae19, indicating that the EAE regulatory effect is not limited to the disease severity, as suggested by the analysis of the congenic strains. The LOD score curves for the different EAE phenotypes are presented in Figure 4. The confidence interval, defined as a drop of 1 in the LOD score, comprises an ∼13-Mb region (D15Mgh4-D15Rat102). The DA allele at the peak marker (D15Rat71) is disease enhancing in an additive fashion. Sequence alignments and map comparisons revealed that Eae19 is syntenic to human 13q22.1–q31.2 (Table 1).
TABLE 1.
Position and LOD scores forEae19 in the (DA × PVG.AV1)F7 advanced intercross line
| LOD scorea
|
||||||||
|---|---|---|---|---|---|---|---|---|
| QTL | Inc | Ons | Max | Cum | Peak marker | Marker interval | CIb (Mb) | Syntenic human regionc |
| Eae19 | 2.9 | 3.2 | 4.7 | 3.2 | D15Rat71 | D15Mgh4–D15Rat102 | 13 | 13q22.1–q31.2 |
LOD scores and thresholds for significance based on 5000 permutations were generated with R/qtl. Significance threshold: 2.3 for Inc (Incidence of EAE); 2.0 for Ons (day of onset), 2.9 for Max (Maximum EAE score) and 2.2 for Cum (Cumulative EAE score).
Confidence intervals (CI) defined as a drop of 1 in the LOD score and the closest corresponding microsatellite markers are reported.
Synteny data derived from http://www.ensembl.org/.
DISCUSSION
The definition of Eae19 in two different strain combinations (DA × ACI and DA × PVG.AV1) strengthens the importance of this locus. Further mapping of genes may also be facilitated by comparisons of genetic polymorphisms among the three different strains, especially since the low polymorphism rate between the DA and the ACI strain may decrease substantially the number of relevant genetic polymorphisms. However, at this stage it is impossible to rule out that Eae19 is composed of several genes and/or genes that differ between the different strains (Morel et al. 2001; Becanovic et al. 2004). Definition of subcongenic strains from the C15R4 strain will be performed to further reduce the size of the congenic fragment contributing to relative disease protection. Positional cloning will then be needed to define the exact genes responsible for the EAE-regulating effect of Eae19. Successful positional cloning through the definition of smaller and smaller congenic fragments has recently been demonstrated in rat experimental arthritis (Olofsson et al. 2003).
A possible problem with gene mapping in congenic strains is the presence of contaminating fragments of DNA from the donor strain, contributing to differences in the disease phenotype between the congenic strains and the parental strains that wrongly would be interpreted as genetically localized to the congenic fragment. The speed congenic strategy applied in the present study is a way to improve the control of contaminating fragments as well as to speed up the process of generating congenic strains. Mapping of the disease expression in recombinant congenic strains is another way to rule out significant contributions from genes outside the investigated congenic fragment. The lack of clinical effects in the C15R1 and C15R3a congenic strains strongly argues against any significant contributions from contaminating ACI DNA fragments outside Eae19, since those strains are expected to share possible contaminating fragments with the C15R3b and the C15R4 strains. Another issue in the analysis of EAE QTL is the stability of the models and the observed genetic effects. In the present study, there were clear differences in the six different experiments regarding disease severity in the parental DA rat strain (Figure 3, a and b) as well as a variable difference between the full-length C15 and the DA strain. Differences in the disease expression in parental/control strains are possible to minimize by applying strict protocols for immunization and environmental monitoring, but in practice it is very difficult to obtain completely stable conditions in EAE. It may also be argued that repeating experiments with different disease severity maximizes the possibility to detect weak genetic effects. It is highly likely that most genetic effects in MOG-EAE (and MS) are relatively weak and/or present only in certain disease subphenotypes (Morel et al. 2000). This may help to explain the relative lack of progress in QTL mapping in EAE since the first whole-genome scan was published in 1995 (Sundvall et al. 1995).
