Abstract
The rRNA gene (rDNA) loci of all arthropod lineages contain non-LTR retrotransposable elements that have evolved to specifically insert into the 28S rRNA genes. Extensive in vitro experiments have been conducted to investigate the mechanism of R2 retrotransposition but little is known of the insertion frequency or cellular factors that might regulate R2 activity. In this article, isofemale lines obtained from a population of Drosophila simulans were surveyed for recent R2 insertions. Within most lines, all individuals showed the same collection of R2 insertions, providing no evidence for recent R2 activity. However, in a few of the isofemale lines, virtually all individuals differed in their R2 insertion profiles. The descendants of individual pairs of flies from these “active lines” rapidly accumulated new insertions. The frequent insertion of new R2 elements was associated with the elimination of old R2 elements from the rDNA locus. The existence of lines in which R2 retrotransposes frequently and lines in which the elements appear dormant suggests that cellular mechanisms that can regulate the activity of R2 exist. Retrotransposition activity was correlated with the number of full-length R2 elements but not with the size of the rDNA locus or the number of uninserted units.
TRANSPOSABLE elements compose a significant proportion of eukaryotic genomes and have played an important role in their evolution. To understand the dynamics of transposable element insertions, attempts have been made, most often in the genus Drosophila, to monitor the insertion of transposable elements over time. Most studies have detected no activity or activity on the order of 10−5–10−3 new insertions per copy per generation (Nuzhdin and Mackay 1995; Dominguez and Albornoz 1996; Maside et al. 2001). Higher rates of new insertion have been identified in dysgenic crosses (Kidwell et al. 1977; Bucheton 1990; Petrov et al. 1995) or in isolated laboratory stocks (Prud'homme et al. 1995; Desset et al. 2003; Houle and Nuzhdin 2004), which suggests that transposable element activity is normally restrained. It is not known what percentage of the transposition events in a population occurs by elements occasionally escaping this regulation and what percentage occurs by the periodic breakdown of regulation in certain individuals, permitting high rates of transposition.
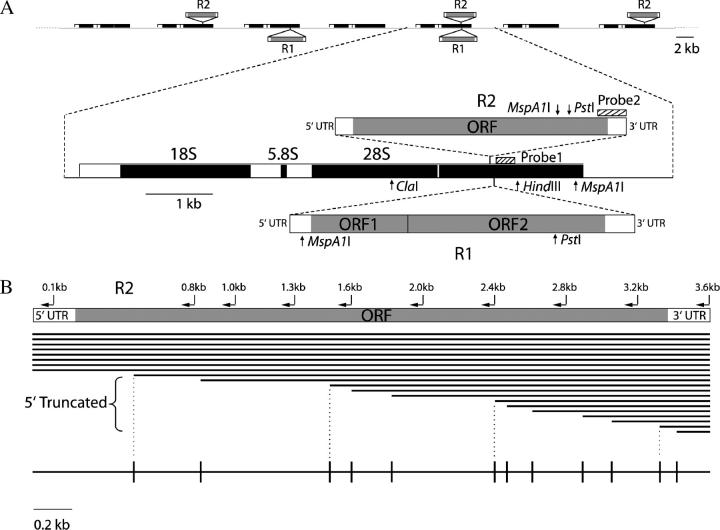
R1 and R2 elements are non-LTR retrotransposable elements that insert specifically into the 28S genes of the ribosomal RNA (rRNA) gene locus (Figure 1A). R1 and R2 elements exist in all lineages of arthropods (Jakubczak et al. 1991; Burke et al. 1998) as well as in other animal phyla (Kojima and Fujiwara 2004). When units within the rDNA array are inserted by R1 and/or R2, they can no longer synthesize functional 28S rRNA (Long and Dawid 1979; Jamrich and Miller 1984; Eickbush and Eickbush 2003). Despite this deleterious effect on the host, R1 and R2 elements appear to have been maintained in arthropods by vertical transmission since the origin of the phylum (Burke et al. 1998, 1999; Gentile et al. 2001). The long-term maintenance of R1 and R2 in arthropods is achieved without close regulation of the fraction of the rDNA units they occupy. The levels of R1 or R2 insertion can vary from only a small percentage to >70% of the total rDNA units in different Drosophila populations (Jakubczak et al. 1992; Malik and Eickbush 1998; Perez-Gonzalez and Eickbush 2001).
Figure 1.—
The rDNA locus and its R1 and R2 insertions. (A) The rDNA locus of D. simulans, as in most eukaryotes, is composed of a tandem array of rDNA units each encoding 18S, 5.8S, and 28S rRNA. Each rDNA unit can be inserted by R1, R2, or both elements. The ORFs and untranscribed region of the R1 and R2 elements are shown. Also shown are the locations of restriction sites for ClaI, HindIII, PstI, MspA1I and the positions of the probes used for the Southern blot experiments to determine the data in Table 1. (B) Sample summary diagram of the collection of 5′-truncated R2 elements present in the rDNA loci of the Paradise lines. The locations of R2 PCR primers used to determine the length of 5′-truncated and full-length R2 elements are shown above the diagram of the R2 element. The R2 copies in any organism are a collection of full-length copies and copies truncated at their 5′ end. The length of each copy is represented by the closely spaced horizontal lines. The 5′-truncated elements are usually present only at a single copy per rDNA locus (each copy has a unique length). Multiple full-length R2 elements of similar length are also present. Differences of these full-length elements can be detected on high-resolution gels (see Figure 4). A summary (truncation profile) of the 5′-truncated R2 copies in an rDNA locus is shown in the diagram at the bottom. The horizontal line represents the length of a full-length R2 and vertical lines represent the different 5′-truncated R2 copies.
Understanding how R1 and R2 are maintained requires information on how often these elements are inserted into and eliminated from the rDNA locus. Sequence analysis from many Drosophila species revealed low levels of R1 and R2 sequence variation, strong selection on their open reading frames, and few examples of elements containing mutations that disrupted their reading frames (Eickbush and Eickbush 1995; Eickbush et al. 1995; Lathe et al. 1995; Lathe and Eickbush 1997; Gentile et al. 2001). Population surveys in Drosophila melanogaster and D. simulans have suggested that recombination eliminates individual copies of R1 and R2 from the rDNA locus more often than it amplifies them (Jakubczak et al. 1992; Perez-Gonzalez and Eickbush 2001). The recombination bias against these elements suggests they must propagate by retrotransposition to remain within the rDNA locus. In the only studies to directly follow the retrotransposition of new R1 and R2 elements and the elimination of old copies, hundreds of new R1 insertions but few R2 insertions were observed in the Harwich mutation accumulation lines of D. melanogaster (Perez-Gonzalez and Eickbush 2002; Perez-Gonzalez et al. 2003). The identification of stocks in which new R2 integrations could be monitored in vivo would be of significant value to understand the dynamics of R2 retrotransposition and to complement our in vitro studies of the R2 integration mechanism (reviewed in Eickbush 2002).
In this article we have returned to a population of D. simulans in which a previous survey had suggested a rapid turnover of R2 insertions (Perez-Gonzalez and Eickbush 2001). Analysis of isofemale lines derived from this population revealed lines in which the R2 elements appeared completely inactive and lines where R2 frequently retrotransposed.
