Abstract
An understanding of the mutational and evolutionary mechanisms underlying the emergence of novel genes is critical to studies of phenotypic and genomic evolution. Here we describe a new example of a recently formed chimeric fusion gene that occurs in Drosophila guanche, D. madeirensis, and D. subobscura. This new gene, which we name Adh-Twain, resulted from an Adh mRNA that retrotransposed into the Gapdh-like gene, CG9010. Adh-Twain is transcribed; its 5′ promoters and transcription patterns appear similar to those of CG9010. Population genetic and phylogenetic analyses suggest that the amino acid sequence of Adh-Twain evolved rapidly via directional selection shortly after it arose. Its more recent history, however, is characterized by slower evolution consistent with increasing functional constraints. We present a model for the origin of this new gene and discuss genetic and evolutionary factors affecting the evolution of new genes and functions.
GENOMES gain and lose genes at surprisingly high rates in both unicellular (Jain et al. 1999; Ochman and Jones 2000; Lynch and Conery 2003) and multicellular organisms (Patthy 1999; Betran et al. 2002; Harrison et al. 2002; Borevitz et al. 2003; Holland 2003; Tian et al. 2003). Identifying the mutational and population genetic mechanisms involved in gene loss and gain is critical to understanding the forces shaping genome variation. The spread of gene duplications, by either drift or natural selection (Wagner 2001; Ohta 2003; reviewed in Wolfe and Li 2003), is one mechanism by which gene number increases (Haldane 1932; Ohno 1970). Gene duplications may, in many cases, evolve new functions or become subfunctionalized simply as a result of amino acid or expression evolution and not as a consequence of large-scale changes in gene organization (Ohno 1970; Lynch and Conery 2000; Lynch et al. 2001; Hughes 2002; Betran and Long 2003; Katju and Lynch 2003).
Occasionally, duplication events lead to radical reorganization of gene structures that likely lead to dramatic and immediate functional divergence. One type of radical reorganization is gene fusion, whereby two previously separate and independent genes are fused to form a single contiguous gene. Such chimeric fusion genes (CFGs) have been identified in several taxa. For example, in plants CFGs are implicated in cytoplasmic male sterility (He et al. 1996). A few CFGs have also been found in vertebrates (Finta and Zaphiropoulos 2000; Rogalla et al. 2000; Thomson et al. 2000; Courseaux and Nahon 2001). Finally, several novel CFGs have been described in Drosophila, such as jingwei in Drosophila tessieri and D. yakuba (Long and Langley 1993), Sdic in D. melanogaster (Nurminsky et al. 1998), and Adh-finnegan (Sullivan et al. 1994; Begun 1997).
Two novel Drosophila genes, jingwei and Adh-finnegan, were previously thought to be Alcohol-dehydrogenase (Adh) pseudogenes (Fischer and Maniatis 1985; Jeffs and Ashburner 1991). In both cases, further analysis showed that these genes were functional genes that acquired protein-coding sequence 5′ of the Adh-derived region of gene (Long and Langley 1993; Begun 1997). jingwei is a fusion of the amino terminus of a gene known as yellow emperor and a retrotrasposed Adh (Wang et al. 2000). The jingwei expression profile has diverged from its Adh ancestors. In D. tessieri expression is now limited to the testes (like ymp), although in D. yakuba it is expressed in other tissues as well (Long and Langley 1993). Adh-finnegan was created by the chromosomal duplication of Adh combined with the recruitment of a new 5′ exon of unknown origin (Begun 1997). Adh-finnegan appears to be expressed broadly in adult tissues (Sullivan et al. 1994). Although these two Adh-derived fusion genes arose via different mechanisms and show dramatically different expression patterns, directional selection appears to have driven rapid amino acid evolution in both genes (Long and Langley 1993; Begun 1997). The fact that two novel Drosophila genes are derived from Adh and share some common aspects of their evolution raises two important questions:
Is Adh overrepresented among novel fly genes? Or, does the discovery of jingwei and Adh-finnegan—both of which were discovered unintentionally—reflect the intense study of Adh in Drosophila?
If Adh frequently participates in radical reorganizations associated with novel function, what general principles of the evolution of novel function are revealed by these examples of repeated evolution?
Given the history of the discovery of jingwei and Adh-finnegan, a report of a third putative Adh pseudogene in the obscura group of Drosophila (Marfany and Gonzalez-Duarte 1992; Luque et al. 1997) attracted our attention. DNA sequencing showed that this putative pseudogene originated by retrotransposition. Results from polytene in situ hybridization showed that this Adh retrosequence had transposed to chromosome arm E from chromosome U, which is the expected location of Adh on the basis of the conservation of Muller elements (Ashburner 1989). This retrotransposed Adh was found in D. subobscura, D. guanche, and D. madeirensis, but not in D. ambigua (Visa et al. 1991; Marfany and Gonzalez-Duarte 1992; Luque et al. 1997), suggesting that the gene likely arose within the past 3 million years. Luque et al. (1997) sequenced six clones harboring this putative Adh retropseudogene, two each from genomic libraries of D. subobscura, D. guanche, and D. madeirensis. Comparisons of putative Adh pseudogenes to Adh for these species revealed frameshift mutations or indels in 5′ and 3′ regions flanking the Adh coding regions. The codon homologous to the ATG initiation codon of the ancestral Adh was CTG in both D. guanche clones. Premature stop codons were evident in one of the two D. guanche clones and one of the two D. subobscura clones, but none of the D. madeirensis clones. These observations suggested that at least some of these Adh sequences were no longer functional. The fact that the duplicate Adh was a retrosequence was also interpreted as support for the pseudogene hypothesis, as retrotransposed sequences potentially lack regulatory elements necessary for proper expression. All three species showed elevated amino acid substitution rates (dN) relative to Adh. None of the putative retropseudogenes, however, showed a nonsynonymous to synonymous substitution rate (dN/dS) close to one, the expectation for a neutrally evolving pseudogene. Moreover, codon bias increased in the putative Adh retropseudogenes, an unexpected result for a nonfunctional gene. Overall, the data presented a conflicting picture of the Adh retrosequence. Some aspects of the data supported the pseudogene hypothesis, yet others were strangely inconsistent with the hypothesis and were similar to the situation previously observed in the repleta group Adh duplication (Sullivan et al. 1994; Begun 1997). We present here our analysis of this retrotransposed Adh. We show that this putative Adh retropseudogene is actually part of a new chimeric fusion gene that is the result of an Adh mRNA inserting into the Gapdh-like gene, CG9010. This fusion gene is actively and widely transcribed. While the 5′ promoters and transcription patterns of this gene are similar to those of CG9010, the protein-coding region has diverged for both the CG9010 and the Adh-like regions. Population genetic and phylogenetic analyses suggest that this amino acid evolution resulted from directional selection shortly after the chimeric fusion gene was formed.
MATERIALS AND METHODS
Stocks:
D. subobscura stocks were obtained from the Species Stock Center and from A. Davis. D. guanche, D. hydei, D. pseudoobscura, D. melanogaster, and D. yakuba stocks were originally obtained from the Species Stock Center and the Bloomington Stock Center. A D. madeirensis stock was kindly provided by M. Aguadé. All stocks were reared on standard Drosophila medium at room temperature.
DNA sequencing:
PCR products were sequenced directly using an ABI 377 automated sequencer and BigDye Terminator chemistry (Applied BioSystems, Foster City, CA).
RNA extraction, cDNA preparation, and RT-PCR:
Poly(A+) RNA was prepared from whole flies or larvae using a MicroPoly(A) kit (Ambion, Austin, TX). cDNA for reverse transcriptase-PCR and rapid amplification of cDNA ends (RACE) was prepared from this RNA using the SMART RACE cDNA amplification kit (CLONTECH, Palo Alto, CA). SuperScript II reverse transcriptase (GIBCO BRL, Rockville, MD) was used for all RT reactions. Gene-specific primers were used to assay gene expression by RT-PCR on cDNA isolated from larvae (first, second, and third instar), whole adult males, and whole adult females.