Linkage mapping in the (DA × PVG.AV1)F7 AIL localized Eae19 within a 13-Mb confidence interval, which overlaps with the congenic fragment defined by C15R4. The linkage analysis in the AIL both increased the LOD score and decreased the confidence interval of Eae19 compared to previous F2 analysis. This region contains only 32 confirmed and predicted genes, including genes such as immune-responsive gene 1 and neuronal ceroid lipofuscinose gene 5. A current list of genes mapped to the interval can be retrieved at http://www.ensembl.org. There were few obvious candidate genes, which may be explained by the presence of yet-unmapped genes, by the presence of regulatory elements altering the expression of genes mapping outside Eae19, or by complex interactions. Eae19 also overlaps with adjuvant-induced arthritis QTL 4 (Aia4) (Kawahito et al. 1998), serum cholesterol level QTL 1 (Kato et al. 2000), blood pressure QTL cluster 12 (Stoll et al. 2000), and gastric cancer susceptibility QTL 1 (Ushijima et al. 2000). Aia4 may be the most interesting of these QTL, since we previously demonstrated EAE regulatory effects in rat strains congenic for arthritis-regulating QTL (Becanovic et al. 2003). It is highly likely that some genetically regulated disease mechanisms are shared between arthritis and EAE (Becker et al. 1998). Given the emerging evidence for the importance of immune mechanisms in cardiovascular diseases and cancer, a shared genetic regulation with these conditions, as suggested by the overlapping QTL, is another intriguing possibility. However, due to the large numbers of QTL described and the usual size of the confidence intervals, a certain degree of overlap among QTL is expected by chance. Eae19 is syntenic to the human chromosome 13q22.1–q31.2 that has not shown evidence of linkage to MS. An explanation, in addition to the lack of power to exclude gene regions in human linkage and association studies, is that pathways but not the regulating genes are shared between the animal model and the human disease.
In conclusion, a new EAE-regulating locus on rat chromosome 15, Eae19, is mapped in congenic and recombinant congenic strains in combination with linkage analysis in an AIL. Further dissection of Eae19 is possible by the creation of congenic strains with increasingly smaller congenic fragments. The identification of genetic polymorphisms regulating autoimmune neuroinflammation will reveal disease-relevant mechanistic pathways and thereby provide new targets for therapeutic interventions.
References
- Adelmann, M., J. Wood, I. Benzel, P. Fiori, H. Lassmann et al., 1995. The N-terminal domain of the myelin oligodendrocyte glycoprotein (MOG) induces acute demyelinating experimental autoimmune encephalomyelitis in the Lewis rat. J. Neuroimmunol. 63: 17–27. [DOI] [PubMed] [Google Scholar]
- Akesson, E., A. Oturai, A. Svejgaard, P. Holmans, A. Compston et al., 2002. A genome-wide screen for linkage in Nordic sib-pairs with multiple sclerosis. Genes Immun. 3: 279–285. [DOI] [PubMed] [Google Scholar]
- Ban, M., G. Stewart, B. Bennetts, R. Heard, R. Simmons et al., 2002. A genome screen for linkage in Australian sibling-pairs with multiple sclerosis. Genes Immun. 3: 464–469. [DOI] [PubMed] [Google Scholar]
- Becanovic, K., L. Backdahl, E. Wallstrom, F. Aboul-Enein, H. Lassmann et al., 2003. Paradoxical effects of arthritis-regulating chromosome 4 regions on myelin oligodendrocyte glycoprotein-induced encephalomyelitis in congenic rats. Eur. J. Immunol. 33: 1907–1916. [DOI] [PubMed] [Google Scholar]
- Becanovic, K., M. Jagodic, E. Wallstrom and T. Olsson, 2004. Current gene mapping strategies in experimental models of multiple sclerosis. Scand. J. Immunol. 60: 39–51. [DOI] [PubMed] [Google Scholar]
- Becker, K. G., R. M. Simon, J. E. Bailey-Wilson, B. Freidlin, W. E. Biddison et al., 1998. Clustering of non-major histocompatibility complex susceptibility candidate loci in human autoimmune diseases. Proc. Natl. Acad. Sci. USA 95: 9979–9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley, S., 2001. A genome screen for multiple sclerosis in Italian families. Gene Immun. 2: 205–210. [DOI] [PubMed] [Google Scholar]
- Broman, K., H. Wu, S. Sen and G. Churchill, 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 1: 889–890. [DOI] [PubMed] [Google Scholar]
- Chataway, J., R. Feakes, F. Coraddu, J. Gray, J. Deans et al., 1998. The genetics of multiple sclerosis: principles, background and updated results of the United Kingdom systematic genome screen. Brain 121: 1869–1887. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coraddu, F., 2001. A genome screen for multiple sclerosis in Sardinian multiplex families. Eur. J. Hum. Genet. 9: 621–626. [DOI] [PubMed] [Google Scholar]