MATERIALS AND METHODS
Fly stock, PCR amplification, and R2 5′-truncation profiles:
The D. simulans isofemale lines were the gift of M. Turelli and have been previously described (Perez-Gonzalez and Eickbush 2001). Fifteen of the lines in the previous study were still in existence and were used in this report. The PCR protocol was the same as described in Perez-Gonzalez and Eickbush (2001) except that the Taq DNA polymerase and PCR buffer were from Promega (Madison, WI). Genomic DNA was extracted from single flies and prepared for PCR as described by Gloor et al. (1993). A forward primer, 5′-TGCCCAGTGCTCTGAATGTC-3′, which anneals to the 28S gene 80 bp upstream of the R2 insertion site, was used in combination with one of the following reverse primers: 5′-ATACCCACGCAGGTTCCGC-3′, 5′-CGCTGGACGACAGCATACTGC-3′, 5′-GAAGACGGTTCTGGCCAGTC-3′, 5′-TCGAATGCCTTGCTTACATC-3′, 5′-GATAGAAAATCCAACGTTCTGTC-3′, 5′-GTGCATGAATGAATGCTATC-3′, 5′-CCCCTTGTAGTACGAGACTTC-3′, 5′-GTCACCTGCGGCTTCGAATC-3′, and 5′-GTATGGAAATCTATCGAAAGATACT-3′, which bind, respectively, to R2 sequences 0.8, 1.0, 1.3, 1.6, 2.0, 2.4, 2.8, 3.2, and 3.6 kb from the 5′ end of a canonical full-length R2 element (see Figure 1B). To construct an R2 5′-truncation profile, DNA extracted from a single fly was PCR amplified with the above primer pairs and the PCR products were separated on 8.75% polyacrylamide gels. Lengths of the R2 5′ truncations were calculated on the basis of the sizes of the PCR products and the positions of the reverse primers. To avoid scoring spurious bands that are occasionally produced by PCR, every band was confirmed by two adjacent primers before being scored as an authentic R2 5′-truncated copy.
Determination of the number of full-length R2 elements:
To score full-length R2 elements, the primer (5′-GACTTGAGTAAAGGAGAGACT-3′) that annealed to the R2 sequence 123 bp downstream of the 5′ end of a full-length R2 was 32P-end-labeled (Perez-Gonzalez and Eickbush 2002) and used in combination with the primer complementary to the 28S gene 80 bp upstream of the R2 insertion site. The products were separated on an 8% high-voltage denaturing polyacrylamide gel, exposed in a PhosphorImager cassette, and quantitated in a Molecular Dynamics (Sunnyvale, CA) Storm Analyzer using ImageQuant version 1.2. Within each lane, the weakest bands were defined as representing single-copy elements; copy numbers of the remaining bands were estimated by dividing the signal of each band by the average signal of the single-copy bands.
Southern blots and the size of the rDNA locus:
Southern blots were conducted with genomic DNA extracted from 30 adults of each Paradise line. To determine the proportions of different types of rDNA units, genomic DNA was digested with ClaI, HindIII, and PstI, electrophoresed on a 1% agarose gel, transferred to a nitrocellulose membrane and probed with a 280-bp segment from the 28S gene (as described in Jakubczak et al. 1992) immediately downstream of the R1 insertion site (see Figure 1A for the position of the probe and the locations of restriction sites). Uninserted rDNA units, R1-inserted units (including units that have both R1 and R2 insertions), and R2-inserted units gave rise to 1.9-, 1.1-, and 0.9-kb fragments, respectively. Using the PhosphorImager and ImageQuant to analyze the gel, the percentages of these three rDNA unit types were calculated for each line. To determine the proportions of double-inserted rDNA units, the genomic DNA was digested with MspA1I and probed with a sequence from the 3′ end of the R2 element (as described in Perez-Gonzalez and Eickbush 2001). rDNA units inserted by R2 alone gave rise to a 2.3-kb fragment and units inserted with both a full-length R1 and either a full-length or a 5′-truncated R2 gave rise to a 1.2-kb fragment (see Figure 1A). The ratio of double- to single-inserted units was again quantitated with a PhosphorImager. By combining data from these two Southern blots, the proportions of uninserted units, units inserted by R1 alone, units inserted by R2 alone, and units inserted by both elements were calculated. The total number of rDNA units in the locus of each line was estimated on the basis of the number of R2 elements directly counted by virtue of their different 5′ ends (see above) divided by the fraction of the rDNA locus containing an R2 insertion.
RESULTS
New R2 insertions within the rDNA locus of an organism can be characterized even though all elements insert into the same site of the 28S gene because the mechanism of R2 insertion generates extensive sequence variants at their 5′ junction with the 28S gene. Variation is generated because after initiating cDNA synthesis at the 3′ end of the R2 RNA transcript, the R2 reverse transcriptase does not need to polymerize to the 5′ end for a complete integration reaction to occur (Luan et al. 1993). Thus the R2 copies in any rDNA locus are a collection of “full-length” copies and copies that are 5′ truncated at various positions along the length of the element (see Figure 1B). The different 5′-truncated R2 elements can be scored by PCR using one primer that binds to the 28S gene upstream of the R2 insertion site and a series of second primers that bind to different regions of R2 (Perez-Gonzalez and Eickbush 2001).
On the basis of an analysis of isofemale lines established from a population of D. simulans in Paradise, California, it was found that a majority of the 5′-truncated R2 insertions within this population were of unique length (i.e., correspond to a single copy per X chromosome) (Perez-Gonzalez and Eickbush 2001). In addition, the 5′-truncated copies found in each line were not extensively shared among the different isofemale lines. Thus the R2 truncation patterns in these lines indicated that most R2 insertions were sufficiently recent that they had neither spread by recombination to other chromosomes in the population nor been amplified by recombination to higher numbers on the same chromosome. Such rapid turnover of R2 elements can explain the <0.5% nucleotide sequence divergence found in the full-length and 5′-truncated R2 elements of all Drosophila species studied to date (Eickbush and Eickbush 1995; Lathe and Eickbush 1997).
If R2 insertions and eliminations continued while these isofemale lines were maintained as inbred stocks within the laboratory, then changes in the R2 profile over time would be expected. Therefore, ∼75 generations after the initial characterization of the lines, the PCR analysis was repeated.
