Abstract
The semisterile meiotic mutant mei-352 alters the distribution of meiotic exchanges without greatly affecting their total frequency. We show that the mei-352 mutation is an allele of the klp3A gene, which encodes a kinesin-like protein of the Kinesin-4 family. The semisterility observed in mei-352 females results from a known defect of klp3A oocytes in mediating pronuclear fusion. Interestingly, other klp3A alleles also exhibit defects in meiotic recombination similar to those of mei-352. Finally, we show that the Klp3A protein localizes within the oocyte nucleus during meiotic prophase, the time at which exchange distribution is established, and extensively colocalizes with DNA. The parallel of the klp3A phenotype with a meiotic defect observed for kar3 mutants in yeast suggests a role for kinesins in early meiosis and might reflect a previously suggested role for this class of kinesins in chromosome condensation.
MEIOTIC exchanges occur at nonrandom locations along chromosome arms. In Drosophila melanogaster females, meiotic exchange occurs most frequently in medial regions of chromosomal arms and less often in proximal or distal regions (Lindsley and Sandler 1977; McKim et al. 2002). This regulation of exchange distribution is necessary for ensuring at least one exchange per bivalent. Failures in establishing an exchange and aberrant exchange distributions along bivalents have been associated with nondisjunction (Hassold and Hunt 2001; Lamb et al. 2005). A desire to understand the mechanism by which exchanges are distributed and the biological importance of this mechanism have fueled extensive efforts to characterize mutants that alter the regional distribution of exchange.
Mutants that alter the distribution of exchange have been isolated in a number of genetic screens in Drosophila (Sandler et al. 1968; Baker and Carpenter 1972; Sekelsky et al. 1999) and are often described as precondition mutants (Carpenter and Sandler 1974; Lindsley and Sandler 1977; Bhagat et al. 2004). Most such mutants decrease the total amount of meiotic recombination per chromosome and also make the exchange distribution within the euchromatin more proportional to physical distance (Baker and Hall 1976; Lindsley and Sandler 1977). Exchange does not occur in the heterochromatin in wild type, and none of these mutants allows exchange within the heterochromatin (Carpenter and Baker 1982). The mei-352 mutant is unique among precondition mutants because it does not decrease the overall frequency of exchange despite altering the distribution of those exchanges. This phenotype makes the Mei-352 protein a candidate for a function that is specifically required for mediating the distribution of exchanges.
mei-352 was identified in a screen for ethyl methanesulfonate (EMS)-induced meiotic mutants (Baker and Carpenter 1972). Although the total frequency of exchanges is unchanged in mei-352 females compared to wild type, the locations of those exchanges differ. Exchanges are increased near the centromere and decreased near the telomere of a chromosome arm, while exchange remains prohibited in the heterochromatin (Baker and Carpenter 1972; Carpenter and Baker 1982). In addition, mei-352 females display a modest meiotic nondisjunction phenotype and are semisterile.
We show here that mei-352 is an allele of klp3A, which encodes kinesin-like protein at 3A (Klp3A). Drosophila Klp3A is a member of the Kinesin-4 family within the kinesin protein superfamily, whose members play multiple roles in mitosis and meiosis (Vernos and Karsenti 1995; Lawrence et al. 2004). Studies of Drosophila embryos microinjected with antibodies or mutant proteins that exert a dominant-negative effect have shown Klp3A to be involved in the organization of interpolar microtubules and in anaphase B spindle elongation during mitosis (Brust-Mascher et al. 2004; Kwon et al. 2004). RNA interference depletion of Klp3A in S2 cells resulted in numerous spindle defects and in abnormal prometaphase chromosome alignment (Goshima and Vale 2003; Kwon et al. 2004). In Drosophila spermatocytes, Klp3A is necessary for contractile ring assembly during meiotic cytokinesis (Williams et al. 1995; Giansanti et al. 1998), although evidence for a role in mitotic cytokinesis has not been found (Somma et al. 2002). Female meiotic spindle formation and chromosome segregation proceed relatively normally in klp3A mutants, although a low frequency of spindle defects was reported (Williams et al. 1997). Immediately following fertilization, Klp3A is involved in the separation of the female pronucleus from the polar bodies or in the migration of the female pronucleus toward the male pronucleus. In the absence of functional Klp3A, the majority of embryos fail to undergo pronuclear fusion and arrest prior to the first gonomeric division (Williams et al. 1997).
Our data suggest an additional function for Klp3A in regulating the distribution of exchanges during meiosis. We also show that the Klp3A protein localizes within the oocyte nucleus during meiotic prophase, the time at which exchange distribution is established. These findings, which are similar to observations made for the yeast Kar3 kinesin-like protein (Bascom-Slack and Dawson 1997), suggest a novel function for Klp3A and perhaps other kinesin-like proteins during meiotic prophase.
MATERIALS AND METHODS
Genetics:
Genetic markers and chromosomes used in this study are described in FlyBase (http://www.flybase.org) (Drysdale et al. 2005). Flies were reared on a standard Drosophila medium at 25°. Genetic analysis of recombination along the left arm of chromosome 2 was performed as described previously (Page et al. 2000). Deficiencies Df(1)ED6582 and Df(1)ED6579 were constructed using FLP-mediated mitotic recombination between FRT-bearing P elements as described by Ryder et al. (2004). The pairs of P elements used were P{RS3}CB-5899-3 and P{RS5}5-SZ-3093 [Df(1)ED6582] and P{RS3}UM-8329-3 and P{RS5}5-SZ-3093 [Df(1)ED6579].
Egg hatch assay:
Females of the stated genotypes were mated to males for 1 or more days and allowed to lay eggs on grape juice agar plates supplemented with wet yeast paste (Rothwell and Sullivan 2000) for a period of 14 hr at 25°. Parental flies were then removed and the plates containing eggs were incubated at 25° for an additional 24 hr. The numbers of hatched eggs (empty eggshells) and unhatched eggs were then counted under a dissecting microscope to determine the frequency of hatched eggs among all eggs laid.
Antibodies:
Mouse antihistones antibody (MAB052, Chemicon International, Temecula, CA) was used at a dilution of 1:500. Rat antitubulin, clones YL1/2 (Chemicon) and YOL1/34 (Serotec, Oxford), were used together with each at a dilution of 1:250. Mouse monoclonal anti-C(3)G, clone 1A8-1G2 (Anderson et al. 2005) was used at a dilution of 1:500. Affinity-purified anti-Klp3A (Kwon et al. 2004) was used at a dilution of 1:750. Mouse monoclonal antilamin, clone ADL101, was used at a dilution of 1:50. Primary antibodies were detected with Alexa 488-conjugated anti-mouse IgG and anti-rabbit IgG (Molecular Probes, Eugene, OR) and Cy3-conjugated anti-rat IgG and anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). Secondary antibodies were each used at a dilution of 1:1000.
Immunofluorescence and microscopy:
To examine embryonic phenotype by immunofluorescence, embryos were collected by the method described in Egg hatch assay, above. Embryos were collected over a period of 2.5 hr at 25°, fixed in methanol, and rehydrated prior to immunostaining essentially as described by Rothwell and Sullivan (2000). Antibody staining of embryos was performed as described previously for ovarioles (Page and Hawley 2001). Following antibody staining, embryos were cleared in 2:1 benzyl benzoate:benzyl alcohol and mounted on slides as described by Theurkauf (1994). For immunofluorescence of ovarioles, ovaries were dissected, fixed, and immunostained as described in Page and Hawley (2001). Ovarioles were mounted in either Prolong or Prolong Gold antifade mountant (Molecular Probes). Immunofluorescence data were collected using a Leica TCS SP2 confocal microscope.