Genomic library construction and screening:
D. subobscura genomic DNA was isolated from adult flies, partially digested with Sau3aI (New England BioLabs, Beverly, MA), and then dephosphorylated with CIAP (Promega, Madison, WI). These fragments were ligated into the Lambda DASH II vector according to the manufacturer's instructions (Stratagene, La Jolla, CA; T4 ligase was from GIBCO BRL), followed by packaging using Gigapack III Gold packaging reactions (Stratagene, La Jolla, CA). The library was amplified once on plates using XL-1Blue MRA [P2] cells.
Primary and secondary plaque lifts were carried out on Nytran Nylon membranes (Schleicher & Schuell, Keene, NH). The library was screened with a 1-kb CG9010 probe that was PCR amplified from D. melanogaster. Because this probe cross-hybridizes to plaques harboring Gapdh, we used PCR with primers designed for D. subobscura Gapdh to rule out false positives. Phage containing CG9010 were digested with EcoRI and PstI, resolved on a 0.7% agarose gel, Southern blotted, and probed with CG9010. The fragment containing D. subobscura CG9010 was subcloned into pBluescript.
Genomic Southern blot analysis of CG9010:
Southern blot analysis was used to infer copy number of CG9010 in D. subobscura. Genomic DNA (5 μg) was purified from D. melanogaster, D. pseudoobscura, D. guanche, and D. subobscura. These samples were digested with PstI or HindIII (GIBCO BRL), electrophoresed on a 0.7% gel, and Southern blotted to Nytran nylon membranes. These blots were then probed with PCR-amplified 1-kb fragments of D. melanogaster CG9010 and then D. subobscura CG9010.
Protein analyses:
One gram of tissue (whole adults and larvae) was homogenized in 2 ml ice-cold homogenization buffer and then centrifuged. Protein concentration in the supernatant was determined using the Bradford method (Bio-Rad, Hercules, CA). We then applied SDS-PAGE to our samples (10% acrylamide resolving gels, 4% acrylamide stacking gels). Typically, 5 μg of sample was boiled and then loaded in each lane. Gels were run at 70 mA constant current for 30–40 min. Gels were electroblotted on nitrocellulose at 0° for 30–60 min at 100 V constant voltage followed by blocking for 1 hr in a TBS-milk solution. Blots were incubated overnight (at 4°) with goat anti-D. melanogaster ADH (courtesy of C. Benyanjati) diluted in TBS-milk solution. Blots were placed in fresh TBS-milk and incubated with the secondary antibody (anti-goat HRP) for 1 hr followed by washing with TBS-Tween. The secondary antibody was visualized with ECL Plus (Amersham Biosciences, Piscataway, NJ) followed by autoradiography.
This approach repeatedly worked well for ADH proteins in all species we assayed (D. melanogaster, D. suboscura, D. yakuba, D. pseudoobscura, and D. hydei). Overexposure of the film to the blot would visualize a number of minor bands, but it was impossible to determine which, if any, would correspond to the band of interest.
We also used allozyme gels to look for residual ADH activity in the D. subobscura CFG. We adapted the protocol of Batterham et al. (1983). Although we tried a variety of conditions, we observed only a single band of activity. This was consistent with what was reported in the literature (see results).
DNA sequence analysis:
BLASTN and TBLASTX were used to identify similar sequences from the NCBI databases (Altschul et al. 1997). DNAstar (DNASTAR, Madison, WI) was used for sequence alignments, contig assembly, and restriction mapping. Accession numbers of previously published data used in this analysis are X55390, X55391, M55545, X60112, U68470, U68469, X60113, U68472, U68471, AF175211, and AE003805.
Basic population genetic analyses were done using either DNAsp (Rozas and Rozas 1999) or software written by C.D.J. We limited our analysis to regions of high sequence quality. Typically, insertion/deletion polymorphism was ignored in our calculations of population genetics statistics.
Promoter prediction was accomplished using NNPP (Reese 2001) and McPromoter (Ohler et al. 2002). As noted in results, a threshold of 0.8 was used (which is predicted to give a false positive rate of 0.4% for NNPP). Signal peptide prediction used SignalP (Nielsen et al. 1997). No signal peptides were detected.
Phlyogenetic analysis:
PAML provides a suite of maximum likelihood-based tools for combining DNA sequence and phylogenetic data to test molecular evolutionary hypotheses (Yang 1997; Yang and Bielawski 2000). We used the phylogeny of Ramos-Onsins et al. (1998). There are three major steps to using PAML: (1) choice of appropriate model, (2) parameterization of that model, and (3) sequential comparison using log-likelihood ratio tests of simpler to more complex models to evaluate if a more complex model provides a significantly better fit to the data. For clarity, steps 1 and 2 will be described here and step 3 will be presented in results.
Evolution of protein-coding regions of CG9010 and Adh-derived sequences were analyzed independently using the codon model (Codeml; Goldman and Yang 1994; Yang 1997). In the following sections, the difference in the log likelihood (Δlnl), for the relevant degrees of freedom, implied a P-value <0.05 and was typically <0.001. Unless noted otherwise, model comparisons involving multiple tests remained significant after Bonferroni corrections. F3X4 codon model fit the CG9010 data the best of the codon models; the estimated codon table fit the Adh data the best. When appropriate and when a significantly better fit to the data was produced, κ, ω, and α were estimated (see Yang 1997). For the analyses discussed in results, we a priori hypothesize that the lineage created by the formation of the CFG (Adh-Twain) will be undergoing more rapid evolution than the Adh or CG9010 lineages [e.g., hypothesis generation is independent of the data used to test it; see Yang (1997) p. 23]. In several cases, convergence to maximum likelihood estimates was verified by changing the “small difference” parameter (see Yang 1997, p. 19). Reconstruction of ancestral sequences was done using both joint and marginal reconstruction. Ancestral states were identical regardless of method. Note that all amino acid positions are in terms of their position in D. subobscura, not D. melanogaster.
RESULTS
Open reading frame extends 5′ of Adh retrosequence and is homologous to CG9010:
Analysis of previously published D. subobscura genomic sequences revealed an open reading frame (ORF) 5′ of the Adh retrosequence. This ORF was contiguous and in the same reading frame as the Adh restrosequence ORF. TBLASTX of the 5′ ORF to the D. melanogaster reference sequence showed that the predicted amino acid sequence from this region was homologous to the translated D. melanogaster protein for CG9010, a broadly expressed, intronless gene that is similar to Gapdh. This region, roughly 300 bases 5′ of the Adh sequence, is homologous to CG9010 in all three species. In D. subobscura, the inferred amino acid sequence of the region from 135 to 441 bases 5′ of the putative retrosequence start codon shows ∼60% identity with the amino terminus of the D. melanogaster predicted protein CG9010 (TBLASTX E-value = e−15). A similar region was found in D. madeirensis. The more distantly related D. guanche also contains a homologous region showing ∼67% identity with D. melanogaster CG9010 (TBLASTX E-value = e−18).
As mentioned in Luque et al. (1997), the region immediately upstream of the putative retrosequence initiation codon and downstream of the CG9010-like region is similar to obscura group Adh 5′-UTR. It is not clear from sequence comparisons whether this region represents the larval or adult leader variant. The Adh-like region of the retrosequence retained the Adh 3′-UTR and a viable polyadenylation signal. The region 3′ of the Adh-like region 3′-UTR showed no significant similarity to any known gene, although it clearly harbors GEM elements (Vivas et al. 1999). Given the location of CG9010 in D. melanogaster, the D. subobscura CG9010 would be located on chromosome arm E, which is also the location of the Adh retrosequence.
These results suggest that the Adh pseuodogene described by Luque et al. (1997) is a chimeric fusion gene that resulted from the retrotransposition of an Adh transcript into a D. subobscura CG9010. Our analysis, however, did not resolve the issue of the frameshift and nonsense mutations associated with at least some copies of the Adh retrosequence. To address this issue, we sequenced DNA encompassing 97% of the potential open reading frame of the chimeric fusion gene and some 5′-flanking sequence from 15 lines of D. subobscura, one line of D. guanche, and one line of D. madeirensis. Complete coding sequences were obtained for one D. subobscura line and one D. guanche line. All sequences had a contiguous open reading frame that included both the CG9010-like region and the Adh-like region. None of the frameshift insertion/deletions observed by Luque et al. were observed in our data. Nor were any premature stop codons found in any of the regions surveyed. This suggests that these indels and stop codons may have been sequencing errors in the original article. (If these stop codons and frameshifts do indeed exist, our data suggest that they are rare variants in D. subobscura.) The substitution of the canonical Adh start codon (ATG) by a leucine (CTG) reported by Luque et al. (1997) was also observed in our D. guanche sequence. From these data, we conclude that this Adh “pseudogene” is likely a novel Adh-derived fusion gene, which we have tentatively named “Adh-Twain.” (Mark Twain's famous statement, “The rumors of my demise have been greatly exaggerated,” was the inspiration for the name of this gene. We propose the abbreviation AdhT for Adh-Twain.)