Variation in the R2 5′-truncation profiles of the Paradise lines:
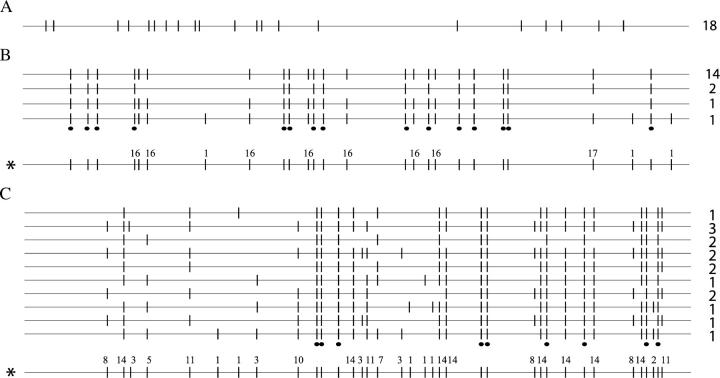
It is straightforward to score the R2 profile of individual rDNA loci in D. simulans because the rDNA units exist exclusively on the X chromosome; i.e., there are no units on the Y chromosome, unlike most other Drosophila species (Lohe and Roberts 1990; Mecheva and Semionov 1992). Therefore the R2 elements scored in individual D. simulans males from each Paradise line represented the R2 insertions on individual X chromosomes in that line. As examples of the variation found in the different Paradise lines, the profiles of the 5′-truncated R2 elements from three lines are shown in Figure 2. In line 1 (Figure 2A), the same 21 PCR fragments were found in all 18 males surveyed. The 5′ ends of the truncated R2 copies that gave rise to these fragments are indicated by the vertical lines on the horizontal line representing the length of the complete R2 element (see Figure 1B). All PCR products corresponding to 5′-truncated copies were of similar intensity, suggesting that they each correspond to a single R2 copy. Thus there was no evidence of recent R2 insertions, deletions, or amplifications in line 1.
Figure 2.—
R2 5′-truncation profiles of the X chromosome rDNA loci from Paradise lines 1, 31, and 58. rDNA units are only on the X chromosome of D. simulans; thus the R2 profiles of individual males represent the profiles of individual X chromosomes in that population. As diagrammed in Figure 1B, the horizontal line represents the length of the complete R2 element, with vertical lines representing the different 5′-truncated copies. (A) Line 1. All 18 males had the same profile containing 21 5′-truncated elements. (B) Line 31. Four different R2 truncation profiles were found in males from this line. The number to the right of each R2 truncation profile is the number of males in which the particular profile was found. The 5′-truncated elements marked with dots at the bottom represent R2 elements found in all males. (Bottom) A composite truncation profile, representing all R2 5′ truncations seen in this line, is shown. Polymorphic truncations (truncations that are not present in all males sampled) are marked with the number of males in which the particular truncation was found. (C) Line 58. Ten different R2 truncation profiles were identified. Nine truncation copies were present in all males while 27 truncated copies were polymorphic.
Unlike line 1, variation within the R2 insertion pattern was found in D. simulans line 31 (represented by the four horizontal lines in Figure 2B). While 14 males sampled had the same profile consisting of 23 5′-truncated copies, 2 males were missing 7 R2 copies found in all other males, and 1 male had 1 R2 copy missing. Finally, 1 male had 3 R2 copies that were not found on any other chromosome. Thus new R2 insertions and/or eliminations had occurred in this line since the last X chromosome fixation event. A composite R2 truncation profile is shown at the bottom of Figure 2B with the number of males in which each unfixed R2 copy was found indicated above the vertical line.
An even higher level of variation in R2 profiles was detected in line 58 (Figure 2C). Ten different X chromosomes were found with no chromosome type present in more than three of the males sampled. The number of 5′-truncated R2 copies on the X chromosomes in this line ranged from 14 to 26, while the number of differences in R2 elements between any two X chromosomes within this line ranged from 1 to 16. As shown by the composite R2 truncation profile, only 9 of 36 total R2 length variants were present in all males. This level of variation suggested many recent insertions and/or deletions in line 58.
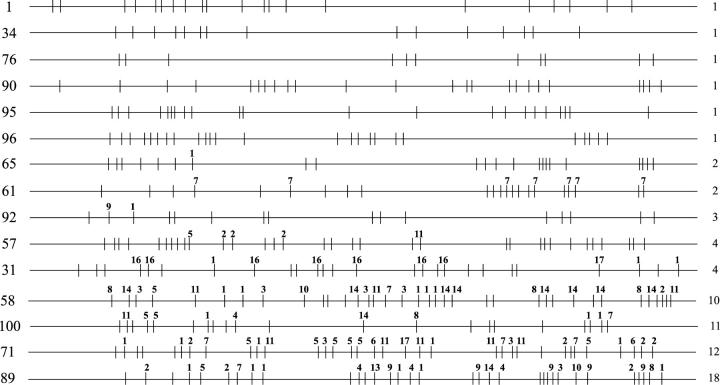
A similar analysis was done for the remaining 12 Paradise lines, and the composite R2 truncation profiles for all 15 lines are shown in Figure 3. The 15 lines could be divided into three groups on the basis of the level of R2 5′-truncation variation. The first group comprised 6 lines (top), in which all males within a line had identical R2 truncation profiles on their X chromosomes. The 11 (line 76) to 25 (line 96) 5′-truncated copies matched the profiles scored 75 generations earlier. Therefore, there was no evidence in this group for insertions or deletions over a 3-year period.
Figure 3.—
R2 truncation profiles of all Paradise lines. Numbers at left are isofemale line numbers. Numbers at right are the number of X chromosome R2 truncation profile types found in that line. In the first six Paradise lines shown, all males sampled from each line had identical 5′-truncation profiles. For the remaining lines the profiles presented are composite R2 truncation profiles. Polymorphic R2 truncations are marked with the number of males in which the particular truncation was found. The number of males sampled in each line was 18 except for the sampling of 11 males in line 65, 13 males in line 61, and 16 males in line 58.
The five paradise lines shown in the middle of Figure 3 composed a group in which two to four different X chromosome profiles were found among the males sampled from each line. These lines had from 14 to 28 5′-truncated R2 copies present on all X chromosomes and from 1 to 11 copies present on only a subset of the X chromosomes. Most of the variation in two lines (lines 31 and 61) could be explained by one chromosome in each line undergoing a deletion, which eliminated 7 R2 copies. Because the analysis in the R2 profile in the original study (Perez-Gonzalez and Eickbush 2001) had been conducted with DNA derived from a pool of flies in each line, deletions as well as any insertions present in only a few flies would have gone undetected. Therefore while suggestive, this group of lines did not provide definitive evidence for recent insertion or deletion events.
Finally, the four lines in the third group (Figure 3, bottom) had 10 or more different X chromosomes in each line. In line 89, all 18 males had a different R2 5′-truncation profile. In all four lines less than half of the R2 copies were present in all males sampled, and the R2 truncation profiles had dramatically changed compared to the previous study (Perez-Gonzalez and Eickbush 2001). These stocks had not become contaminated because the R1 truncation profiles showed little variation from the profiles seen 75 generations earlier (data not shown). The high level of variation seen within these lines suggested the recent and rapid turnover of R2 elements.