RESULTS
Mapping of mei-352:
A single allele of the mutant mei-352 was isolated by Baker and Carpenter (1972) in their screen for X-linked meiotic mutants. During the initial analysis of mei-352, it was noted that homozygous females produced a reduced number of progeny, and this defect in fertility was more severe than would be predicted, given the frequency of nondisjunction observed among the surviving progeny (Baker and Carpenter 1972). (As described in detail below, we have shown that this semisterility is a consequence of maternal-effect lethality.) The original report did not determine whether the meiotic and semisterility phenotypes were due to a single mutation or to mutations in two separate loci on the X chromosome (Baker and Carpenter 1972). To map the mei-352 locus and determine whether the two phenotypes were separable, we first undertook classical recombination mapping of the semisterility defect exhibited by mei-3521 homozygotes. Analysis of recombinants showed that this defect mapped to the vicinity of the white locus, which is located in polytene map region 3C (data not shown).
To further map mei-352, we tested 16 deficiencies and two duplications in the 2F6–4F11 interval for complementation of the semisterility of mei-352. The series of aberrations tested is listed in Table 1. The interval containing mei-352 was further defined by the construction of two new deficiencies, Df(1)ED6582 and Df(1)ED6579, for which the breakpoints are defined at the sequence level (see materials and methods). Both Df(1)ED6582 and Df(1)ED6579 failed to complement mei-352 (Table 1). Thus, the mutation responsible for the semisterility of mei-352 was localized to between positions 2345982 and 2399458 on the X chromosome of the Drosophila Genome, Release 3.1 (Celniker et al. 2002), the interval defined by the smaller of these two deficiencies, Df(1)ED6579.
TABLE 1.
Deficiencies and duplications for the 2F6–4F11 interval tested for complementation of the semisterility defect ofmei-3521
| Aberration | Breakpoints | Complementation of mei-3521 semisterility |
Reference for aberration |
|---|---|---|---|
| Df(1)JC19 | 2F6; 3C5 | Fails to complement | Craymer and Roy (1980) |
| Df(1)X12 | 2F6; 3B5 | Fails to complement | Judd et al. (1972) |
| Df(1)62g18 | 3A1; 3A4 | Complements | Judd et al. (1972) |
| Df(1)65j26 | 3A1; 3A4-6 | Complements | Judd et al. (1972) |
| Df(1)54 | <3A1; 3A9 | Fails to complement | Goldberg et al. (1989) |
| Df(1)HC194 | 3A1;3C3-4 | Fails to complement | Pauli et al. (1995) |
| Df(1)w258-11 | 3A2; 3C4 | Fails to complement | Slizynska (1938) |
| Df(1)ED6582 | 3A4; 3A8 | Fails to complement | This study |
| Df(1)ED6579 | 3A6; 3A8 | Fails to complement | This study |
| Df(1)w-N71a | 3A6:3C10 | Complements | Craymer and Roy (1980) |
| Df(1)64j4 | 3A9; 3B2 | Complements | Smith and Konopka (1981) |
| Df(1)wrJ2 | 3A9; 3C2-3 | Complements | Judd et al. (1972) |
| Df(1)w258-45 | 3B3-4; 3C2 | Complements | Demerec (1940) |
| Df(1)N-8 | 3C1; 3D6 | Complements | Slizynska (1938) |
| Df(1)GA102 | 3D5; 3F7-8 | Complements | Pauli et al. (1995) |
| Df(1)HC244 | 3E8; 4F11 | Complements | Craymer and Roy (1980) |
| Dp(1;3)w+67k | 3A5; 3E8 | Complements | Judd et al. (1972) |
| Dp(1;2)w+70h | 3A7-8; 3C2-3 | Fails to complement | Judd et al. (1972) |
To determine whether the semisterility and exchange distribution defects of mei-352 mapped together, we assayed exchange along chromosome arm 2L in mei-3521/Df(1)ED6579 females (Figure 1). The results indicated a defect in exchange distribution that was similar to that observed in mei-3521 homozygotes. This result suggested that the meiotic phenotype and semisterility mapped either to the same locus or to closely spaced loci positioned within a region of ∼54 kb.
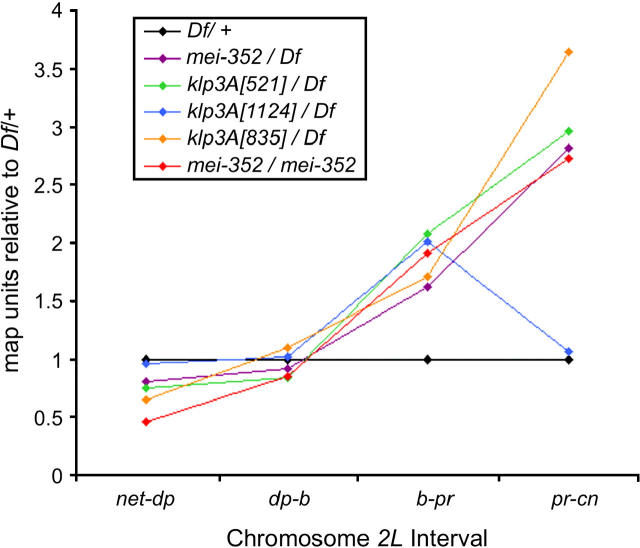
Figure 1.—
Alterations of meiotic exchange distribution are a general feature of klp3A mutants. Genetic map distances relative to wild type were calculated by dividing the map distance by the map distance for the Df/+ for the same interval. These data are plotted on the y-axis for four intervals on the left arm of chromosome 2 (x-axis). Df, Df(1)ED6579.
The mei-352 mutant is an allele of the klp3A gene:
Several point mutations in the region were tested for complementation with the mei-352 semisterility phenotype. Female double heterozygotes for mei-3521 and each of the following mutants were generated: wds, egh, klp3A, mit(1)15, sgg, l(1)3Ad, l(1)3Ag, and l(1)3Ah. With the exception of klp3A, the fertility of double heterozygote (i.e., mutant +/+ mei-3521) females for each genotype was equivalent to that of mutant/+ controls, indicating complementation. In contrast, mei-3521/klp3A521 and mei-3521/klp3Ae4 females were only weakly fertile in comparison to klp3A/+ or mei-3521/+ females. klp3A521 is a point mutation and klp3Ae4 is a small deletion that disrupts two genes, klp3A and egghead (egh). klp3Ae4 failed to complement mei-3521 in the presence of an egh+ transgene (Williams et al. 1995), which ruled out egh as a candidate for mei-352. These results demonstrated that the klp3A521 and klp3Ae4 mutations failed to complement mei-352.
To confirm that mei-352 is a klp3A allele, we tested whether a transgene carrying a wild-type copy of the klp3A gene, P{SCA9} (Williams et al. 1995, 1997), could rescue the semisterility of mei-3521. As shown in Table 2, the egg hatch rate for mei-3521; P{SCA9}/+ mothers is increased >10-fold over that for mei-3521 females without the transgene, indicating that the mei-352 semisterility phenotype is ameliorated by the presence of klp3A+. The lack of full rescue of the mutant phenotype to wild type may be due to poor expression from the transgene, which was not the same insertion used in other studies (Williams et al. 1995, 1997). These data support the view that mei-352 is an allele of the klp3A gene.