CG9010 exists in D. subobscura and D. guanche:
If CG9010 function was necessary for the ancestor of D. subobscura, D. guanche, and D. madeirensis (which is likely given its similarity to Gapdh and its conservation across Drosophila), then CG9010 must have duplicated in this lineage. In other words, if insertion of the retrosequence into CG9010 abolished CG9010 function, then an alternative functional copy of CG9010 should exist in the subobscura clade. We used Southern analysis to determine if CG9010 is present in more than one copy in D. subobscura. Digestion of genomic DNA with several restriction enzymes followed by blotting and hybridization with the 5′ end of CG9010 showed that CG9010 is single copy in D. melanogaster and D. pseudoobscura. The CG9010 probe, however, consistently produced two bands in D. subobscura (data not shown), supporting the idea that CG9010 duplicated in this lineage. BLAST searches of D. pseudoobscura suggest the presence of only one copy CG9010 in the strain used for the genome sequence.
To confirm that our Southern detected a surviving full copy of CG9010, we cloned CG9010. We used several methods to obtain the DNA sequence for the CG9010 homolog in D. subobscura and D. guanche (see materials and methods). The amino acid sequence of the D. subobscura homolog of CG9010 shares 91% amino acid identity with D. pseudoobscura and 82% amino acid identity with D. melanogaster. ESTs from D. melanogaster show that CG9010 is transcribed, as do our RT-PCR data from D. guanche and D. subobscura (Figure 1).
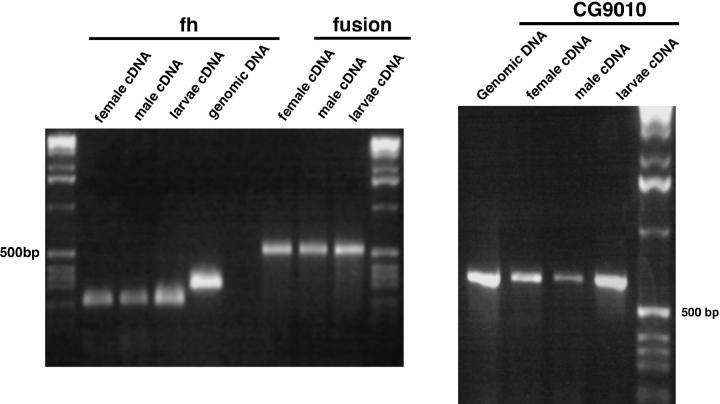
Figure 1.—
Transcription patterns of Adh-Twain and CG9010 are similar. We used RT-PCR to amplify a fragment of fh, Adh-Twain (fusion), and CG9010 from cDNA made from poly(A+) RNA extracted from D. subobscura adult females, adult males, and larvae. The same cDNA prep was used for all three amplifications. Adh-Twain is actively transcribed in all three samples. Qualitatively, the expression of Adh-Twain is similar to that of CG9010. The fh gene was a control to show that our cDNA did not contain genomic DNA. The fh amplicon spans an intron; thus, if the cDNA were contaminated with genomic DNA we would observe a second band in the fh lanes.
Adh-Twain is transcribed:
RT-PCR using Adh-Twain-specific primers showed that it is transcribed in D. subobscura larvae, adult males, and adult females (Figure 1). The gene is also expressed in D. guanche (data not shown). 5′-RACE data from D. subobscura and D. guanche definitively show that the CG9010-like region of the fusion gene is part of a longer transcript that includes the entire Adh-like region in both D. subobscura and D. guanche. The RACE data also allowed us to experimentally identify the 5′-UTR. Our 3′-RACE data confirmed that the Adh-Twain transcript terminates near the polyadenylation site of the Adh-like 3′-UTR region.
We used RT-PCR to compare transcription between D. subobscura Adh-Twain and D. subobscura CG9010. Figure 1 suggests no significant differences in the expression patterns of Adh-Twain and CG9010, although CG9010 maybe slightly less abundant in males.
Adh-Twain-predicted protein characteristics:
Basic characteristics of the predicted D. subobscura ADH-TWAIN protein are presented in Table 1. The predicted protein is ∼40% larger than ADH. Other than size, the most notable difference between ADH-TWAIN protein vs. ADH and CG9010 is that ADH-TWAIN is much more positively charged at pH 7.0 than either CG9010 or ADH. The ADH-derived portion of ADH-TWAIN also appears to have increased in molecular weight relative to ADH.
TABLE 1.
Characteristics ofAdh-Twain protein
| Adh-Twain- D. subobscura | Adh-D. subobscura |
CG9010-D. subobscura (for region conserved) |
|
|---|---|---|---|
| Molecular weight (Da) | 39664.22 | 27566.79 | 10805.75 |
| Amino acids | 356 | 254 | 96 |
| Isolectric point | 9.524 | 8.349 | 9.313 |
| Charge at pH 7.0 | 19.495 | 2.523 | 5.983 |
The predicted amino acid sequence of ADH-TWAIN is substantially different from that of its ancestors, in both the ADH-derived and the CG9010-derived parts of the protein. For example, the ADH-derived portion of ADH-TWAIN is roughly 50% more diverged from ADH than D. subobscura ADH in comparisons vs. D. melanogaster ADH. Indeed, the ADH-TWAIN ADH-derived sequence is more diverged from D. melanogaster than any of the published Drosophila ADH proteins we have surveyed. These data suggest that it is unlikely that ADH-TWAIN retains significant ADH-like activity. In fact, prior allozyme studies of ADH in D. subobscura provided no evidence of a protein other than ADH that oxidized ethanol (Loukas et al. 1979; Balanya et al. 1994; Castro et al. 1999). Our own allozyme analysis confirmed these previous results and is consistent with the idea that ADH-TWAIN has reduced ability to catalyze the oxidation of ethanol. Of course, this experiment does not rule out the possibility that ADH-TWAIN can oxidize some other alcohol or related molecule (e.g., aldehydes).
Western blots (data not shown) revealed a single band of the size expected for ADH in D. melanogaster, D. subobscura, D. guanche, D. yakuba, D. pseudoobscura, D. virilis, and D. hydei. However, we observed no strong secondary band of the size expected for the predicted ADH-TWAIN. [Similarly, jingwei, which is known to produce a protein in vitro (Zhang et al. 2004), was not visible in our Western blot.] Overexposure of the Western blot revealed a number of minor bands, but it was impossible to determine which, if any, would correspond to the band associated with the predicted ADH-TWAIN. The failure of the ADH antibody to react with ADH-TWAIN is not unexpected, given the very high protein divergence between the ADH-derived portion of ADH-TWAIN and known ADH proteins in Drosophila.
5′ regulatory region of Adh-Twain shows similarity to that of CG9010:
If the 5′ end of Adh-Twain is derived from a chromosomal duplication of CG9010, then we expect the 5′ region of Adh-Twain to harbor regulatory elements, such as promoters, derived from those of CG9010 (expression patterns of Adh-Twain and D. subobscura CG9010 are similar; Figure 1). To investigate this possibility, we gathered sequence data for ∼600 bases 5′ of the D. subobscura Adh-Twain start codon and ∼400 bp of 5′-flanking sequence from D. subobscura CG9010. These sequences were aligned to a sequence from the 5′-flanking region of D. pseudoobscura CG9010. We also used the VISTA browser (http://pipeline.lbl.gov/pseudo/) to qualitatively compare these sequences to D. melanogaster CG9010.