- Dahlman, I., L. Jacobsson, A. Glaser, J. C. Lorentzen, M. Andersson et al., 1999. a Genome-wide linkage analysis of chronic relapsing experimental autoimmune encephalomyelitis in the rat identifies a major susceptibility locus on chromosome 9. J. Immunol. 162: 2581–2588. [PubMed] [Google Scholar]
- Dahlman, I., E. Wallstrom, R. Weissert, M. Storch, B. Kornek et al., 1999. b Linkage analysis of myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis in the rat identifies a locus controlling demyelination on chromosome 18. Hum. Mol. Genet. 8: 2183–2190. [DOI] [PubMed] [Google Scholar]
- Dahlman, I., E. Wallstrom, H. Jiao, H. Luthman, T. Olsson et al., 2001. Polygenic control of autoimmune peripheral nerve inflammation in rat. J. Neuroimmunol. 119: 166–174. [DOI] [PubMed] [Google Scholar]
- Darvasi, A., and M. Soller, 1995. Advanced intercross lines: an experimental population for fine genetic mapping. Genetics 141: 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi, A., and M. Soller, 1997. A simple method to calculate resolving power and confidence interval of QTL map location. Behav. Genet. 27: 125–132. [DOI] [PubMed] [Google Scholar]
- De Rosbo, N. K., R. Milo, M. B. Lees, D. Burger, C. C. Bernard et al., 1993. Reactivity to myelin antigens in multiple sclerosis. Peripheral blood lymphocytes respond predominantly to myelin oligodendrocyte glycoprotein. J. Clin. Invest. 92: 2602–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebers, G. C., 1996. Genetic epidemiology of multiple sclerosis. Curr. Opin. Neurol. 9: 155–158. [DOI] [PubMed] [Google Scholar]
- Ebers, G. C., D. E. Bulman, A. D. Sadovnick, D. W. Paty, S. Warren et al., 1986. A population-based study of multiple sclerosis in twins. N. Engl. J. Med. 315: 1638–1642. [DOI] [PubMed] [Google Scholar]
- Ebers, G. C., A. D. Sadovnick, N. J. Risch and C. C. S. Group, 1995. A genetic basis for familial aggregation in multiple sclerosis. Nature 377: 150–151. [DOI] [PubMed] [Google Scholar]
- Ebers, G. C., K. Kukay, D. E. Bulman, A. D. Sadovnick, G. Rice et al., 1996. A full genome search in multiple sclerosis. Nat. Genet. 13: 472–476. [DOI] [PubMed] [Google Scholar]
- Haines, J. L., M. Ter-Minassian, A. Bazyk, J. F. Gusella, D. J. Kim et al., 1996. A complete genomic screen for multiple sclerosis underscores a role for the major histocompatibility complex. Nat. Genet. 13: 469–471. [DOI] [PubMed] [Google Scholar]
- Haines, J., Y. Bradford, M. Garcia, A. Reed, E. Neumeister et al., 2002. Multiple susceptibility loci for multiple sclerosis. Hum. Mol. Genet. 11: 2251–2256. [DOI] [PubMed] [Google Scholar]
- Hedrich, H. J., 1990 Genetic Monitoring of Inbred Strains of Rats. Gustav Fischer Verlag, New York.
- Jacob, H. J., D. M. Brown, R. K. Bunker, M. J. Daly, V. J. Dzau et al., 1995. A genetic linkage map of the laboratory rat, Rattus norvegicus. Nat. Genet. 9: 63–69. [DOI] [PubMed] [Google Scholar]
- Jagodic, M., B. Kornek, R. Weissert, H. Lassmann, T. Olsson et al., 2001. Congenic mapping confirms a locus on rat chromosome 10 conferring strong protection against myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis. Immunogenetics 53: 410–415. [DOI] [PubMed] [Google Scholar]
- Jagodic, M., K. Becanovic, J. R. Sheng, X. Wu, L. Backdahl et al., 2004. An advanced intercross line resolves Eae18 into two narrow quantitative trait loci syntenic to multiple sclerosis candidate loci. J. Immunol. 173: 1366–1373. [DOI] [PubMed] [Google Scholar]
- Jersild, C., T. Fog, G. S. Hansen, M. Thomsen, A. Svejgaard et al., 1973. Histocompatibility determinants in multiple sclerosis, with special reference to clinical course. Lancet 2: 1221–1225. [DOI] [PubMed] [Google Scholar]
- Johns, T. G., N. Kerlero de Rosbo, K. K. Menon, S. Abo, M. F. Gonzales et al., 1995. Myelin oligodendrocyte glycoprotein induces a demyelinating encephalomyelitis resembling multiple sclerosis. J. Immunol. 154: 5536–5541. [PubMed] [Google Scholar]
- Kato, N., T. Tamada, T. Nabika, K. Ueno, T. Gotoda et al., 2000. Identification of quantitative trait loci for serum cholesterol levels in stroke-prone spontaneously hypertensive rats. Arterioscler. Thromb. Vasc. Biol. 20: 223–229. [DOI] [PubMed] [Google Scholar]
- Kawahito, Y., G. W. Cannon, P. S. Gulko, E. F. Remmers, R. E. Longman et al., 1998. Localization of quantitative trait loci regulating adjuvant-induced arthritis in rats: evidence for genetic factors common to multiple autoimmune diseases. J. Immunol. 161: 4411–4419. [PubMed] [Google Scholar]