TABLE 2.
Frequency of hatching among eggs fromklp3A mutant females
| Eggs hatched
|
||
|---|---|---|
| Maternal genotype | % | n |
| klp3Amei-352/FM7w | 90.21 | 1175 |
| Df(1)ED6579/+ | 55.52 | 996 |
| klp3Amei-352/Df(1)ED6579 | 2.02 | 1239 |
| klp3Amei-352 | 3.70 | 675 |
| klp3Amei-352/klp3A521 | 1.90 | 1053 |
| klp3Amei-352/klp3A835 | 1.46 | 1303 |
| klp3Amei-352/klp3A1124 | 10.29 | 1399 |
| klp3A521/Df(1)ED6579 | 0.34 | 1470 |
| klp3Amei-352 ; P{SCA9}/+ | 43.33 | 2804 |
mei-352 bears a missense mutation in the klp3A gene:
To further investigate the possibility that mei-352 is an allele of klp3A, we sequenced the klp3A gene from mei-352 and from three previously identified alleles of klp3A. The klp3A alleles klp3A521, klp3A835, and klp3A1124 were originally recovered in an EMS screen for female sterile mutants (Mohler 1977; Mohler and Carroll 1984; Williams et al. 1995). Similarly, mei-352 was isolated during an EMS screen for meiotic mutants (Baker and Carpenter 1972), and thus all four mutations were expected to be single-base changes.
Sequencing of the klp3A gene from mei-3521 revealed two base substitutions that would be predicted to result in amino acid changes within the protein. One of these changes the isoleucine at position 609 to a threonine residue. Although this is a nonconservative change, the identical substitution was found in the klp3A gene present on the parental chromosome used in the original mutagenesis, so it is most likely a polymorphism. The second amino acid change was not present on the parental chromosome and results in a glutamic acid-to-lysine change at residue 321, located within the kinesin-like motor domain (Figure 2). A glutamic acid residue at this position is highly conserved throughout the kinesin superfamily, and replacement of the corresponding residue of kinesin heavy chain (E311) with alanine results in reduced ATPase and motor activity (Woehlke et al. 1997). This suggests that the E321K change in mei-352 could result in decreased Klp3A function. Given the nature of this mutation, its absence on the parental chromosome, and the noncomplementation demonstrating allelism with klp3A, we will henceforth refer to mei-352 as klp3Amei-352.
Figure 2.—
Schematic of the 1212-amino-acid Klp3A protein, showing domain structure and locations of identified mutations. Klp3A contains an N-terminal kinesin-like motor domain (aa 1–342, hatched area), a coiled-coil-rich stalk domain (aa 343–996, open area), and a C-terminal tail domain (aa 997–1212, stippled area). The locations and nature of four point mutations in klp3A identified in this study (klp3A521, klp3Amei-352, klp3A835, and klp3A1124) are indicated above the protein.
We also determined the molecular lesion present in three additional klp3A alleles (Figure 2). Comparison of the sequence of the klp3A gene from klp3A521, klp3A835, and klp3A1124 with the klp3A sequence from the parental chromosome allowed the identification of a single-base change in each allele compared to the parental sequence. In klp3A521, a C-to-T transition results in the replacement of arginine at residue 16 with a cysteine residue. This residue corresponds to the second arginine within the conserved motif RXRP, which is involved in ATP binding (Sack et al. 1999). The missense mutation in klp3A1124 is a G-to-A change resulting in a glutamic acid-to-lysine change at residue 829. Although a role for this residue in protein function has not been determined, it is conserved between Klp3A and other Kinesin-4 proteins and is located within a section predicted to form a coiled-coil structure. The G-to-A base change in the klp3A835 allele changes the tryptophan at position 683 to a stop codon that is expected to result in the expression of a truncated protein consisting of the motor domain and approximately half of the stalk domain. Previously, Williams et al. (1995) demonstrated a lack of full-length Klp3A protein in klp3A835 by Western blot analysis but reported the presence of a band of smaller size and weaker intensity that may represent the predicted truncated protein.
The semisterility of the klp3Amei-352 mutation is due to maternal-effect lethality, a known phenotype of klp3A:
The semisterility phenotype of klp3Amei-352 could result from a defect in oogenesis or from early embryonic lethality, but it is not thought to be simply due to the death of aneuploid embryos (Baker and Carpenter 1972). Mutations in klp3A are known to exhibit a maternal-effect lethality phenotype that is not rescuable by fertilization by klp3A+ sperm and results in a low frequency of egg hatching (Williams et al. 1997). Thus, it is possible that maternal-effect lethality could also be the cause of the semisterility in klp3Amei-352 females. To determine whether the semisterility of klp3Amei-352 results from maternal-effect lethality, we determined the frequency of egg hatching for klp3Amei-352 homozygotes and klp3Amei-352/klp3A trans-heterozygotes (Table 2). Testing of heterozygous control females (klp3Amei-352/FM7w) indicated hatching of ∼90% of eggs laid. In comparison, <4% of eggs laid by klp3Amei-352 homozygous females hatched. A similar decrease in hatching frequency was observed for klp3Amei-352/Df(1)ED6579 compared to Df(1)ED6579/+ controls. A comparably low frequency of egg hatching occurs for klp3A521/Df(1)ED6579 females (0.34%). We then measured hatch rates for eggs from klp3Amei-352/klp3A521, klp3Amei-352/klp3A835, and klp3Amei-352/klp3A1124 females. In each case, the egg hatch rate was severely decreased in comparison to the klp3Amei-352/FM7w control. These data support the view that the semisterility defect in klp3Amei-352 is the result of maternal-effect lethality.
The maternal-effect lethality in mei-352 resembles that of klp3A mutants:
Mutations in klp3A result in an early arrest of embryonic development prior to pronuclear fusion (Williams et al. 1997). In most of the embryos from klp3A mutant females, the female pronucleus fails to separate from polar body chromosomes, indicating a defect in female pronuclear specification or migration. The resulting phenotype is an embryo containing a large mass of condensed chromatin representing the products of female meiosis surrounded by an array of microtubules and a second, smaller chromatin mass, the male pronucleus, located on a bipolar, metaphase-like spindle.
To determine whether the maternal-effect lethality observed for klp3Amei-352 results from a defect similar to that for other klp3A mutations, embryos from klp3Amei-352 homozygote females and heterozygous controls, as well as klp3A homozygotes and klp3Amei-352/klp3A trans-heterozygotes, were analyzed by immunofluorescence. Staining the klp3Amei-352 embryos with antihistone and antitubulin antibodies revealed a phenotype quite similar to that previously described for klp3A (Table 3 and Figure 3, B and C). The development of the majority of embryos appeared to have arrested very early, with only one small nucleus plus a mass of polar body chromatin. A small percentage of embryos apparently escape this early arrest and continue development, which is consistent with the few adult progeny that are recovered from klp3Amei-352 homozygote mothers. In contrast, most of the embryos from klp3A+ or klp3Amei-352/FM7w control females progress beyond this stage (Table 3). Trans-heterozygotes between klp3Amei-352 and klp3A1124, klp3A521, or klp3A835 also display this early arrest phenotype (Figure 3, D and E; Table 3). In addition, centrosome abnormalities were frequently observed. In contrast to centrosomes from wild-type embryos, which are associated with the spindle poles in early embryos (Figure 3A), centrosomes in embryos derived from mutant females often appeared to have detached from spindles and to have undergone cycles of centrosome replication independent of mitosis, resulting in multiple centrosomes (Figure 3, C and E). The same types of centrosome defects were previously reported for klp3A mutants (Williams et al. 1997). Like Williams et al. (1997), we were unable to obtain sufficient numbers of embryos from klp3A521 and klp3A1124 homozygotes for analysis, but we were able to analyze embryos from homozygotes for klp3A835, which was thought to be a weak hypomorphic allele. However, embryos from klp3A835 displayed an early arrest phenotype that was essentially identical to that which was previously reported (Williams et al. 1997), although we noted a much higher frequency of early-arrested embryos (Table 3).