Figure 2 shows a multiple alignment of the 5′ regions of CG9010. It is immediately obvious that there are several highly conserved regions. Using a combination of our 5′-RACE data, published cDNA data, interspecific sequence comparisons, and promoter prediction methods (see materials and methods), we identified the putative 5′-UTRs and putative promoter elements of Adh-Twain and CG9010. Of particular note is the conserved sequence between base 177 and base 357 of the Adh-Twain 5′ sequence in Figure 2. This region is very highly conserved in D. guanche Adh-Twain (data not shown), well conserved in D. pseudoobscura CG9010, and weakly conserved in D. melanogaster. The methods of Reese (2001) and Ohler et al. (2002) both suggest a promoter element in this region of Adh-Twain and CG9010 in D. subobscura. This region shows some similarity to TATA-less promoters seen in D. melanogaster (e.g., Burke and Kadonaga 1997). This conserved region may be important for regulation of Adh-Twain and CG9010 and could potentially contribute to their similar expression patterns. The Adh-Twain 5′-UTR is much larger than the CG9010 5′-UTR. Using parsimony, we infer that the Adh-Twain 5′-UTR represents the derived state and that these sequence insertions arose after duplication of CG9010. The potential biological consequences of this larger UTR are not known, although 5′-UTRs are known to play a critical role in translational regulation (Gray and Wickens 1998). Most other insertions/deletions relative to D. pseudoobscura are shared by D. subobscura CG9010 and Adh-Twain.
Figure 2.—

5′-UTR and promoters are similar for Adh-Twain (D. sub Fusion) and CG9010 and conserved across taxa.
DNA polymorphism and divergence in Adh-Twain:
We surveyed most of the Adh-Twain open reading frame and ∼330 bases of the 5′-UTR and regulatory regions from 15 iso-female lines of D. subobscura. We also collected sequence data from iso-female lines of D. guanche (n = 1) and D. madeirensis (n = 1). These data are summarized in Figure 3 and Table 2.
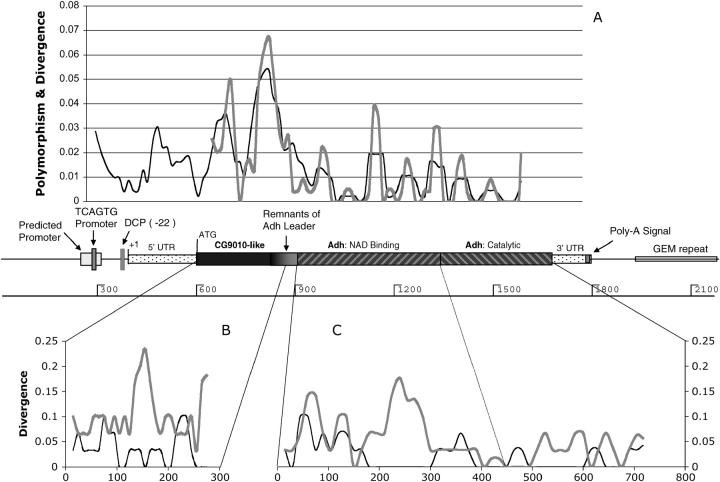
Figure 3.—
Major sequence features of Adh-Twain compared to patterns of polymorphism and divergence. (A) Comparison of the nucleotide polymorphism observed at Adh-Twain in our population of D. subobscura (black line) and the nucleotide divergence between D. subobscura and D. guanche Adh-Twain (gray line). (B) Divergence between D. subobscura and D. guanche CG9010 genes (black line) and divergence between D. subobscura Adh-Twain and CG9010 (gray line). (C) Divergence between D. subobscura and D. guanche Adh genes (black line) and divergence between D. subobscura Adh-Twain and Adh (gray line). Both B and C suggest that, relative to Adh, a distinctly different set of nucleotides are under constraint in Adh-Twain.
TABLE 2.
Nucleotide polymorphism statistics forAdh-Twain
| Category | Result |
|---|---|
| No. of sequences | 15 |
| No. of bases surveyed | 1428 |
| Total sites (excluding gaps) | 1381 |
| No. of polymorphic (segregating) sites, S | 73 |
| No. of haplotypes, NHap | 14 |
| Haplotype (gene) diversity | 0.99 |
| Polymorphism | |
| Nucleotide diversity, π | 0.01378 |
| θ (per site) | 0.01626 |
| Average no. of nucleotide differences among D. subobscura alleles |
19.029 |
| Divergence | |
| No. of fixed differences between D. subobscura and D. guanche |
44 |
Several interesting patterns can be seen. First, polymorphism in the 5′-UTR is relatively low for a noncoding region, especially near regions hypothesized to play important regulatory roles. The most variable region is near the junction of CG9010-like and Adh-like sequences, in the area presumably derived from the Adh leader sequence. In contrast, DNA derived from the coding region of Adh has much lower levels of polymorphism. Patterns of polymorphism and divergence are correlated in Adh-Twain, consistent with the hypothesis of differing levels of functional constraint across the gene (see Figure 3A).
There are two in-frame indel polymorphisms in D. subobscura Adh-Twain, both of which are located near the junction of the CG9010-like and Adh-like regions (D1, bases 772–787 and D2, 814–820 on Figure 3). We used parsimony to infer that these were deletions relative to the ancestral CG9010 in D. subobscura and D. guanche (these amino acids are also present in CG9010 in D. melanogaster and D. pseudoobscura). Among the surveyed D. subobscura Adh-Twain alleles were three deletion haplotypes: (1) no deletions (1/15); (2) D2 only (3/15); and (3) D1 and D2 (11/15). Our D. madeirensis allele has both the D1 and the D2 deletions. D. guanche has neither of these deletions, but does have an in-frame 6-base deletion from base 889 to 895. All of these deletions occur in a region of high nucleotide polymorphism. This suggests that relative to most of the CG9010-like region and most of the Adh-like region, the intersection of these two regions is under relatively low constraint.
We also compared nucleotide divergence between the D. suboscura Adh-Twain and the ancestral D. suboscura CG9010 and Adh genes (gray lines in Figure 3, B and C). We looked at patterns of divergence between the D. subobscura and the D. guanche homologs of CG9010 and Adh (black lines in Figure 3, B and C). Comparison of these two sets of data (black vs. gray lines in Figure 3, B and C) shows a clear disconnect between the sites that tend to diverge between species in the parental genes and those that diverge between the Adh-Twain regions and their parental genes. This result, combined with the correlation between the polymorphism and divergence in Adh-Twain, suggests a shift in the sites that are conserved in Adh-Twain.
As initially reported by Luque et al. (1997), the Adh region of Adh-Twain shows more codon bias than does Adh in D. subobscura. Table 3 shows that the entire Adh-Twain has substantial codon bias in all three species. Relative to other genes in the obscura group, Adh-Twain appears to be a highly biased gene (Powell 1997). Codon bias, however, has decreased in the CG9010 region of Adh-Twain, relative to CG9010.
TABLE 3.
Codon bias ofAdh-Twain has changed relative to parental genes
| Region | Species | ENC | CBI | Nucleotides |
|---|---|---|---|---|
| CG9010 | D. guanche | 37.3 | 0.679 | 288 |
| D. subobscura | 37.8 | 0.679 | 288 | |
| CG9010 | D. guanche | 41.8 | 0.561 | 288 |
| D. madeirensis | 42.4 | 0.581 | 288 | |
| D. subobscuraa | 38.6 | 0.617 | 288 | |
| Adh | D. guanche | 48.1 | 0.456 | 762 |
| D. madeirensis | 47.9 | 0.449 | 762 | |
| D. subobscura | 46.8 | 0.455 | 762 | |
| Adh | D. guanche | 42.0 | 0.530 | 762 |
| D. madeirensis | 40.6 | 0.533 | 762 | |
| D. subobscuraa | 41.7 | 0.534 | 762 | |
| Allb | D. subobscuraa | 43.7 | 0.479 | 1409 |
| Adh-Twain | D. guanche | 41.5 | 0.496 | 1050 |
| D. madeirensis | 43.7 | 0.485 | 1050 | |
| D. subobscuraa | 43.7 | 0.479 | 1050 |
ENC, effective number of codons; CBI, codon bias index.
Population mean of D. subobscura alleles.
GC content of this region is 53%.