Variation within the full-length R2 elements of line 58:
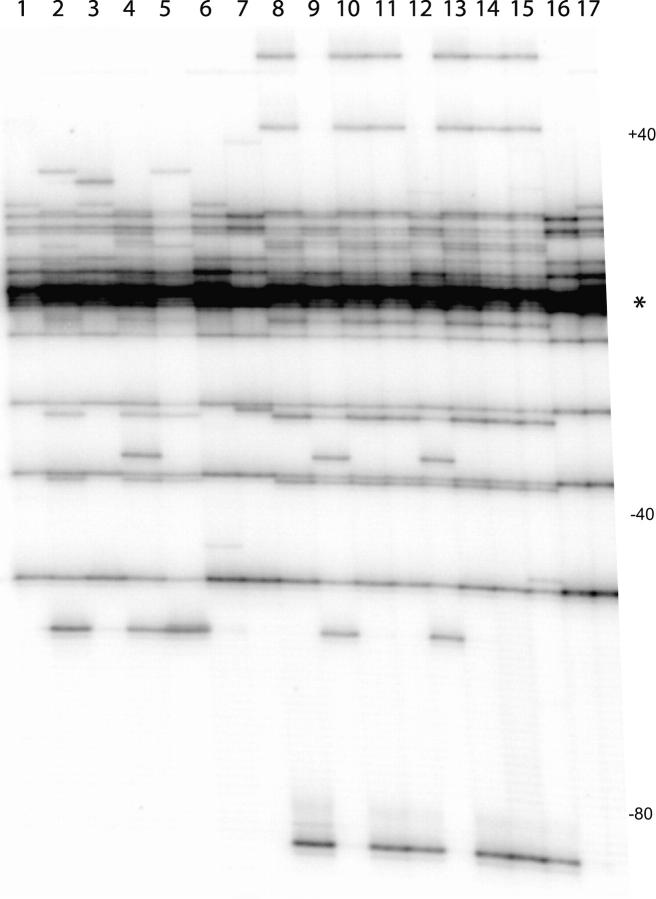
Forty to 80% of the R2 elements in the Paradise isofemale lines do not have large 5′ truncations and are thus classified as full length (see below for the exact percentage of full-length elements in each line). Fortunately, even individual full-length R2 insertions can be identified because the mechanism used by R2 to initiate second-strand DNA synthesis is imprecise, frequently generating small deletions, insertions, and duplications at the 5′ junction of the element (George et al. 1996). To determine if the many differences in the 5′-truncated R2 elements scored among males from Paradise line 58 were also true of full-length elements, a PCR primer that anneals to R2 sequences 123 bp from the 5′ end of full-length elements was end-labeled and used in combination with the primer that anneals to the 28S gene 80 bp upstream of the R2 insertion site. The labeled PCR products were then electrophoresed on a high-resolution denaturing gel to resolve small differences in product lengths. Figure 4 shows the results of individual males sampled from line 58. The full-length R2 profile of the males differed in the number of bands as well as in the intensity of some bands. The canonical full-length R2 element as determined in the original study (Perez-Gonzalez and Eickbush 2001) is the intense band identified with an asterisk. Many other R2 copies were up to 80 bp shorter or longer than this canonical length. Nine different X chromosome types could be identified solely on the basis of the combination of R2 copy lengths present. The intensity of some bands also differed between X chromosomes, indicating an increase or decrease in the number of elements of these specific lengths. By quantitating the amount of signal in each PCR band on a PhosphorImager, virtually all X chromosomes sampled from line 58 could be resolved as different. Clearly the extensive variation in R2 profiles in line 58 involved both 5′-truncated and full-length elements.
Figure 4.—
5′ variation in the full-length R2 elements of D. simulans line 58. DNA from 17 males of line 58 was individually PCR amplified with an end-labeled primer that annealed to the 28S rRNA gene 80 bp upstream of the R2 insertion site and the 0.1-kb R2 primer shown in Figure 1B. The labeled PCR products were run on a high-voltage, denaturing 8% polyacrylamide gel and the signal was quantitated on a PhosphorImager screen (see materials and methods). The asterisk indicates the length of canonical full-length R2 elements as described in Perez-Gonzalez and Eickbush (2001). Base positions relative to this canonical full-length R2 are shown at right.
R2 insertions and deletions rapidly accumulated in line 58:
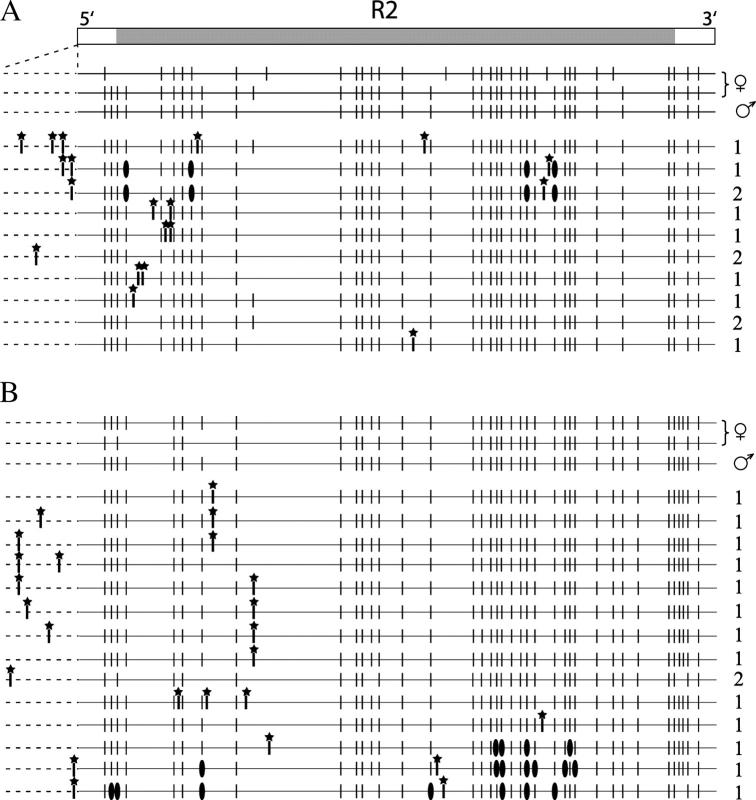
To determine how quickly variation in the R2 profile could accumulate in these D. simulans lines, a subline of line 58 was started with one pair of flies to reduce the initial level of variation. Single pairs of flies from this subline were mated and their progeny were maintained as separate lineages. R2 5′-truncation profiles as well as full-length R2 profiles of individual male descendents after five generations were compared (Figure 5). Because the R2 truncation profiles of the original pair were determined (top of each panel), new insertions could be distinguished from deletions. In lineage A, 16 new insertions (5 full length and 11 5′ truncated) generated nine X chromosome profiles that differed from those in the original pair. Only two males tested in generation 5 contained an X chromosome identical to one of the parental X chromosome profiles. Three X chromosomes of the descendents, in addition to gaining new insertions, had lost four R2 elements present in the original pair.
Figure 5.—
R2 insertions and deletions rapidly accumulated in line 58. Single pairs of flies from line 58 were mated and their progeny were maintained as separate lineages for five generations. Data from two lineages are presented. Shown at the top of A and B are the R2 profiles of the two X chromosomes from the original female and the single X chromosome from the original male used to establish the lineage. The profiles of the two X chromosomes of the female were determined by analysis of the R2 profiles of a collection of her sons. Below these progenitor profiles are the profiles of males collected at the fifth generation. The dotted line to the left of each R2 5′-truncation profile represents an expanded scale of differences in the full-length R2 elements. Because there are many full-length R2 elements, only full-length elements not present in parental X chromosomes are shown. New R2 insertions are in bold and marked with a star. Deletions of the parental R2 copies are marked by solid ovals. Numbers on the right are the number of generation 5 males in which this profile type was found.