TABLE 3.
Development of embryos produced byklp3A mutant females
| Developmental stage (%)
|
|||||||
|---|---|---|---|---|---|---|---|
| Maternal genotype | One nucleus |
Two nuclei |
Cycles 2–9 |
Cycles 10–13 |
Cellular blastoderm |
Later | N |
| klp3Amei-352 | 62.6 | 25.2 | 7.3 | 3.3 | 0 | 1.6 | 123 |
| klp3Amei-352/klp3A1124 | 67.4 | 4.7 | 10.2 | 12.6 | 1.9 | 3.3 | 215 |
| klp3Amei-352/klp3A521 | 89.4 | 9.7 | 1.0 | 0 | 0 | 0 | 207 |
| klp3Amei-352/klp3A835 | 85.7 | 5.9 | 2.1 | 4.2 | 2.1 | 0.8 | 238 |
| klp3A835 | 68.8 | 10.2 | 17.6 | 1.7 | 1.7 | 0 | 176 |
| klp3Amei-352/FM7w | 3.1 | 7.3 | 45.8 | 26.6 | 10.9 | 6.3 | 192 |
| w1118 | 3.8 | 5.9 | 44.6 | 30.1 | 12.4 | 3.2 | 186 |
Embryos at 0–2.5 hr were collected from females of the stated maternal genotypes on grape juice agar plates at 25° prior to fixation and immunofluorescence analysis (see materials and methods).
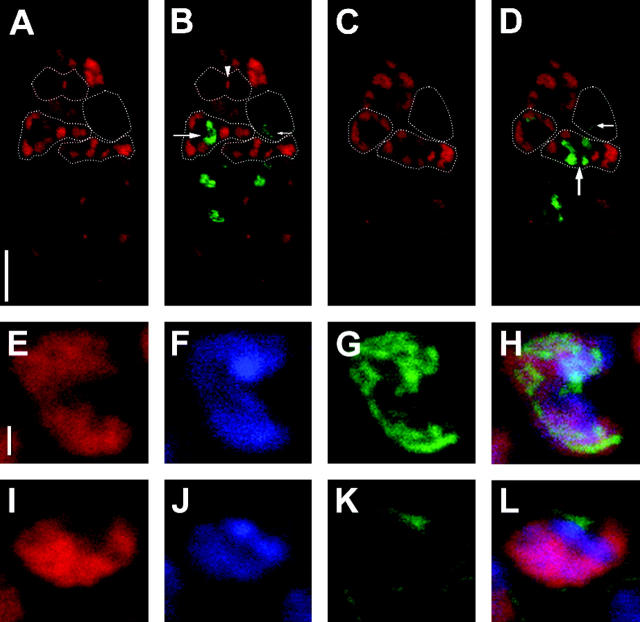
Figure 3.—
Phenotype of embryos from klp3A females. In A–E, embryos are stained with antihistones (green) to visualize chromatin and antitubulin (red) to visualize microtubules. (A) Embryo from a klp3Amei-352/FM7w female in mitotic cycle 2. Two zygotic nuclei are present on mitotic spindles with centrosomes at each pole (small arrows). The polar body chromatin mass is visible at left (large arrow). (B) Low-magnification image of an embryo from a klp3Amei-352 homozygous female showing that only two masses of chromatin are present: a large mass of polar body chromatin and a smaller nucleus on a spindle. (C) Higher-magnification image of the embryo from B. The mass of polar body chromatin is associated with a circular concentration of microtubules (large arrow) and the small nucleus sits on a spindle (small arrow). However, the spindle appears to lack asters associated with the spindle poles but several free-lying centrosomes are present (arrowheads). (D) Embryo from a klp3Amei-352/klp3A521 female. The only antihistone signals are the polar body chromatin mass (large arrow) and a small nucleus residing on a spindle (small arrow). Centrosomes (arrowheads) appear to have dissociated from the spindle and additional centrosomes are visible in the embryo. (E) Embryo from a klp3Amei-352/klp3A1124 female. Five independent centrosomes are visible (arrowheads), but none appear associated with the single spindle (small arrow) or polar body chromatin mass (large arrow).
These results indicate that the maternal-effect lethality in both klp3Amei-352 and other klp3A mutants occurs at a very early stage of embryogenesis and that this phenotype is not complemented in klp3Amei-352/klp3A heterozygotes. The lack of pronuclear fusion, due to either failure of female pronucleus specification or movement of pronuclei toward each other (Williams et al. 1997), most likely underlies this early arrest phenotype. The degree to which mutations in klp3A manifest this phenotype varies depending on the allele. Our results show the greatest frequency of early arrest among embryos from klp3Amei-352/klp3A521 trans-heterozygotes, and the lowest frequency for klp3Amei-352/klp3A1124 (Table 3). This agrees with a previous analysis of these alleles performed using Df(1)54 and klp3Ae4 trans-heterozygotes (Williams et al. 1997).
Alterations of meiotic exchange distribution are a general feature of klp3A mutants:
To determine whether disruptions in meiotic exchange distribution are specific to the klp3Amei-352 allele or a general phenotype of klp3A mutants, we analyzed meiotic exchange in females carrying mutations in klp3A (Figure 1 and Table 4). Although the maternal-effect lethality among progeny of these females results in near sterility, we were able to measure exchange in the minority of progeny that escape this phenotype.
TABLE 4.