Patterns of recent molecular evolution in Adh-Twain:
The McDonald-Kreitman (MK) test is commonly used to detect directional selection acting on amino acid sequences (McDonald and Kreitman 1991). We used polymorphism data from D. subobscura and divergence data from all three species in our tests. Table 4 shows that most analyses do not reject neutral evolution. The unpolarized MK test of D. subobscura and D. guanche for the Adh-derived region of Adh-Twain is significant at a P < 0.05 threshold without correcting for multiple tests. If we restrict our analysis to only fixations that occurred along the D. subobscura lineage—as estimated by PAML as well as by parsimony (see materials and methods)—the MK test is no longer significant. In short, comparisons of the patterns of divergence and polymorphism among extant Adh-Twain homologs shows no evidence of recent adaptive evolution.
TABLE 4.
Tests functional constraint and adaptation inAdh-Twain
| Fixed nonsynonymous |
Nonsynonymous polymorphism |
Fixed synonymous |
Synonymous polymorphism |
Fisher exact P-value |
G-test | |
|---|---|---|---|---|---|---|
| Unpolarized MK tests | ||||||
| Adh region of Adh-Twain | ||||||
| D. subobscura vs. D. guanche | 22 | 6 | 13 | 15 | 0.0159 | 0.0130 |
| D. subobscura vs. D. madeirensis | 4 | 6 | 6 | 15 | 0.6854 | 0.5248 |
| CG9010 region | ||||||
| D. subobscura vs. D. guanche | 17 | 20 | 5 | 8 | 0.7510 | 0.6400 |
| D. subobscura vs. D. madeirensis | 7 | 20 | 2 | 8 | 0.9999 | 0.7866 |
| Whole gene | ||||||
| D. subobscura vs. D. guanche | 39 | 26 | 18 | 23 | 0.1149 | 0.1055 |
| D. subobscura vs. D. madeirensis | 11 | 26 | 8 | 23 | 0.7905 | 0.7195 |
| Polarized MK tests | ||||||
| D. subobscura Adh region vs. D. guanche Adh | 7 | 6 | 7 | 15 | 0.2882 | 0.1987 |
| D. subobscura CG9010 vs. D. guanche CG9010 | 12 | 20 | 1 | 8 | 0.2283 | 0.1328 |
| D. subobscura Adh-Twain vs. D. guanche Adh-Twain | 19 | 26 | 8 | 23 | 0.1544 | 0.1417 |
| Tests of Adh-Twain vs. ancestors | ||||||
| Adh region vs. Adh | 26 | 6 | 18 | 15 | 0.0332 | 0.0214 |
| CG9010 region vs. CG9010 | 29 | 20 | 8 | 8 | 0.5702 | 0.5195 |
| Polarized | ||||||
| Adh region vs. Adh | 21 | 6 | 7 | 15 | 0.0017 | 0.0012 |
| CG9010 region vs. CG9010 | 25 | 20 | 4 | 8 | 0.2070 | 0.1713 |
Kern et al. (2002) recently proposed a method for detecting directional selection using polymorphism data combined with two outgroup sequences. We used PAML to reconstruct ancestral sequences for Adh-Twain (see materials and methods). Using this approach we could analyze all nucleotide fixations that had occurred along the D. subobscura lineage since the D. subobscura-D. madeirensis speciation event. Consistent with the MK test data, we found no evidence for recent directional selection driving these fixations (analysis not shown).
Rapid early evolution of Adh-Twain:
Given the striking differences in the spatial patterns of divergence between Adh-Twain and its CG9010 and Adh ancestors (Figure 3, B and C), we also applied a contingency table analysis to compare the Adh-Twain divergence and polymorphism to that of its ancestor genes (Table 4). This is not a canonical MK test as it compares paralogous loci within a species, rather than homologous loci between species. A significant disconnect between polymorphism and divergence does not mean we can reject the neutral model, as we cannot rule out a fundamental shift in the neutral or nearly neutral mutation rate for one or more of these genes after the duplication events. Nevertheless, this is a useful way to detect a substantial change in the substitution rate at nonsynonymous or synonymous sites. The data indicate that there is no significant difference between the CG9010 ancestor and the CG9010-derived part of Adh-Twain. The Adh region, however, is marginally significant in the unpolarized comparison and highly significant in the polarized comparison. In the polymorphism data, there are ∼2.5 synonymous polymorphisms per nonsynonymous polymorphism. In contrast, in the polarized divergence data we estimate only 0.333 synonymous fixations per nonsynonymous fixation—an eightfold difference. By comparison, Adh is estimated to have 2.6 synonymous fixations for every nonsynonymous fixation. Fisher's exact test shows that the ratio of synonymous to nonsynonymous fixations in Adh is not significantly different from the ratio of synonymous to nonsynonymous polymorphisms in Adh-Twain (P = 0.999). Both of these ratios are strikingly different from the ratio of synonymous to nonsynonymous fixations in the Adh-like regions of Adh-Twain (Adh-Twain fixations vs. Adh-Twain polymorphism, P = 0.0017; Adh-Twain fixations vs. Adh fixations, P = 0.0024). These highly significant results suggesting rapid amino acid divergence of Adh-Twain from its paralog Adh contrasts with our earlier comparisons of orthologous Adh-Twain sequences. From these results, we hypothesize that adaptive protein evolution in Adh-Twain occurred early in its history, prior to the speciation events leading to D. subobscura/D. guanche/D. madeirensis.
As the comparison of paralogous genes differs from the comparison of orthologous genes in many ways (e.g., time, genomic location), we applied the more suitable phylogenetic approach implemented in PAML (Yang 1997) to test the hypothesis that Adh-Twain evolved rapidly shortly after its formation (solid bars in Figure 4). Specifically, we estimated the nonsynonymous to synonymous rate ratio (dN/dS) for various branches of the Adh-Twain gene tree. In these analyses we separately investigated CG9010 and Adh, the corresponding regions of Adh-Twain.
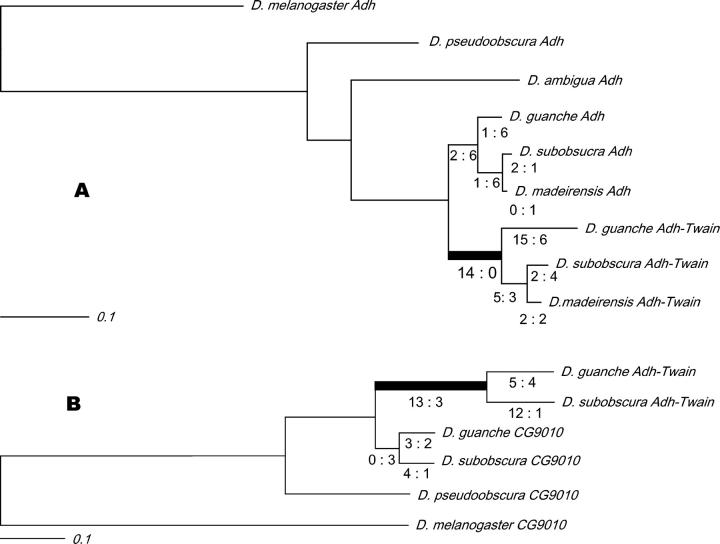
Figure 4.—
Nucleotide substitutions in Adh and CG9010 regions of Adh-Twain and their parental genes. These results are based on our PAML analysis. The solid bars represent the time period during which we hypothesize that directional selection acted on Adh-Twain. Estimates of the number of nonsynonymous and synonymous changes are beneath the branches of the Adh-Twain lineages. In both the Adh-derived and the CG9010-derived regions of the fusion gene there has been an increase in the rates of amino acid substitution, but not synonymous substitutions, relative to the ancestral genes.
We followed Yang (1998) and Yang et al. (2000) to test for rate heterogeneity between CG9010 and the CG9010-derived region of Adh-Twain (see materials and methods). Specifically, we compared the fit of the data to three hypotheses: (1) all branches were evolving at the same rate (“one-ratio” model 0 in PAML), (2) all branches are evolving at different rates (“free-ratio” model 1), and (3) the branches after the formation of Adh-Twain are evolving differently from the rest (“branch-specific” model 2). The free-ratio model fit the data better than the one-ratio model (model 0, ln l = −951.5; model 1, lnl = −937.1; 2Δlnl = 28.8, d.f. = 9, P = 0.0007). Clearly, the model of a single rate for dN/dS does not fit the CG9010 data.