In the B lineage, 16 new insertions (7 full length and 9 5′ truncated) generated 14 different X chromosome types. This lineage had three X chromosomes with 4–7 deletions that involved in total 12 R2 copies. Interestingly 2 of the deleted R2 copies had also been deleted in the A lineage. None of the males tested in generation 5 contained an X chromosome identical to one of the parental types.
Finally in a third lineage (data not shown), new insertions were not detected but five R2 copies were deleted from one male. These deleted copies were among the same R2 copies deleted from lineages A and B, indicating that certain R2 copies (or locations within the rDNA locus) were prone to elimination. The failure to find new insertions in this lineage suggested that all X chromosomes in line 58 did not contain equally active R2 elements.
Characterization of the rDNA loci of the Paradise lines:
We next asked whether the R2 activity detected in some of the D. simulans lines could be correlated with the number of R2 elements, the fraction of the rDNA units inserted with R2, or the size of the rDNA locus (i.e., the total number of rDNA units). The number of R2 insertions present within individuals of a line can be accurately determined because of the extensive sequence variation present at the R2 5′ ends. We have previously shown that the many 5′-truncated bands seen by PCR amplification of individual D. simulans males or D. melanogaster males are of equal intensity, and thus correspond to one copy per chromosome (Perez-Gonzalez and Eickbush 2001, 2002; Eickbush and Eickbush 2003; Perez-Gonzalez et al. 2003). Therefore the number of 5′-truncated copies can be determined by counting the number of distinct PCR bands detected in our 5′-truncation profiles (Figure 4). Polymorphic truncations (i.e., those not present in all X chromosomes sampled from each line) were counted as the number of chromosomes in which the truncations were observed divided by the number of X chromosomes sampled. Because we have not shown that all 5′-truncated elements are single copy, these counts may represent an underestimation of the number of 5′-truncated R2 elements in each line. The number of full-length R2 insertions was determined by using the end-labeled PCR primer and separating the products on high-resolution denaturing gels (see Figure 4). Many of the full-length copies were present at one copy per chromosome. The number of elements corresponding to the more intense PCR bands was estimated by dividing the total 32P signal in each band by the signal present in the single-copy bands (see materials and methods).
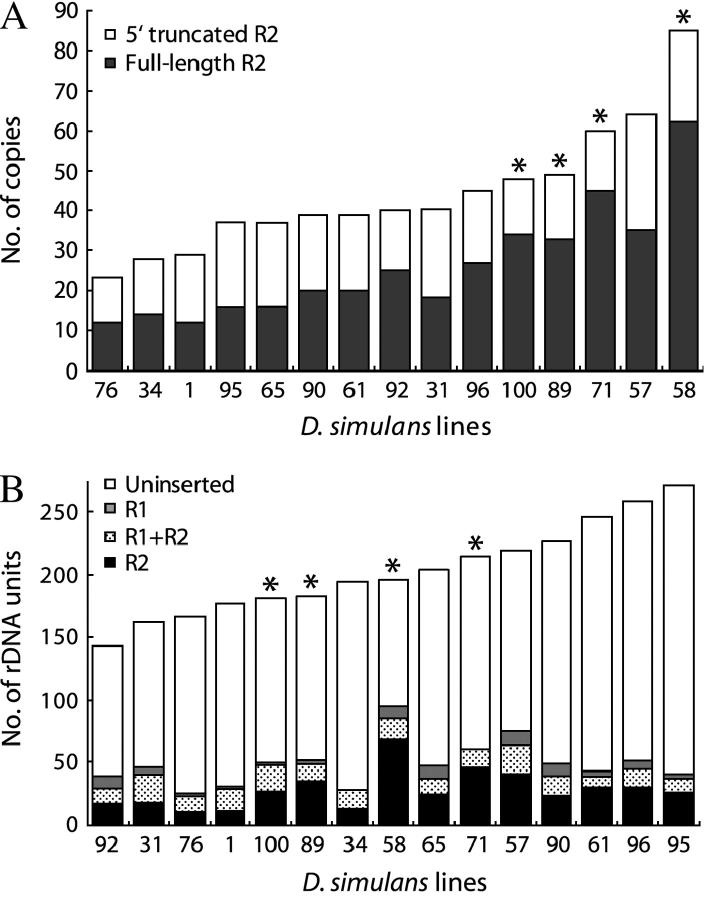
Shown in ascending order in Figure 6A are the total number of 5′-truncated R2 elements (open area of the bar) and full-length R2 elements (solid area of the bar) present in each of the D. simulans lines. The number of R2 elements varied from a low of 23 in line 76 to a high of 85 in line 58 (3.7-fold range). Most of the differences between lines were due to the variation in the number of the full-length R2 elements (range 12–62).
Figure 6.—
R2 copy number and size of the rDNA locus in each Paradise line. (A) Average R2 copy number in each Paradise line. The open portion of each bar represents the number of distinct 5′-truncated R2 elements scored in the PCR analysis (Figure 3), and the solid portion of each bar represents the number of full-length elements as determined by a PCR analysis similar to that in Figure 4. Polymorphic R2 insertions were scored by the number of males containing the element divided by the number of total males sampled. (B) Average size of the rDNA locus in each Paradise line. The estimated number of uninserted, R1-inserted, R2-inserted, and double-inserted (R1 + R2) rDNA units are represented, respectively, by the open, shaded, solid, and stippled portions of each bar. These values were determined by genomic DNA blots and using the calculated total number of R2 insertions in each line divided by the fraction of the total rDNA units that were inserted by R2 to determine the total number of units (Table 1) (see materials and methods for details). In A and B, lines with high R2 activity, as indicated by the level of variation in the R2 profiles (see Figure 3), are indicated with an asterisk.
To determine the fraction of the rDNA locus inserted with R1 and R2 elements, genomic DNA from 30 adults of each line was extracted and two Southern blots were conducted. In the first Southern blot, restriction enzymes were used that cleaved the rDNA units into three fragments: uninserted, R1 inserted (including R1 and R2 double inserts), and R2 inserted. The genomic blot was probed with a fragment of the 28S gene downstream of the R1 insertion site. In the second Southern blot, a restriction enzyme was used that cleaved the R2-inserted units into two fragment sizes depending upon whether they were or were not also inserted with an R1 element. This blot was probed with sequences from the 3′ end of the R2 element (see Figure 1A and materials and methods for a more detailed description of these blots). By determining the relative intensity of the bands from these two Southern blots, the percentages of uninserted, total R1-inserted, total R2-inserted, and R1 + R2 double-inserted rDNA units were calculated (Table 1). The fraction of the rDNA locus inserted with R2 elements averaged 22% and varied from a low of 14% in line 95 to a high of 44% in line 58. The fraction of the locus inserted with R1 elements averaged 10% and also showed significant variation among the lines. The fraction of the rDNA units inserted with R1 showed no relationship to the fraction inserted with R2. The fraction of the R1 and R2 double-inserted rDNA units varied from 3 to 14%, which, as we have noted previously (Perez-Gonzalez and Eickbush 2001), is significantly higher than what would be expected from the random insertion of R1 and R2 elements into the rDNA units.