Results of crosses of females of the genotypeX/X;net ho dp Sp b pr cn/+ + + + + + + carrying the indicatedX chromosomes by +/Y;net ho dp b pr cn/net ho dp b pr cn males
| Maternal genotype
|
||||||
|---|---|---|---|---|---|---|
| Progeny typea | Df(1)ED6579/+ | klp3Amei-352/Df(1)ED6579 | klp3A521/Df(1)ED6579 | klp3A1124/Df(1)ED6579 | klp3A835/Df(1)ED6579 | klp3Amei-352 |
| Noncrossover | 1139 | 856 | 132 | 374 | 34 | 156 |
| Single crossover | ||||||
| 1 | 235 | 128 | 19 | 75 | 4 | 13 |
| 2 | 546 | 361 | 50 | 191 | 17 | 52 |
| 3 | 66 | 73 | 18 | 46 | 3 | 14 |
| 4 | 17 | 42 | 5 | 7 | 1 | 7 |
| Double crossover | ||||||
| 1, 2 | 11 | 7 | 1 | 5 | 0 | 1 |
| 1, 3 | 3 | 9 | 0 | 4 | 0 | 0 |
| 1, 4 | 2 | 3 | 1 | 0 | 1 | 0 |
| 2, 3 | 7 | 9 | 0 | 4 | 1 | 4 |
| 2, 4 | 7 | 9 | 3 | 3 | 1 | 2 |
| 3, 4 | 1 | 1 | 0 | 0 | 0 | 0 |
| Triple crossover | 0 | 1 | 0 | 0 | 0 | 0 |
| Total progeny | 2034 | 1499 | 229 | 709 | 62 | 249 |
| Map distances | ||||||
| 1 (net-dp) | 12.34 | 9.87 | 9.17 | 11.85 | 8.07 | 5.62 |
| 2 (dp-b) | 28.07 | 25.82 | 23.58 | 28.63 | 30.65 | 23.69 |
| 3 (b-pr) | 3.79 | 6.14 | 7.86 | 7.62 | 6.45 | 7.23 |
| 4 (pr-cn) | 1.33 | 3.74 | 3.93 | 1.41 | 4.84 | 3.61 |
| Total | 45.53 | 45.56 | 44.54 | 49.51 | 50.00 | 40.16 |
| Map relative to control | ||||||
| 1 (net-dp) | 1 | 0.800 | 0.743 | 0.960 | 0.654 | 0.455 |
| 2 (dp-b) | 1 | 0.920 | 0.840 | 1.020 | 1.092 | 0.844 |
| 3 (b-pr) | 1 | 1.620 | 2.074 | 2.011 | 1.702 | 1.908 |
| 4 (pr-cn) | 1 | 2.812 | 2.955 | 1.060 | 3.639 | 2.714 |
| Total | 1 | 1.001 | 0.978 | 1.087 | 1.098 | 0.882 |
| Exchange rank frequencies | ||||||
| E0 | 0.150 | 0.193 | 0.197 | 0.100 | 0.194 | 0.309 |
| E1 | 0.789 | 0.708 | 0.716 | 0.810 | 0.613 | 0.578 |
| E2 | 0.061 | 0.093 | 0.087 | 0.090 | 0.194 | 0.112 |
| E3 | 0 | 0.005 | 0 | 0 | 0 | 0 |
Female progeny were scored (see Page et al. 2000) except for the klp3Amei-352 cross, in which all progeny were scored.
Region 1 is net-dp; region 2 is dp-b; region 3 is b-pr; and region 4 is pr-cn.
Homozygotes for klp3Amei-352 show little deviation from wild-type controls in terms of total exchange frequency (Table 4). However, the distribution of exchanges is altered. In the most telomeric interval, net-dp, the genetic map distance is decreased to ∼50% of wild type, whereas the b-pr and pr-cn intervals are increased relative to wild type. The map distance for the pr-cn interval, which spans the centromere region of chromosome 2, is >2.5 times the length of wild type. The frequency of exchange is elevated in centromeric regions, which ordinarily have less exchange per unit of physical distance, whereas the reverse occurs in the distal region of the chromosome, in which exchange per unit of physical distance is normally high. The distribution of exchange observed for klp3Amei-352 is thus more random than that in wild type. A similar phenotype is observed for klp3Amei-352/Df(1)ED6579 females. The total frequency of exchange is very close to that measured for Df(1)ED6579/+ control females, yet the distribution of those exchanges is altered. The degree to which the distribution is skewed seems to be increased in klp3Amei-352 homozygotes relative to klp3Amei-352/Df(1)ED6579. This could indicate a dose-dependent effect caused by a higher level of the mutant Klp3A protein encoded by the klp3Amei-352 allele or perhaps the existence of genetic modifiers of the exchange phenotype.
To compare the effects of klp3A mutants on exchange, we analyzed females that were trans-heterozygous for klp3A521, klp3A835, or klp3A1124, and the deficiency Df(1)ED6579 (Figure 1 and Table 4). Similar to the klp3Amei-352 allele, all three mutant genotypes demonstrated a total exchange frequency that was similar to the Df(1)ED6579/+ control. For both klp3A521/Df(1)ED6579 and klp3A835/Df(1)ED6579, the exchange distribution was altered in a manner similar to klp3Amei-352. Interestingly, the klp3A1124/Df(1)ED6579 females revealed a distribution of exchange that was similar to control (with the exception of the b-pr interval). Studies of the maternal-effect lethality phenotype suggested that klp3A1124 is a weaker allele than the others tested (Williams et al. 1997; this study), so the results could represent a very subtle phenotype. Alternatively, the residue affected in the klp3A1124 allele may not be required for the role of Klp3A in exchange distribution.
Klp3A protein is present in germline cysts during early meiotic prophase:
In the Drosophila ovary, germline cells form 16-cell cysts within region 2A of the germarium. Meiotic recombination is thought to be initiated soon after 16-cell cyst formation and completed by the exit of the cyst from the germarium, on the basis of timing of the appearance and disappearance of recombination nodules and phosphorylated histone H2AV (Carpenter 1979; Jang et al. 2003). To better understand the role that Klp3A could be playing in the regulation of exchange, we examined the distribution of Klp3A protein in germaria from wild-type females.
In previous studies, Klp3A displayed a dynamic localization pattern during embryonic mitotic divisions and meiotic divisions in oocytes and spermatocytes (Williams et al. 1995, 1997; Kwon et al. 2004). This pattern is generally characterized by nuclear localization during interphase and prophase and association with the spindle midzone during anaphase and telophase. Although previous studies investigated Klp3A localization on female meiotic spindles from metaphase through anaphase II, the presence of Klp3A at earlier stages of female meiosis was not determined.
To localize Klp3A in germline cysts that were entering meiosis, germaria were co-immunostained with antibodies against Klp3A and the synaptonemal complex (SC) protein C(3)G (Figure 4, A–D). The SC forms between homologous chromosomes at the start of meiotic prophase and can be visualized using anti-C(3)G as a thread-like nuclear staining pattern (Page and Hawley 2001). C(3)G thus acts as a marker for cysts in which meiosis has initiated and also indicates nuclei in which meiotic recombination is occurring, since exchange normally occurs in the context of the SC in Drosophila (Page and Hawley 2001; Jang et al. 2003).