The branch-specific model, which has one dN/dS rate for the fusion gene-related lineages and one rate for all other lineages, is a significantly better fit to the data than the one-ratio model (model 0 vs. model 2, lnl −943.1; 2Δlnl = 16.8, d.f. = 1, P < 0.0001). In contrast, the free-ratio model does not fit the data better than the two-ratio branch-specific model (2Δlnl = 12, d.f. = 8, P = 0.1512). A three-ratio branch-specific model—in which there is one rate from the branch immediately after the duplication of CG9010, one rate from the branches after the D. subobscura-D. guanche speciation, and one rate for all others—does not fit the data better than the two-ratio model (2Δlnl = 0.2, d.f. = 1, P = 0.62). Interestingly, a four-ratio model, where all branches after the CG9010 duplication are free to vary their dN/dS ratio, fits the data marginally better than the two-ratio model (2Δlnl = 7.4, d.f. = 2, P = 0.024). We are cautious of this later result given the large number of tests performed. Figure 4 illustrates these results.
Not surprisingly, dN/dS is small for CG9010 in most lineages (Figure 4). However, the dN/dS ratio of the CG9010-derived part of Adh-Twain early in its history is close to 1 (dN/dS = 0.9919). The D. guanche branch is clearly evolving slowly and is consistent with functional constraint (dN/dS = 0.3313). The only evidence for dN/dS significantly greater than 1 in the CG9010-derived portion of Adh-Twain is found in the D. subobscura lineage, which has a dN/dS of 2.09. This result is consistent with directional selection acting on the CG9010 portion of the D. subobscura Adh-Twain. This result, however, must be interpreted with caution as the dN/dS ratio is still close to 1 and MK tests described above—which also take into account polymorphism data—were not significant.
We repeated the above analysis for the Adh-derived region of Adh-Twain. Again, the free-ratio model fit the data substantially better than the one-ratio model (model 0, lnl = −2128.27; model 1, lnl = −2083.60; 2Δlnl = 89.34, d.f. = 15, P > 0.0001). We compared the one-ratio and the free-ratio models to several different models of sequence evolution for the Adh-Twain lineages. First, we compared a two-ratio model, with one dN/dS ratio for the Adh-Twain branch and one ratio for all other lineages. The two-ratio model fit better than the one-ratio model, but the free-ratio model fit slightly better than the two-ratio model (two-ratio model vs. model 0, two-ratio model lnl = −2097.46; 2Δlnl = 60.62, d.f. = 1, P > 0.0001; two-ratio model vs. model 1, 2Δlnl = 27.7, d.f. = 14, P = 0.0156). A three-ratio model, with rapid evolution after the formation of Adh-Twain and then a subsequent slowing down after the D. subobscura-D. guanche speciation, fit better than the two-ratio model (three-ratio model vs. two-ratio model, three-ratio model lnl = −2093.19; 2Δlnl = 8.54, d.f. = 1, P = 0.0035). Interestingly, the free-ratio model does not fit the data significantly better than the three-ratio model (2Δlnl = 19.18, d.f. = 13, P = 0.1176). We also compared several other models, none of which were significant improvements over the three-ratio model (analysis not shown).
For the three-ratio model, the dN/dS ratio for the branch immediately after the formation of Adh-Twain cannot be calculated, as dS is 0. This implies a dN/dS ratio much greater than 1. In contrast, dN/dS ratio for the Adh branches is 0.0411. The likely nonsynonymous and synonymous substitutions along this branch were inferred with PAML. There were at least 14 nonsynonymous amino acid substitutions and 0 synonymous silent substitutions, strongly suggesting a bout of rapid adaptive amino acid evolution shortly after the formation of Adh-Twain. This is consistent with our earlier contingency table analysis. Moreover, the ratio of nonsynonymous substitutions to synonymous substitutions for this early branch is dramatically different from the ratio of nonsynonymous substitutions to synonymous substitutions observed in the D. subobscura polymorphism data, P < 0.0001. Interestingly, the fact that no silent substitutions occurred between the retrotransposition of the Adh mRNA and the speciation of D. guanche and D. subobscura hints that Adh-Twain may have formed shortly before this speciation event.
dN/dS is 0.3991 for Adh-Twain after the speciation event leading to D. guanche and D. subobscura. This, while greater than is typical for Adh, is not suggestive of adaptive evolution and is consistent with our earlier MK test analysis.
DISCUSSION
Adh-Twain in D. guanche, D. madeirensis, and D. subobscura:
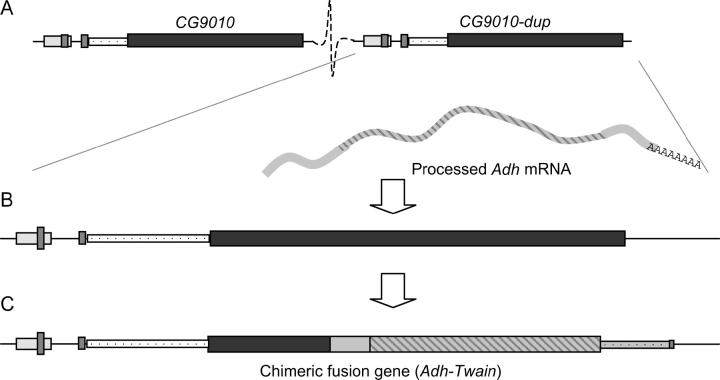
The gene originally hypothesized by Luque et al. (1997) to be an Adh retropseudogene in D. subobscura and two of its close relatives is instead a recently evolved chimeric fusion gene. We have named this gene Adh-Twain. Figure 5 summarizes our model for the origin and evolution of Adh-Twain. A chromosomal duplication of CG9010 and its 5′ regulatory sequence occurred prior to speciation of D. subobscura, D. madeirensis, and D. guanche. A processed Adh mRNA subsequently retrotransposed into one of the copies of CG9010. The CG9010 region and the Adh region were fused into the contiguous open reading frame that exists today as a result of either the original retrotransposition event or this insertion event and subsequent evolution.
Figure 5.—
A model for the evolution of Adh-Twain. (A) Chromosomal duplication of CG9010 in the ancestor of D. subobscura, D. madeirensis, and D. guanche. (B) Retrotransposition of an Adh mRNA into one of the copies of CG9010. Note that the 5′ regulatory regions of CG9010 are conserved. (C) Subsequent formation of a contiguous ORF spanning the remainder of CG9010 and the Adh retrosequence. The 3′ end of CG9010 is lost.
The data suggest that CG9010 duplicated prior to the origin of the fusion gene, but after D. subobscura and its relatives split off from D. pseudoobscura. One of these copies subsequently fused with the Adh retrosequence, to produce the fusion gene, Adh-Twain. We do not know precisely when CG9010 duplicated. The fact that only one copy of CG9010 is found in D. pseudoobscura sets an upper bound for the CG9010 duplication of ∼8.0–12.0 million years ago (MYA) and a lower bound of 1.8–2.8 MYA (Ramos-Onsins et al. 1998). Second, we do not know which copy of CG9010 participated in the fusion event. We found no evidence for DNA sequence homologous to the 3′ end of CG9010 in the sequence data from the fusion gene. This is consistent with two hypotheses. Either the CG9010 target of the Adh retrosequence was a truncated, nonfunctional copy (e.g., Katju and Lynch 2003) or the 3′ end of the CG9010 target is no longer recognizable as a result of extensive molecular evolution.
Several puzzling results from the Luque et al. (1997) original data, including high codon bias and low dN/dS ratio compared to the expectation for a putative pseudogene, are explained by our data. The fact that the putative initiation codon of the D. guanche Adh retrosequence was CTG rather than ATG (which was interpreted as evidence of loss of function in Adh) is now explained by the fact that the actual initiation codon is upstream of this codon and is derived from the duplicated CG9010. Transcription patterns of the CFG were qualitatively similar to those of CG9010. This is consistent with conservation of putative 5′ regulatory elements between Adh-Twain and CG9010. Bioinformatic analysis of Adh-Twain suggests that the fusion protein may have lost or reduced ancestral ADH activity, which is consistent with the negative results of our experimental work. The function of the Adh-Twain protein, however, remains unknown.