TABLE 1.
Percentages of uninserted, R1-inserted, R2-inserted, and double-inserted rDNA units in the Paradise lines
| Fly line | Uninserted (%) |
R1 (%)a | R2 (%)a | R1 + R2 (%) |
|---|---|---|---|---|
| 1 | 83 | 11 | 16 | 10 |
| 31 | 71 | 17 | 25 | 14 |
| 34 | 86 | 8 | 14 | 8 |
| 57 | 66 | 15 | 29 | 10 |
| 58 | 51 | 14 | 44 | 9 |
| 61 | 83 | 5 | 16 | 3 |
| 65 | 77 | 11 | 18 | 6 |
| 71 | 72 | 7 | 28 | 7 |
| 76 | 85 | 9 | 14 | 7 |
| 89 | 72 | 9 | 27 | 8 |
| 90 | 79 | 11 | 17 | 6 |
| 92 | 73 | 15 | 20 | 8 |
| 95 | 85 | 5 | 14 | 4 |
| 96 | 80 | 8 | 17 | 6 |
| 100 | 73 | 12 | 27 | 11 |
Includes both single- and double-inserted (R1 + R2) units.
Finally, to determine the size of the rDNA locus (the total number of rDNA units) in each line, the total number of R2 elements (5′ truncated and full length, Figure 6A) was divided by the fraction of the units that contained R2 insertions (Table 1). The size of the locus averaged ∼200 units varying from an estimated 140 in line 92 to 270 units in line 95 (shown in Figure 6B). The calculated numbers of uninserted, R1 only inserted, R2 only inserted, and R1 + R2 double-inserted rDNA units are represented by the open, shaded, solid, and stippled areas of the bars, respectively. Lines with high R2 activity are indicated with an asterisk. As will be described in the discussion, R2 activity correlated with the number of full-length R2 elements but not with the size of the rDNA locus.
DISCUSSION
All laboratory stocks and geographical lines of D. melanogaster and D. simulans characterized to date have contained significant numbers of R2 elements (Jakubczak et al. 1992; Eickbush and Eickbush 1995; Eickbush et al. 1995; Perez-Gonzalez and Eickbush 2001). The various copies of these elements contained <0.5% nucleotide sequence variation and few had mutations that disrupted the R2 open reading frame. These observations suggest R2 retrotransposition occurs on a continuing basis in a population, but do not indicate the frequency of such events. To monitor the rate of retrotranspositions, replicate sublines of one D. melanogaster stock were scored after 350 generations (Perez-Gonzalez and Eickbush 2002; Perez-Gonzalez et al. 2003). Only 16 retrotransposition events were found randomly distributed over 19 sublines, suggesting a uniform low rate of new insertions.
The isofemale lines of D. simulans characterized in this report offered a different indication of the rate and pattern of R2 retrotransposition. Most lines were similar to the previously reported D. melanogaster lines, and provided little evidence of active retrotransposition. The insertion profiles of R2 elements in these lines were similar or identical to the profiles scored 75 generations earlier. On the other hand, virtually every rDNA locus sampled from a few isofemale lines had differences in their R2 insertion profiles, suggesting retrotransposition. Direct evidence for frequent retrotransposition was demonstrated by showing that the descendants of individual pairs of flies from the “active” lines rapidly accumulated new R2 insertions.
The characterization of naturally occurring active and inactive lines of D. simulans provides new insights into the dynamics of R2 element insertions and deletions. The data suggest that there are stable genetic conditions under which new retrotransposition events can occur frequently and stable genetic conditions under which events are rare or absent. Because some of the lines contained intermediate levels of R2 variation, it will be important to determine whether these lines have lower rates of retrotransposition or whether the current variation in these populations represents either remnants of former activity or the recent initiation of activity.
Genetic conditions under which quiescent mobile elements actively generate new copies have most often been found associated with hybrid dysgenic crosses (Kidwell et al. 1977; Bucheton 1990; Petrov et al. 1995). In these crosses, mobile elements are activated when an individual that has copies of a mobile element mates with an individual with no copies of that element. Element activity continues in the descendents of this cross for a number of generations until eventually element activity is turned off. The mechanisms that suppress the continued activity of these elements are believed to be cellular systems designed to detect the expression of any sequence present in multiple copies (Chaboissier et al. 1998; Roche and Rio 1998; Jensen et al. 1999; Boivin et al. 2003). The R2 activity we have detected in the D. simulans isofemale lines clearly differs from that of hybrid dysgenesis in that both males and females from all lines contain many copies of R2 elements, and R2 activity is maintained for many generations. We do not know how long the R2 elements have been active in these lines but the rapid accumulation of new insertions in the descendants of single pairs has been observed over a 1-year period (∼25 generations) in several independent trials with two of the active lines (58 and 89) (Figure 5; data not presented).
R2 activity is more similar to the activity associated with the LTR retrotransposons copia, gypsy, Zam, and Idefix. The activity of these elements is associated with one or more alleles present at specific “control” loci in the genome (Prud'homme et al. 1995; Nuzhdin et al. 1998; Desset et al. 2003), although a more general control mechanism may be involved (Sarot et al. 2004). Element activity continues for generations and does so irrespective of the number of mobile element copies present in the organism.
One likely locus that might control the activity of R2 elements is the rDNA locus itself. In Figure 7A the number of full-length and, thus, potentially active R2 elements has been plotted vs. the amount of R2 variation that was present in each subline (scored as the number of X chromosome R2 profiles identified in 18 males). The six lines with no R2 activity had on average 17 full-length R2 elements, while the four lines with high activity (10 and more X chromosome R2 profiles) averaged 44 full-length elements or over 2.5 times that of the no activity lines. The five lines with 2–4 different R2 profiles averaged higher numbers of full-length elements but overlapped extensively with the no activity lines. In contrast, there was no correlation between R2 activity and the size of the rDNA locus. The four active lines had on average ∼195 rDNA units while the six inactive lines averaged ∼215 units (see Figure 6B). However, because the active lines had more R2 elements there was a positive correlation between R2 activity and the percentage of the locus that was inserted with R2 (average ∼15% for the inactive lines and ∼31% for the active lines, Figure 7B).