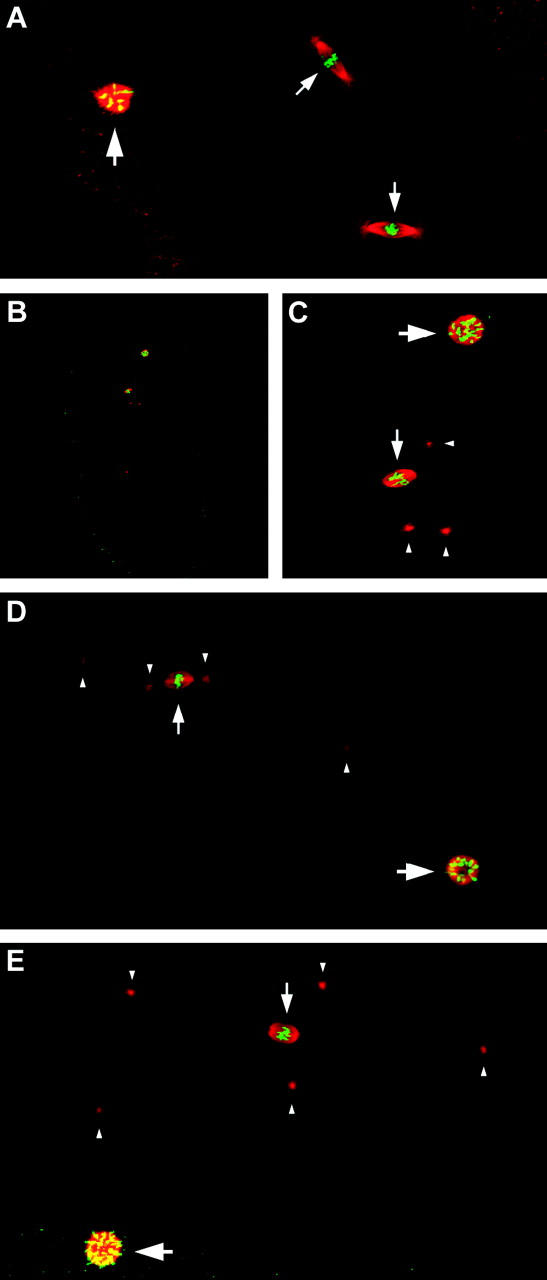
Figure 4.—
Localization of Klp3A protein during early meiotic prophase. (A–D) Two optical sections from the same w1118 germarium stained with anti-Klp3A (red) and anti-C(3)G (green). The germarium is oriented with the anterior tip at the top. As developing cysts (dotted outlines) move toward the posterior, the initiation of meiosis is marked by the formation of the SC, of which C(3)G is a component. Mitotically dividing germline cells display a variable localization of Klp3A, including concentration at the spindle midbody in telophase (arrowhead in B), similar to Klp3A localization in embryos (Kwon et al. 2004). Klp3A level is low in zygotene cysts, in which the SC is just beginning to form and the anti-C(3)G signal is weak and punctate (small arrows in B and D). Two pachytene 16-cell cysts that show full accumulation of C(3)G protein (large arrows in B and D) also show distinct, strong nuclear localization of Klp3A. The level of Klp3A apparently decreases in older 16-cell cysts as prophase continues, although Klp3A can be observed in the follicle cells surrounding these cysts. Bar, 10 μm. (E–H) A cell from a 16-cell cyst stained with anti-Klp3A (red, E), DAPI (blue, F), and anti-C(3)G (green, G). A merged image is shown in H. The anti-Klp3A signal shows extensive colocalization with DAPI-stained DNA and C(3)G, suggesting that the Klp3A protein may be associated with chromatin. Bar, 1 μm. (I–L) A cell from a 16-cell cyst stained with anti-Klp3A (red, I), DAPI (blue, J), and antilamin (green, K). A merged image is shown in L. The strong anti-Klp3A signal is contained within the nuclear envelope, as detected by antilamin, whereas Klp3A protein in the cytoplasm is not detected above background levels. Bar, 1 μm.
In the germarium, developing cysts travel from the anterior tip toward the posterior. The most-anterior cysts that contain cells expressing C(3)G protein are therefore newly formed 16-cell cysts in which meiosis has initiated. Within these cysts, Klp3A is initially present in low levels that rise as SC formation proceeds (Figure 4, A–D). In cysts that have reached full SC formation, Klp3A protein shows strong nuclear localization in all cells of the cyst, including the pro-oocytes that contain the highest levels of C(3)G. Klp3A protein could also be observed in younger cysts that had not yet entered meiosis and were undergoing mitotic divisions. The nuclear or cytoplasmic localization of Klp3A within the younger cysts appeared to vary both within and between germaria, as might be expected for cells undergoing mitosis and therefore in different phases of the cell cycle. A fraction of these were in telophase and displayed Klp3A localization to midbodies between cells (Figure 4, A and B). In contrast, the robust nuclear localization in 16-cell cysts was consistent among germaria. The strong nuclear localization of Klp3A was present in approximately one to three 16-cell cysts per germarium, which always coincided with the most-anterior cysts in which a pachytene level of C(3)G was also present. In older cysts that were in more posterior positions in the germarium, the anti-Klp3A signal was dramatically reduced, possibly indicating a rapid reduction of Klp3A protein level.
In pachytene meiotic cells, Klp3A appears to be present throughout the nucleus and shows extensive colocalization with DNA. Comparison of anti-Klp3A staining with nuclear DNA visualized using DAPI (Figure 4, E and H) showed that Klp3A is found throughout the nucleus. Much of the anti-Klp3A signal overlaps with DNA, although in some nuclei (as shown in Figure 4, I–L), Klp3A was clearly less intensely localized in regions of heterochromatin, which appear as regions that are more brightly stained with DAPI in the nucleus. Co-immunostaining germaria with anti-Klp3A and antilamin indicated that the majority of the Klp3A signal is located within the interior of the nucleus (Figure 4, I–L). Klp3A protein in the cytoplasm was not detected above background levels in these cells.
The intense localization of Klp3A in early meiotic prophase nuclei suggests that Klp3A could act prior to or during the initiation of meiotic recombination or perhaps during the processing of recombination intermediates to influence the final distribution of exchanges during meiosis. Klp3A contains a cysteine-rich zinc-finger-like motif in its tail region that might mediate interactions with chromatin. The action of Klp3A in the recombination process may be limited to early meiotic prophase, however, since the level of Klp3A protein in meiotic cysts appears to decrease rapidly following SC formation.
DISCUSSION
Many species exhibit a nonrandom distribution of meiotic exchanges along the length of a chromosome, but little is known about the factors that determine the nonrandom exchange distribution. These determinants may differ among species as well. Human, mouse, Drosophila, and maize chromosomes generally show increased rates of exchange as distance from the centromere increases and distance from the telomeres decreases (Lindsley and Sandler 1977; Froenicke et al. 2002; McKim et al. 2002; Anderson et al. 2003; Kong et al. 2004). This does not seem to be the case for yeast chromosomes, although recombination rates are highly variable along each chromosome in both yeast and mammals (Kaback et al. 1989; Baudat and Nicolas 1997; Froenicke et al. 2002; Kong et al. 2004; Malkova et al. 2004). In yeast, recombination rates are correlated with sites of frequent double-strand break (DSB) formation (Baudat and Nicolas 1997). Mapping of these recombination hotspots in yeast has suggested that the distribution of hotspots is related to G/C content and transcriptional activity rather than position relative to centromere or telomeres (Gerton et al. 2000). The phenomenon of interference, in which exchanges are inhibited from occurring near each other, also affects their distribution along a chromosome arm. Interference is thought to result from either the physical properties of the structural axis of the chromosome or regulation of the periodicity of resolving recombination intermediates as crossovers (Kleckner et al. 2004; Stahl et al. 2004).
Along the euchromatin of Drosophila chromosome arms, exchange per unit of physical distance increases with distance from the centromeric heterochromatin and peaks in the distal half of the chromosome, sometimes with a slight decrease near the telomere (Lindsley and Sandler 1977; McKim et al. 2002). Lindsley and Sandler (1977) proposed that the distribution of exchanges is determined both by cis-acting chromosomal features and by trans-acting proteins. Studies of exchange along chromosomes bearing inversions that reposition euchromatic sequences on the chromosome arm relative to the centromere and/or heterochromatin showed that the pattern of exchanges generally remains associated with the sequence intervals, regardless of orientation or distance from the centromere or heterochromatin (Szauter 1984). A series of cis-acting sites located along the euchromatin appear to have a role in establishing this pattern (Hawley 1980; Sherizen et al. 2004). Putative trans-acting factors that affect exchange distribution have been identified among a class of exchange-defective meiotic mutants in Drosophila that reduce the total frequency of exchange while altering its distribution. This group has been called “precondition” mutants because their phenotype was thought to result from a defective ability for chromosome regions to establish the capability to undergo exchange (Carpenter and Sandler 1974). However, analyses of the molecular function of many of the genes identified through these mutations support the contention that alterations in exchange distribution may result from defects in any of several stages of meiotic recombination (Bhagat et al. 2004).