Adaptive protein evolution:
In general, patterns of polymorphism and divergence at Adh-Twain provide no strong evidence for recent directional selection in D. subobscura. Our analysis of substitution patterns along branches of the genealogy suggests that a “three-ratio” model has the greatest likelihood. In this model, Adh is normally very evolutionarily constrained. Shortly after Adh-Twain was formed, but prior to the splitting of the lineages leading to D. subobscura and D. guanche, there was a burst of adaptive substitutions in the Adh region. Subsequently, the Adh region of the fusion gene has slowed in its rate of evolution, although not nearly as slow as Adh and not along all lineages (e.g., D. guanche).
A strikingly similar pattern of rapid amino acid evolution shortly after the formation of jingwei was reported by Long and Langley (1993). Interestingly, one of the early, likely adaptive, amino acid changes in Adh-Twain (H190 to Q) occurs at a residue known to affect Adh activity in D. melanogaster mutants (http://www.flybase.org). This result is consistent with the Adh allozyme data from the obscura group, which provided no evidence for canonical Adh activity associated with the fusion gene. This may mean that the enzymatic activity of Adh-Twain has shifted away from secondary alcohols typically catalyzed by Drosophila Adh.
The history of the CG9010-derived region of Adh-Twain is less clear. We showed that, unlike the Adh region, a “two-ratio” model was most likely, given our data. There is strong evidence for rapid evolution early in the history of Adh-Twain. There is also weak evidence that the CG9010-derived region is adaptively evolving along the D. subobscura lineage. However, the strong signal of directional selection—many more amino acid changes than silent changes—observed in the early evolution of the Adh region is obscured in the CG9010 region. Instead, the rates of both nonsynonymous and synonymous substitutions were elevated early in the history of this gene, followed by increasing constraint. The simplest interpretation is that the copy of CG9010 that ultimately became part of Adh-Twain evolved quickly under reduced functional constraint right after the initial duplication event, but then became more constrained once it was fused with the Adh retrogene.
Retrogene evolution:
If a retrogene is to preserve an ancestral function or acquire a new function, it must avoid premature termination codons and be properly expressed. For CFGs originating by retrotransposition of a “donor” gene into a preexisting “acceptor” gene, reading frame preservation implies an insertion site between acceptor gene codons. Furthermore, it implies removal or avoidance of in-frame stop codons in the 5′-UTR of the donor retrosequence. It is also possible that some ultimately successful fusion genes originating by retrotransposition go through an early stage of loss-of-function prior to restoration of function by new mutations. Distinguishing between these possibilities is problematic as we cannot accurately reconstructing the details of the ancestral state of the insertional mutation (e.g., the retrotransposition of the Adh mRNA in the case of Adh-Twain) in all but the most recently derived CFGs. Nevertheless, the fact that Adh-Twain showed early rapid amino acid evolution and no silent substitutions suggests that this gene was functional immediately after it was formed (Long and Langley 1993 reached a similar conclusion for jingwei).
Adh and the origins of new genes:
Recent studies in Drosophila and mammals have shown that retrotransposition of spliced mRNA's is a potent source of gene duplications (Eichler 2001; Betran et al. 2002; Devor and Moffat-Wilson 2003; Long et al. 2003a,b; Emerson et al. 2004). Long and Langley (1993) showed that retrotransposition events such as these can result in the formation of new CFGs, e.g., jingwei. The discovery of three novel Adh-derived Drosophila genes, Adh-Twain, jingwei, and Adh-Finnegan, strongly suggests that Adh-derived novel genes are common in flies.
What aspects of Adh biology might contribute to this phenomenon? Adh in D. melanogaster and other Drosophila can catalyze a variety of substrates ranging from various alcohols to a number of aldehydes (Ashburner 1998). Perhaps the frequency of Adh duplications and Adh-derived CFGs reflects the flexibility and evolutionary potential of the Adh protein or the fact that Adh is one of the most abundant transcripts in Drosophila. Other factors relating to the structure and molecular biology of Adh may make it more likely to participate in retrotransposition-mediated novel gene formation. For example, Goncalves et al. (2000) identified common characteristics of genes that were the source of retropseudogenes in humans. The coding sequences of the parental genes tended to be relatively short, expressed in a variety of tissues, and have a low G/C content. Their amino acid sequences were highly conserved. Nearly a quarter of these genes produced more than one retropseudogene. Consistent with these observations, the 23 recent retrotransposed genes in D. melanogaster identified by Betran et al. (2002) are shorter than the average D. melanogaster gene (mean retrotransposed gene length, 320 amino acids (aa); mean gene length, 522 aa; median retrotransposed gene length, 230 aa; median gene length, 421 aa). The parental genes of these retrotransposed genes are also often expressed in multiple tissues (Betran et al. 2002). Adh in Drosophila qualitatively fits several of the above patterns as it is relatively short, widely expressed, and highly conserved and has duplicated several times in different Drosophila lineages (Yum et al. 1991; Nurminsky et al. 1996; Powell 1997; Amador and Juan 1999). In any event, the discovery of the obscura Adh-derived chimeric fusion gene strongly motivates the search for additional novel Adh genes and opens up the possibility of discovering general principles governing the evolution of novel genes.
Acknowledgments
We thank C. Benyajati (and Uncle Billy) for the Adh antibody. We also are grateful to A. Davis, M. Aguadé, and the Drosophila Species Stock Center for flies. We thank S. Shih for technical assistance and D. Barbash for answering many technical questions. We thank M. Long for discussing his unpublished data with us. We also thank I. Dworkin, A. Kern, C. Langley, M. Lawniczak, S. Nuzhdin, T. Schlenke, and B. Wagstaff for discussion. We are grateful to A. Kern and I. Dworkin for comments on the manuscript. We also thank two thoughtful reviewers for many helpful suggestions. D. subobscura cDNA and genomic libraries are freely available from C.D.J. This work was funded by National Science Foundation grants to C.D.J. and D.J.B.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador, A., and E. Juan, 1999. Nonfixed duplication containing the Adh gene and a truncated form of the Adhr gene in the Drosophila funebris species group: different modes of evolution of Adh relative to Adhr in Drosophila. Mol. Biol. Evol. 16: 1439–1456. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., 1989 Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ashburner, M., 1998. Speculations on the subject of alcohol dehydrogenase and its properties in Drosophila and other flies. BioEssays 20: 949–995. [DOI] [PubMed] [Google Scholar]
- Balanya, J., C. Segarra, A. Prevosti and L. Serra, 1994. Colonization of America by Drosophila subobscura: the founder event and a rapid expansion. J. Hered. 85: 427–432. [DOI] [PubMed] [Google Scholar]
- Batterham, P., J. A. Lovett, W. T. Starmer and D. T. Sullivan, 1983. Differential regulation of duplicate alcohol dehydrogenase genes in Drosophila mojavensis. Dev. Biol. 96: 346–354. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., 1997. Origin and evolution of a new gene descended from alcohol dehydrogenase in Drosophila. Genetics 145: 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran, E., and M. Long, 2003. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics 164: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran, E., K. Thornton and M. Long, 2002. Retroposed new genes out of the X in Drosophila. Genome Res. 12: 1854–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz, J. O., D. Liang, D. Plouffe, H. S. Chang, T. Zhu et al., 2003. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 13: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, T. W., and J. T. Kadonaga, 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11: 3020–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, J. A., M. Ramon, A. Picornell and A. Moya, 1999. The genetic structure of Drosophila subobscura populations from the islands of Majorca and Minorca (Balearic Islands, Spain) based on allozymes and mitochondrial DNA. Heredity 83: 271–279. [DOI] [PubMed] [Google Scholar]
- Courseaux, A., and J. L. Nahon, 2001. Birth of two chimeric genes in the Hominidae lineage. Science 291: 1293–1297. [DOI] [PubMed] [Google Scholar]
- Devor, E. J., and K. A. Moffat-Wilson, 2003. Molecular and temporal characteristics of human retropseudogenes. Hum. Biol. 75: 661–672. [DOI] [PubMed] [Google Scholar]
- Eichler, E. E., 2001. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 17: 661–669. [DOI] [PubMed] [Google Scholar]
- Emerson, J. J., H. Kaessmann, E. Betran and M. Long, 2004. Extensive gene traffic on the mammalian X chromosome. Science 303: 537–540. [DOI] [PubMed] [Google Scholar]
- Finta, C., and P. G. Zaphiropoulos, 2000. The human cytochrome P450 3A locus. Gene evolution by capture of downstream exons. Gene 260: 13–23. [DOI] [PubMed] [Google Scholar]
- Fischer, J. A., and T. Maniatis, 1985. Structure and transcription of the Drosophila mulleri alcohol dehydrogenase genes. Nucleic Acids Res. 13: 6899–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, N., and Z. Yang, 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11: 725–736. [DOI] [PubMed] [Google Scholar]
- Goncalves, I., L. Duret and D. Mouchiroud, 2000. Nature and structure of human genes that generate retropseudogenes. Genome Res. 10: 672–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, N. K., and M. Wickens, 1998. Control of translation in animals. Annu. Rev. Cell Dev. Biol. 14: 399–458. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1932 The Causes of Evolution. Longmans Green & Co., London.