Figure 7.—
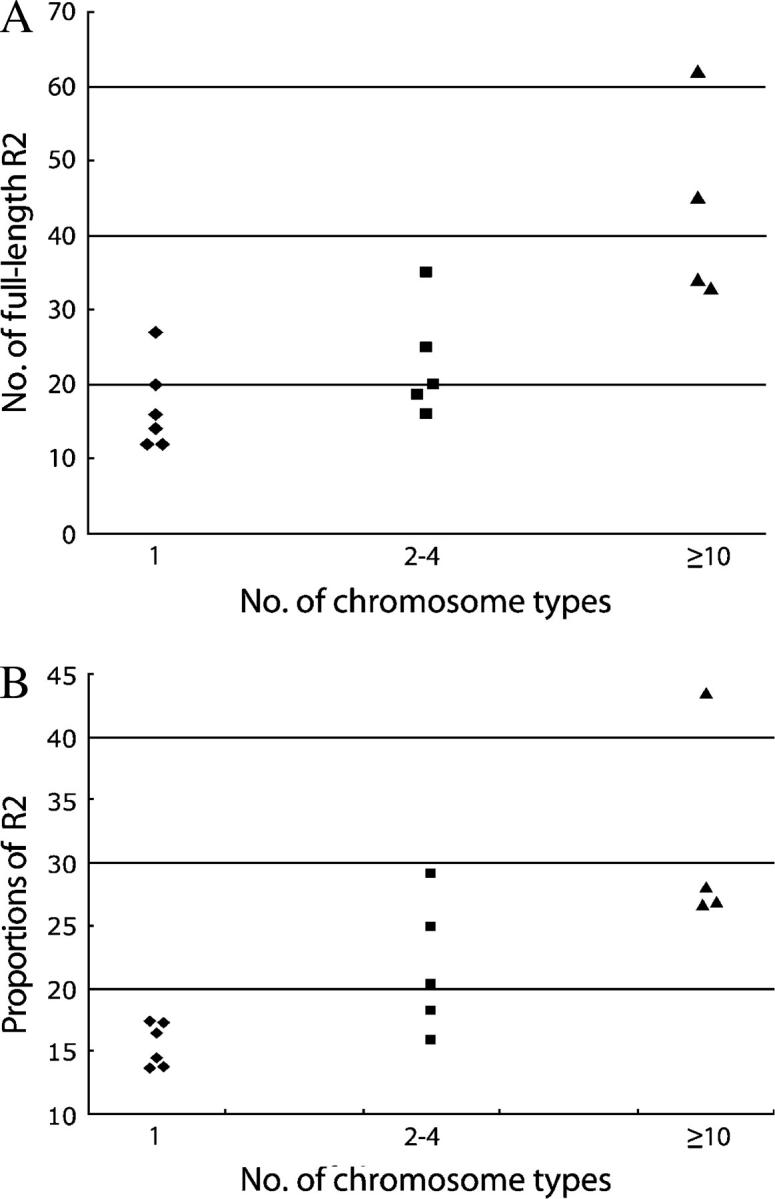
Correlation between R2 activity and the number of full-length R2 elements or the fraction of the rDNA locus inserted with R2. The level of R2 activity was scored as the level of variation within each line (i.e., the number of X chromosome variant profiles from Figure 3). The Paradise lines are divided into three groups on the basis of the number of chromosome types found in sampled males from that line and plotted vs. (A) the number of full-length R2 elements found in the Paradise lines and (B) the fraction (as percentage) of the rDNA units in each line inserted with R2.
Transposable element insertion rates have previously been correlated with the number of elements (Nuzhdin et al. 1996; Pasyukova et al. 1998). Thus one possible model to explain the correlation of R2 activity with element number is that the larger number of R2 elements or the greater fraction of inserted rDNA units is able to produce a high level of the retrotransposition intermediates, which overwhelms cellular control mechanisms. However, such a correlation between R2 copy number and R2 activity was not exact. Line 96 contained 26 full-length R2 copies and no detectable R2 activity, while line 89 had 32 full-length elements and the highest level of R2 activity (as measured by the variation within the line), even higher than line 58, which had 65 full-length elements.
An alternative model to explain R2 activity is that the rate of retrotransposition is unrelated to element copy number or the fraction of the locus inserted with R2; rather the larger numbers of R2 copies present in active lines is the result (not the cause) of R2 activity. In this model, R2 activity could be controlled by a genetic locus other than the rDNA locus itself. We should be able to resolve between these two models by conducting crosses between the active and inactive Paradise lines and genetically mapping control over R2 activity.
Another important finding of this report is that the insertion of many new R2 elements resulted in the elimination of preexisting R2 elements from the locus. Non-LTR retrotransposable elements have never been shown to excise, thus these eliminations presumably involved recombination events that deleted the R2-inserted rDNA units. These recombination events appeared to involve more than single rDNA units because four to seven R2 copies were eliminated per event (Figure 5). The deletions frequently involved the same regions of the rDNA locus because the same subset of R2 elements was often eliminated (Figure 5). Hot spots for the elimination of R1 and R2 elements within the rDNA locus have also been seen in our studies of the D. melanogaster Harwich lines (Perez-Gonzalez et al. 2003). In addition, deletions within the rDNA locus in the presence of active R2 insertion machinery are associated with our in vivo R2 integration system (Eickbush and Eickbush 2003). In this system the injection of purified R2 protein and RNA into Drosophila embryos results in the insertion of marked R2 elements in the rDNA locus. These insertions are frequently accompanied by large deletions of rDNA. Finally, retrotransposition of another non-LTR transposable element, L1, has been shown to cause large deletions (>70 kb) of host genomic DNA (Gilbert et al. 2002).
The elimination of R2-inserted rDNA units in the active lines can explain why the rDNA loci in these lines are not extensively filled with R2 insertions. However, because the total number of rDNA units in the active lines is similar to that of the inactive lines (Figure 6B), the active lines must also be undergoing selection for a compensating expansion of uninserted rDNA units.
Finally there was no evidence for R1 activity in the four Paradise lines with R2 activity. This finding confirms previous suggestions that R1 and R2 elements are separately activated. For example, Jakubczak et al. (1992) found that the levels of R1 and R2 elements were unrelated in various geographical lines of D. melanogaster. In the Harwich mutation accumulation lines, those lines with the highest numbers of new R1 insertions did not have higher numbers of new R2 insertions (Perez-Gonzalez et al. 2003). Separate control over R1 and R2 activity would minimize the direct competition between these elements, thus helping to explain their coexistence in the rDNA locus since the origin of arthropods (Burke et al. 1998, 1999; Gentile et al. 2001).
Acknowledgments
We thank Bill Burke, Danna Eickbush, and Karin Tetzlaff for discussions and comments on the manuscript. We especially thank Naina Phadnis whose initial resurvey of the Paradise lines renewed our interest in these lines. This research was supported by National Science Foundation grant MCB-9974606 and National Institutes of Health grant GM-42790.
References
- Boivin, A., C. Gally, S. Netter, D. Anxolabehere and S. Ronsseray, 2003. Telomeric associated sequences of Drosophila recruit polycomb-group proteins in vivo and can induce pairing-sensitive repression. Genetics 164: 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton, A., 1990. I transposable elements and I-R hybrid dysgenesis in Drosophila. Trends Genet. 6: 16–21. [DOI] [PubMed] [Google Scholar]
- Burke, W. D., H. S. Malik, W. C. Lathe, III and T. H. Eickbush, 1998. Are retrotransposons long-term hitchhikers? Nature 392: 141–142. [DOI] [PubMed] [Google Scholar]
- Burke, W. D., H. S. Malik, J. P. Jones and T. H. Eickbush, 1999. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol. Biol. Evol. 16: 502–511. [DOI] [PubMed] [Google Scholar]
- Chaboissier, M.-C., A. Bucheton and D. J. Finnegan, 1998. Copy number control of a transposable element, the I factor, a LINE-like element in Drosophila. Proc. Natl. Acad. Sci. USA 95: 11781–11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desset, S., C. Meignin, B. Dastugue and C. Vaury, 2003. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 164: 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, A., and J. Albornoz, 1996. Rates of movement of transposable elements in Drosophila melanogaster. Mol. Gen. Genet. 251: 130–138. [DOI] [PubMed] [Google Scholar]
- Eickbush, D. G., and T. H. Eickbush, 1995. Vertical transmission of the retrotransposable elements R1 and R2 during the evolution of the Drosophila melanogaster species subgroup. Genetics 139: 671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush, D. G., and T. H. Eickbush, 2003. Transcription of endogenous and exogenous R2 elements in the rRNA gene locus of Drosophila melanogaster. Mol. Cell. Biol. 23: 3825–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush, D. G., W. C. Lathe, III, M. P. Francino and T. H. Eickbush, 1995. R1 and R2 retrotransposable elements of Drosophila evolve at rates similar to those of nuclear genes. Genetics 139: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush, T. H., 2002 R2 and related site-specific non-long terminal repeat retrotransposons, pp. 813–835 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellart and A. M. Lambowitz. American Society for Microbiology, Washington, DC.