In contrast to most mutants that affect the distribution of exchange, the total frequency of exchange is unaffected by mutations in klp3A. The lack of an exchange reduction in klp3A mutants suggests that Klp3A may be required specifically for establishing the distribution of exchange along the chromosomes. Since meiotic exchange is measured among progeny that escape from the maternal-effect lethality that also results from mutation of klp3A, it is possible that the phenotype results from selection against embryos derived from oocytes in which the chromosomes experienced medially or distally located exchanges or lack exchanges completely. This could occur if oocytes lacking Klp3A were impaired in their ability to segregate chromosomes lacking an exchange or with particular exchange configurations. However, few abnormalities in meiotic spindle function and chromosome segregation were observed for klp3A mutants (Williams et al. 1997), and the frequency of meiotic nondisjunction is low for both the X and always achiasmate 4th chromosomes (Baker and Carpenter 1972). Moreover, the distribution of crossovers in progeny from klp3A1124/Df(1)ED6579 mothers differs from the distributions observed in other klp3A mutant genotypes, even though the maternal-effect lethality phenotype is quite similar, suggesting that the defect resulting in maternal-effect lethality does not determine the ability to recover certain recombinant progeny types. Thus, it is unlikely that the specific defect in exchange distribution is the result of a selection bias.
The Klp3A protein could regulate exchange distribution in several ways. First, Klp3A may regulate the nonrandom localization of DSB formation along the chromosome. In the absence of Klp3A, the DSB sites may be redistributed more in proportion with physical distance. If, as in yeast, the frequency of exchange is largely determined by the frequency of DSB formation, the recombination machinery acting on the DSBs would distribute exchanges similarly. Second, Klp3A could influence the decision between crossover and noncrossover fates for DSBs or recombination intermediates. In wild type, Klp3A would act to promote crossover fate in the distal region of chromosome arms and/or promote noncrossover fate in proximal regions. The loss of this regulation in klp3A mutants could then result in the observed redistribution of crossovers. Third, Klp3A may be necessary for an aspect of chromosome organization that normally enables the wild-type distribution of exchange. The abnormal distribution of exchange caused by klp3A mutants would arise through a regional misregulation of recombination resulting from the defective organization of homologous sequences during meiotic prophase.
Evidence for a role for kinesin-like motor proteins in chromosome organization has been found in other systems. In yeast, Kar3p was proposed to regulate chromosomal interactions during early meiotic prophase (Bascom-Slack and Dawson 1997). Mutations in the kar3 gene result in defects in SC and DSB formation, and Trelles-Sticken et al. (2003) demonstrated a defect in the release of telomere clustering in kar3-deficient haploid meiosis. Similar to Klp3A, Kar3p has been shown to localize to the nucleus and to the spindle poles during meiotic prophase (Shanks et al. 2001). Immunolocalization of the SC protein C(3)G in klp3Amei-352 mutant ovaries has not indicated a defect in SC formation, however (data not shown).
Alternatively, Klp3A could be required for normal chromatin condensation during meiotic prophase. We have demonstrated that Klp3A localizes within pachytene nuclei, where it seems to preferentially colocalize with euchromatin. Another Kinesin-4 protein, human KIF4, has been shown to interact with the condensin complex, and depletion of KIF4 results in hypercondensation of mitotic chromosomes (Mazumdar et al. 2004). The condensation state of meiotic chromatin is likely to affect the ability of meiotic DSBs to form (Baudat and Nicolas 1997; Reddy and Villeneuve 2004; Yamada et al. 2004). Thus, disruption of the normal degree or rate of chromatin condensation in meiotic prophase in the absence of Klp3A could result in an altered distribution of exchanges. Although no evidence of a condensation defect has been reported in Klp3A-depleted mitotic cells (Goshima and Vale 2003; Brust-Mascher et al. 2004; Kwon et al. 2004), this role may be accomplished by a redundant kinesin when Klp3A is eliminated.
In conclusion, we have identified mei-352 as an allele of klp3A and uncovered a previously unrecognized function for the kinesin-like protein Klp3A in regulating the distribution of exchanges during meiosis. Loss of Klp3A function results in an increase in exchange in centromere-proximal regions of chromosomes and a decrease in exchange in distal regions of chromosome arms. The meiotic defect is specific to the distribution of exchanges, as the total frequency of exchange along a chromosome arm is unchanged. The meiotic defect observed for klp3A may reflect a role for this kinesin-like protein in chromosome organization during early meiosis.
Acknowledgments
We thank Ed Van Veen and Jennifer Jeffress for technical assistance. We are grateful to Byron Williams and Michael Goldberg for providing klp3A stocks and reagents and to Jon Scholey for affinity-purified anti-Klp3A. We also thank members of the DrosDel Isogenic Deficiency Project (Ed Ryder, Åsa Rasmuson-Lestander, and Karin Ekström) and the Bloomington and Szeged Drosophila Stock Centers for providing fly stocks. The monoclonal antibody ADL101 was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by The University of Iowa Department of Biological Sciences. This work was supported by a grant from the National Institutes of Health (GMO51444) to R.S.H.
References
- Anderson, L. K., G. G. Doyle, B. Brigham, J. Carter, K. D. Hooker et al., 2003. High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165: 849–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L. K., S. M. Royer, S. L. Page, K. S. McKim, A. Lai et al., 2005. Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. USA 102: 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, B. S., and A. T. C. Carpenter, 1972. Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. Genetics 71: 255–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, B. S., and J. C. Hall, 1976 Meiotic mutants: genetic control of meiotic recombination and chromosome segregation, pp. 351–434 in The Genetics and Biology of Drosophila, Vol. 1a, edited by M. Ashburner and E. Novitski. Academic Press, New York.