- Harrison, P. M., A. Kumar, N. Lang, M. Snyder and M. Gerstein, 2002. A question of size: the eukaryotic proteome and the problems in defining it. Nucleic Acids Res. 30: 1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, S., A. R. Abad, S. B. Gelvin and S. A. Mackenzie, 1996. A cytoplasmic male sterility-associated mitochondrial protein causes pollen disruption in transgenic tobacco. Proc. Natl. Acad. Sci. USA 93: 11763–11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, P. W., 2003. More genes in vertebrates? J. Struct. Funct. Genomics 3: 75–84. [PubMed] [Google Scholar]
- Hughes, A., 2002. Adaptive evolution after gene duplication. Trends Genet. 18: 433–434. [DOI] [PubMed] [Google Scholar]
- Jain, R., M. C. Rivera and J. A. Lake, 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA 96: 3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffs, P., and M. Ashburner, 1991. Processed pseudogenes in Drosophila. Proc. R. Soc. Biol. Sci. 44: 151–159. [DOI] [PubMed] [Google Scholar]
- Katju, V., and M. Lynch, 2003. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics 165: 1793–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern, A. D., C. D. Jones and D. J. Begun, 2002. Genomic effects of nucleotide substitutions in Drosophila simulans. Genetics 162: 1753–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, M., and C. H. Langley, 1993. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260: 91–95. [DOI] [PubMed] [Google Scholar]
- Long, M., M. Deutsch, W. Wang, E. Betran, F. G. Brunet et al., 2003. a Origin of new genes: evidence from experimental and computational analyses. Genetica 118: 171–182. [PubMed] [Google Scholar]
- Long, M., E. Betran, K. Thornton and W. Wang, 2003. b The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 4: 865–875. [DOI] [PubMed] [Google Scholar]
- Loukas, M., C. B. Krimbas, P. Mavragani-Tsipidou and C. D. Kastritsis, 1979. Genetics of Drosophila subobscura populations. VIII. Allozyme loci and their chromosome maps. J. Hered. 70: 17–26. [DOI] [PubMed] [Google Scholar]
- Luque, T., G. Marfany and R. Gonzalez-Duarte, 1997. Characterization and molecular analysis of Adh retrosequences in species of the Drosophila obscura group. Mol. Biol. Evol. 14: 1316–1325. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2003. The origins of genome complexity. Science 302: 1401–1404. [DOI] [PubMed] [Google Scholar]
- Lynch, M., M. O'Hely, B. Walsh and A. Force, 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159: 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfany, G., and R. Gonzalez-Duarte, 1992. Evidence for retrotranscription of protein-coding genes in the Drosophila subobscura genome. J. Mol. Evol. 35: 492–501. [DOI] [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., J. Engelbrecht, S. Brunak and G. von Heijne, 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8: 581–599. [DOI] [PubMed] [Google Scholar]
- Nurminsky, D. I., E. N. Moriyama, E. R. Lozovskaya and D. L. Hartl, 1996. Molecular phylogeny and genome evolution in the Drosophila virilis species group: duplication of the alcohol dehydrogenase gene. Mol. Biol. Evol. 13: 132–149. [DOI] [PubMed] [Google Scholar]
- Nurminsky, D. I., M. V. Nurminskaya, D. De Aguiar and D. L. Hartl, 1998. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature 396: 572–575. [DOI] [PubMed] [Google Scholar]
- Ochman, H., and I. B. Jones, 2000. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 19: 6637–6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler, U., G. C. Liao, H. Niemann and G. M. Rubin, 2002 Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3: RESEARCH0087. [DOI] [PMC free article] [PubMed]
- Ohno, S., 1970 Evolution by Gene Duplication. Springer, Berlin.
- Ohta, T., 2003. Evolution by gene duplication revisited: differentiation of regulatory elements versus proteins. Genetica 118: 209–216. [PubMed] [Google Scholar]
- Patthy, L., 1999. Genome evolution and the evolution of exon-shuffling: a review. Gene 238: 103–114. [DOI] [PubMed] [Google Scholar]
- Powell, J. R., 1997 Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford University Press, Oxford.
- Ramos-Onsins, S., C. Segarra, J. Rozas and M. Aguade, 1998. Molecular and chromosomal phylogeny in the obscura group of Drosophila inferred from sequences of the rp49 gene region. Mol. Phylogenet. Evol. 9: 33–41. [DOI] [PubMed] [Google Scholar]
- Reese, M. G., 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26: 51–56. [DOI] [PubMed] [Google Scholar]
- Rogalla, P., B. Kazmierczak, A. M. Flohr, S. Hauke and J. Bullerdiek, 2000. Back to the roots of a new exon—the molecular archaeology of a SP100 splice variant. Genomics 63: 117–122. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Sullivan, D. T., W. T. Starmer, S. W. Curtiss, M. Menotti-Raymond and J. Yum, 1994. Unusual molecular evolution of an Adh pseudogene in Drosophila. Mol. Biol. Evol. 11: 443–458. [DOI] [PubMed] [Google Scholar]
- Thomson, T. M., J. J. Lozano, N. Loukili, R. Carrio, F. Serras et al., 2000. Fusion of the human gene for the polyubiquitination coeffector UEV1 with Kua, a newly identified gene. Genome Res. 10: 1743–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., M. B. Traw, J. Q. Chen, M. Kreitman and J. Bergelson, 2003. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77. [DOI] [PubMed] [Google Scholar]
- Visa, N., G. Marfany, L. Vilageliu, R. Albalat, S. Atrian et al., 1991. The Adh in Drosophila: chromosomal location and restriction analysis in species with different phylogenetic relationships. Chromosoma 100: 315–322. [DOI] [PubMed] [Google Scholar]
- Vivas, M. V., J. Garcia-Planells, C. Ruiz, G. Marfany, N. Paricio et al., 1999. GEM, a cluster of repetitive sequences in the Drosophila subobscura genome. Gene 229: 47–57. [DOI] [PubMed] [Google Scholar]
- Wagner, A., 2001. Birth and death of duplicated genes in completely sequenced eukaryotes. Trends Genet. 17: 237–239. [DOI] [PubMed] [Google Scholar]
- Wang, W., J. Zhang, C. Alvarez, A. Llopart and M. Long, 2000. The origin of the Jingwei gene and the complex modular structure of its parental gene, yellow emperor, in Drosophila melanogaster. Mol. Biol. Evol. 17: 1294–1301. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., and W. H. Li, 2003. Molecular evolution meets the genomics revolution. Nat. Genet. 33: 255–265. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15: 568–573. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and J. P. Bielawski, 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15: 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., W. J. Swanson and V. D. Vacquier, 2000. Maximum-likelihood analysis of molecular adaptation in abalone sperm lysin reveals variable selective pressures among lineages and sites. Mol. Biol. Evol. 17: 1446–1455. [DOI] [PubMed] [Google Scholar]
- Yum, J., W. T. Starmer and D. T. Sullivan, 1991. The structure of the Adh locus in Drosophila mettleri; an intermediate in the evolution of the Adh locus in the repleta group of Drosophila. Mol. Biol. Evol. 8: 857–867. [DOI] [PubMed] [Google Scholar]
- Zhang, J., A. M. Dean, F. Brunet and M. Long, 2004. Evolving protein functional diversity in new genes of Drosophila. Proc. Natl. Acad. Sci. USA 101: 16246–16250. [DOI] [PMC free article] [PubMed] [Google Scholar]