- Gentile, K. L., W. D. Burke and T. H. Eickbush, 2001. Multiple lineages of R1 retrotransposable elements can coexist in the rDNA loci of Drosophila. Mol. Biol. Evol. 18: 235–245. [DOI] [PubMed] [Google Scholar]
- George, J., W. D. Burke and T. H. Eickbush, 1996. Analysis of the 5′ junctions of R2 insertions with the 28S gene: implications for non-LTR retrotransposition. Genetics 142: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, N., S. Lutz-Prigge and J. V. Moran, 2002. Genomic deletion created upon LINE-1 retrotransposition. Cell 110: 315–325. [DOI] [PubMed] [Google Scholar]
- Gloor, G. B., C. R. Preston, D. M. Johnson-Schlitz, N. A. Nassif, R. W. Phillis et al., 1993. Type I repressors of P-element mobility. Genetics 135: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle, D., and S. V. Nuzhdin, 2004. Mutation accumulation and the effect of copia insertions in Drosophila melanogaster. Genet. Res. 83: 7–18. [DOI] [PubMed] [Google Scholar]
- Jakubczak, J., W. D. Burke and T. H. Eickbush, 1991. Retrotransposable elements R1 and R2 interrupt the rDNA genes of most insects. Proc. Natl. Acad. Sci. USA 88: 3295–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczak, J., M. Zenni, R. Woodruff and T. H. Eickbush, 1992. Turnover of R1 (type I) and R2 (type II) retrotransposable elements in the ribosomal DNA of Drosophila melanogaster. Genetics 131: 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamrich, M., and O. L. Miller, Jr., 1984. The rare transcripts of interrupted rRNA genes in Drosophila melanogaster are processed or degraded during synthesis. EMBO J. 3: 1541–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S., M. P. Gassama and T. Heidmann, 1999. Taming of transposable elements by homology dependent gene silencing. Nat. Genet. 21: 209–212. [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G., J. F. Kidwell and J. A. Sved, 1977. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility, and male recombination. Genetics 86: 813–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, K. K., and H. Fujiwara, 2004. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: various multicopy RNA genes and microsatellites are selected as targets. Mol. Biol. Evol. 21: 207–217. [DOI] [PubMed] [Google Scholar]
- Lathe, W. C., III, and T. H. Eickbush, 1997. A single lineage of R2 retrotransposable element is an active, evolutionarily stable component of the Drosophila rDNA locus. Mol. Biol. Evol. 14: 1232–1241. [DOI] [PubMed] [Google Scholar]
- Lathe, W. C., III, W. D. Burke, D. G. Eickbush and T. H. Eickbush, 1995. Evolutionary stability of the R1 retrotransposable element in the genus Drosophila. Mol. Biol. Evol. 12: 1094–1105. [DOI] [PubMed] [Google Scholar]
- Lohe, A. R., and P. A. Roberts, 1990. An unusual Y chromosome of Drosophila simulans carrying amplified rDNA spacer without rRNA genes. Genetics 125: 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, E. O., and I. B. Dawid, 1979. Expression of ribosomal DNA insertions in Drosophila melanogaster. Cell 18: 1185–1196. [DOI] [PubMed] [Google Scholar]
- Luan, D. D., M. H. Korman, J. L. Jakubczak and T. H. Eickbush, 1993. Reverse transcription of R2Bm is primed by a nick at the chromosomal target site—a mechanism for non-LTR retrotransposition. Cell 72: 595–605. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and T. H. Eickbush, 1998. Retrotransposable elements R1 and R2 in the rDNA units of Drosophila mercatorum: abnormal abdomen revisited. Genetics 151: 653–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maside, X., C. Bartolome, S. Assimacopoulos and B. Charlesworth, 2001. Rates of movement and distribution of transposable elements in Drosophila melanogaster: in situ hybridization vs. Southern blotting data. Genet. Res. 78: 121–136. [DOI] [PubMed] [Google Scholar]
- Mecheva, I. S., and E. P. Semionov, 1992. Localization of ribosomal DNA insertion elements in polytene chromosomes of Drosophila simulans, Drosophila mauritiana and their interspecific hybrids. Genetica 85: 223–229. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S. V., and T. F. Mackay, 1995. The genomic rate of transposable element movement in Drosophila melanogaster. Mol. Biol. Evol. 12: 180–181. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S. V., E. G. Pasyukova and T. F. Mackay, 1996. Positive correlation between copia transposition rate and copy number in Drosophila melanogaster. Proc. R. Soc. Lond. B Biol. Sci. 263: 823–831. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S. V., E. G. Pasyukova, E. A. Morozova and A. J. Flavell, 1998. Quantitative genetic analysis of copia retrotransposon activity in inbred Drosophila melanogaster lines. Genetics 150: 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasyukova, E. G., S. V. Nuzhdin and D. A. Filatov, 1998. The relationship between the rate of transposition and transposable element copy number for copia and Doc retrotransposons of Drosophila melanogaster. Genet. Res. 72: 1–11. [DOI] [PubMed] [Google Scholar]
- Perez-Gonzalez, C. E., and T. H. Eickbush, 2001. Dynamics of R1 and R2 elements in the rDNA locus of Drosophila simulans. Genetics 158: 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gonzalez, C. E., and T. H. Eickbush, 2002. Rates of R1 and R2 retrotransposition and elimination from the rDNA locus of Drosophila melanogaster. Genetics 162: 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gonzalez, C. E., W. D. Burke and T. H. Eickbush, 2003. R1 and R2 retrotransposition and deletion in the rDNA loci on the X and Y chromosomes of Drosophila melanogaster. Genetics 165: 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D. A., J. L. Schutzman, D. L. Hartl and E. R. Lozovskaya, 1995. Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc. Natl. Acad. Sci. USA 92: 8050–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme, N., M. Gans, M. Masson, C. Terzian and A. Bucheton, 1995. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139: 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche, S. E., and D. C. Rio, 1998. Trans-silencing by P elements inserted in subtelomeric heterochromatin involves the Drosophila polycomb group gene, enhancer of zeste. Genetics 149: 1839–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot, E., G. Payen-Groschene, A. Bucheton and A. Pelisson, 2004. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166: 1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]