- Bascom-Slack, C. A., and D. S. Dawson, 1997. The yeast motor protein, Kar3p, is essential for meiosis I. J. Cell Biol. 139: 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat, F., and A. Nicolas, 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. USA 94: 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat, R., E. A. Manheim, D. E. Sherizen and K. S. McKim, 2004. Studies on crossover-specific mutants and the distribution of crossing over in Drosophila females. Cytogenet. Genome Res. 107: 160–171. [DOI] [PubMed] [Google Scholar]
- Brust-Mascher, I., G. Civelekoglu-Scholey, M. Kwon, A. Mogilner and J. M. Scholey, 2004. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc. Natl. Acad. Sci. USA 101: 15938–15943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A. T. C., 1979. Synaptonemal complex and recombination nodules in wild-type Drosophila melanogaster females. Genetics 92: 511–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A. T. C., and B. S. Baker, 1982. On the control of the distribution of meiotic exchange in Drosophila melanogaster. Genetics 101: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A. T. C., and L. Sandler, 1974. On recombination-defective meiotic mutants in Drosophila melanogaster. Genetics 76: 453–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker, S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern et al., 2002 Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: research0079.0071–0079.0014. [DOI] [PMC free article] [PubMed]
- Craymer, L., and E. Roy, 1980. Report of new mutants—Drosophila melanogaster. Dros. Inf. Serv. 55: 200–204. [Google Scholar]
- Demerec, M., 1940. Genetic behavior of euchromatic segments inserted into heterochromatin. Genetics 25: 618–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale, R. A., M. A. Crosby, W. Gelbart, K. Campbell, D. Emmert et al., 2005. FlyBase: genes and gene models. Nucleic Acids Res. 33(database issue): D390–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froenicke, L., L. K. Anderson, J. Wienberg and T. Ashley, 2002. Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 71: 1353–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown et al., 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti, M. G., S. Bonaccorsi, B. Williams, E. V. Williams, C. Santolamazza et al., 1998. Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev. 12: 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, M. L., R. A. Colvin and A. F. Mellin, 1989. The Drosophila zeste gene is nonessential. Genetics 123: 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and R. D. Vale, 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162: 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold, T., and P. Hunt, 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2: 280–291. [DOI] [PubMed] [Google Scholar]
- Hawley, R. S., 1980. Chromosomal sites necessary for normal levels of meiotic recombination in Drosophila melanogaster. I. Evidence for and mapping of the sites. Genetics 94: 625–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, J. K., D. E. Sherizen, R. Bhagat, E. A. Manheim and K. S. McKim, 2003. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116: 3069–3077. [DOI] [PubMed] [Google Scholar]
- Judd, B. H., M. W. Shen and T. C. Kaufman, 1972. The anatomy and function of a segment of the X chromosome in Drosophila melanogaster. Genetics 71: 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback, D. B., H. Y. Steensma and P. de Jonge, 1989. Enhanced meiotic recombination on the smallest chromosome of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86: 3694–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner, N., D. Zickler, G. H. Jones, J. Dekker, R. Padmore et al., 2004. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA 101: 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, X., K. Murphy, T. Raj, C. He, P. S. White et al., 2004. A combined linkage-physical map of the human genome. Am. J. Hum. Genet. 75: 1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, M., S. Morales-Mulia, I. Brust-Mascher, G. C. Rogers, D. J. Sharp et al., 2004. The chromokinesin, KLP3A, drives mitotic spindle pole separation during prometaphase and anaphase and facilitates chromatid motility. Mol. Biol. Cell 15: 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, N. E., K. Yu, J. Shaffer, E. Feingold and S. L. Sherman, 2005. Association between maternal age and meiotic recombination for trisomy 21. Am. J. Hum. Genet. 76: 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. J., R. K. Dawe, K. R. Christie, D. W. Cleveland, S. C. Dawson et al., 2004. A standardized kinesin nomenclature. J. Cell Biol. 167: 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. L., and L. Sandler, 1977. The genetic analysis of meiosis in female Drosophila melanogaster. Philos. Trans. R. Soc. Lond. B Biol. Sci. 277: 295–312. [DOI] [PubMed] [Google Scholar]
- Malkova, A., J. Swanson, M. German, J. H. McCusker, E. A. Housworth et al., 2004. Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics 168: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar, M., S. Sundareshan and T. Misteli, 2004. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J. Cell Biol. 166: 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K. S., J. K. Jang and E. A. Manheim, 2002. Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet. 36: 205–232. [DOI] [PubMed] [Google Scholar]
- Mohler, J. D., 1977. Developmental genetics of the Drosophila egg. I. Identification of 59 sex-linked cistrons with maternal effects on embryonic development. Genetics 85: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler, J. D., and A. Carroll, 1984. Female sterile mutations in the Iowa collection. Dros. Inf. Serv. 60: 236–241. [Google Scholar]
- Page, S. L., and R. S. Hawley, 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S. L., K. S. McKim, B. Deneen, T. L. Van Hook and R. S. Hawley, 2000. Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics 155: 1757–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli, D., B. Oliver and A. P. Mahowald, 1995. Identification of regions interacting with ovoD mutations: potential new genes involved in germline sex determination or differentiation in Drosophila melanogaster. Genetics 139: 713–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, K. C., and A. M. Villeneuve, 2004. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118: 439–452. [DOI] [PubMed] [Google Scholar]
- Rothwell, W. F., and W. Sullivan, 2000 Fluorescent analysis of Drosophila embryos, pp. 141–157 in Drosophila Protocols, edited by W. Sullivan, M. Ashburner and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ryder, E., F. Blows, M. Ashburner, R. Bautista-Llacer, D. Coulson et al., 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack, S., F. J. Kull and E. Mandelkow, 1999. Motor proteins of the kinesin family. Structures, variations, and nucleotide binding sites. Eur. J. Biochem. 262: 1–11. [DOI] [PubMed] [Google Scholar]
- Sandler, L., D. L. Lindsley, B. Nicoletti and G. Trippa, 1968. Mutants affecting meiosis in natural populations of Drosophila melanogaster. Genetics 60: 525–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky, J. J., K. S. McKim, L. Messina, R. L. French, W. D. Hurley et al., 1999. Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics 152: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks, R. M., R. J. Kamieniecki and D. S. Dawson, 2001. The Kar3-interacting protein Cik1p plays a critical role in passage through meiosis I in Saccharomyces cerevisiae. Genetics 159: 939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherizen, D., J. K. Jang, R. Bhagat, N. Kato and K. S. McKim, 2004. Meiotic recombination in Drosophila females depends on chromosome continuity between genetically defined boundaries. Genetics 169: 767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slizynska, H., 1938. Salivary chromosome analysis of the white-facet region of Drosophila melanogaster. Genetics 23: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R. F., and R. J. Konopka, 1981. Circadian clock phenotypes of chromosome aberrations with a breakpoint at the per locus. Mol. Gen. Genet. 183: 243–251. [DOI] [PubMed] [Google Scholar]
- Somma, M. P., B. Fasulo, G. Cenci, E. Cundari and M. Gatti, 2002. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell 13: 2448–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, F. W., H. M. Foss, L. S. Young, R. H. Borts, M. F. Abdullah et al., 2004. Does crossover interference count in Saccharomyces cerevisiae? Genetics 168: 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szauter, P., 1984. An analysis of regional constraints on exchange in Drosophila melanogaster using recombination-defective meiotic mutants. Genetics 106: 45–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf, W. E., 1994 Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis, pp. 489–505 in Drosophila melanogaster: Practical Uses in Cell and Molecular Biology, edited by L. S. B. Goldstein and E. A. Fryberg. Academic Press, San Diego. [DOI] [PubMed]
- Trelles-Sticken, E., J. Loidl and H. Scherthan, 2003. Increased ploidy and KAR3 and SIR3 disruption alter the dynamics of meiotic chromosomes and telomeres. J. Cell Sci. 116: 2431–2442. [DOI] [PubMed] [Google Scholar]
- Vernos, I., and E. Karsenti, 1995. Chromosomes take the lead in spindle assembly. Trends Cell Biol. 5: 297–301. [DOI] [PubMed] [Google Scholar]
- Williams, B. C., M. F. Riedy, E. V. Williams, M. Gatti and M. L. Goldberg, 1995. The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J. Cell Biol. 129: 709–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B. C., A. F. Dernburg, J. Puro, S. Nokkala and M. L. Goldberg, 1997. The Drosophila kinesin-like protein KLP3A is required for proper behavior of male and female pronuclei at fertilization. Development 124: 2365–2376. [DOI] [PubMed] [Google Scholar]
- Woehlke, G., A. K. Ruby, C. L. Hart, B. Ly, N. Hom-Booher et al., 1997. Microtubule interaction site of the kinesin motor. Cell 90: 207–216. [DOI] [PubMed] [Google Scholar]
- Yamada, T., K. I. Mizuno, K. Hirota, N. Kon, W. P. Wahls et al., 2004. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 23: 1792–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]