- Kuokkanen, S., M. Gschwend, J. D. Rioux, M. J. Daly, J. D. Terwilliger et al., 1997. Genomewide scan of multiple sclerosis in Finnish multiplex families. Am. J. Hum. Genet. 61: 1379–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird, P. W., A. Zijderveld, K. Linders, M. A. Rudnicki, R. Jaenisch et al., 1991. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 19: 4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., and L. Kruglyak, 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11: 241–247. [DOI] [PubMed] [Google Scholar]
- Lassmann, H., W. Bruck and C. Lucchinetti, 2001. Heterogeneity of multiple sclerosis pathogenesis: implications for diagnosis and therapy. Trends Mol. Med. 7: 115–121. [DOI] [PubMed] [Google Scholar]
- Linington, C., M. Bradl, H. Lassmann, C. Brunner and K. Vass, 1988. Augmentation of demyelination in rat acute allergic encephalomyelitis by circulating mouse monoclonal antibodies directed against a myelin/oligodendrocyte glycoprotein. Am. J. Pathol. 130: 443–454. [PMC free article] [PubMed] [Google Scholar]
- Lucchinetti, C. F., W. Bruck, M. Rodriguez and H. Lassmann, 1996. Distinct patterns of multiple sclerosis pathology indicates heterogeneity in pathogenesis. Brain Pathol. 6: 259–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, L., B. P. Croker, K. R. Blenman, C. Mohan, G. Huang et al., 2000. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc. Natl. Acad. Sci. USA 97: 6670–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, L., K. R. Blenman, B. P. Croker and E. K. Wakeland, 2001. The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc. Natl. Acad. Sci. USA 98: 1787–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olerup, O., and J. Hillert, 1991. HLA class II-associated genetic susceptibility in multiple sclerosis: a critical evaluation. Tissue Antigens 38: 1–15. [DOI] [PubMed] [Google Scholar]
- Olofsson, P., J. Holmberg, J. Tordsson, S. Lu, B. Akerstrom et al., 2003. Positional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat. Genet. 33: 25–32. [DOI] [PubMed] [Google Scholar]
- Sadovnick, A. D., H. Armstrong, G. P. Rice, D. Bulman, L. Hashimoto et al., 1993. A population-based study of multiple sclerosis in twins: update. Ann. Neurol. 33: 281–285. [DOI] [PubMed] [Google Scholar]
- Sadovnick, A. D., G. C. Ebers, D. A. Dyment, N. J. Risch, D. Bulman et al., 1996. Evidence for genetic basis of multiple sclerosis. Lancet 347: 1728–1730. [DOI] [PubMed] [Google Scholar]
- Sawcer, S., H. B. Jones, R. Feakes, J. Gray, N. Smaldon et al., 1996. A genome screen in multiple sclerosis reveals susceptibility loci on chromosome 6p21 and 17q22. Nat. Genet. 13: 464–468. [DOI] [PubMed] [Google Scholar]
- Stoll, M., A. E. Kwitek-Black, A. W. J. Cowley, E. L. Harris, S. B. Harrap et al., 2000. New target regions for human hypertension via comparative genomics. Genome Res. 10(4): 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J., H. Link, T. Olsson, B. G. Xiao, G. Andersson et al., 1991. T and B cell responses to myelin-oligodendrocyte glycoprotein in multiple sclerosis. J. Immunol. 146: 1490–1495. [PubMed] [Google Scholar]
- Sundvall, M., J. Jirholt, H. T. Yang, L. Jansson, A. Engstrom et al., 1995. Identification of murine loci associated with susceptibility to chronic experimental autoimmune encephalomyelitis. Nat. Genet. 10: 313–317. [DOI] [PubMed] [Google Scholar]
- Ushijima, T., M. Yamamoto, M. Suzui, T. Kuramoto, Y. Yoshida et al., 2000. Chromosomal mapping of genes controlling development, histological grade, depth of invasion, and size of rat stomach carcinomas. Cancer Res. 60: 1092–1096. [PubMed] [Google Scholar]
- Wakeland, E., L. Morel, K. Achey, M. Yui and J. Longmate, 1997. Speed congenics: a classic technique in the fast lane (relatively speaking). Immunol. Today 18: 472–477. [DOI] [PubMed] [Google Scholar]
- Wallstrom, E., M. Khademi, M. Andersson, R. Weissert, C. Linington et al., 1998. Increased reactivity to myelin oligodendrocyte glycoprotein peptides and epitope mapping in HLA DR2(15)+ multiple sclerosis. Eur. J. Immunol. 28: 3329–3335. [DOI] [PubMed] [Google Scholar]
- Weissert, R., E. Wallstrom, M. K. Storch, A. Stefferl, J. Lorentzen et al., 1998. b MHC haplotype-dependent regulation of MOG-induced EAE in rats. J. Clin. Invest. 102: 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, M., and S. Guo, 1997. Fine-scale mapping of quantitative trait loci using historical recombinations. Genetics 145: 1201–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]