Abstract
The flavonoid pigment pathway in plants has been used as a model system for studying gene regulatory mechanisms. C2-Idf is a stable dominant mutation of the chalcone synthase gene, c2, which encodes the first dedicated enzyme in this biosynthetic pathway of maize. Homozygous C2-Idf plants show no pigmentation. This allele also inhibits expression of functional C2 alleles in heterozygotes, producing a less pigmented condition instead of the normal deeply pigmented phenotype. To explore the nature of this effect, the C2-Idf allele was cloned. The gene structure of the C2-Idf haplotype differs substantially from that of the normal c2 gene in that three copies are present. Two of these are located in close proximity to each other in a head-to-head orientation and the third is closely linked. Previous experiments showed that the lower level of pigmentation in heterozygotes is correlated with reduced enzyme activity and low steady-state mRNA levels. We found that c2 transcription occurs in nuclei of C2-Idf/C2 heterozygotes, but mRNA does not accumulate, suggesting that the inhibition is mediated by RNA silencing. Infection of C2-Idf/C2 heterozygotes with viruses that carry suppressors of RNA silencing relieved the phenotypic inhibition, restoring pigment production and mRNA levels. Finally, we detected small interfering RNAs (siRNAs) in plants carrying C2-Idf, but not in plants homozygous for the wild-type C2 allele. Together, our results indicate that the inhibitory effect of C2-Idf occurs through RNA silencing.
THE maize flavonoid pigment pathway offers an excellent model system for studying the regulation of gene expression. The pathway is genetically well characterized and most of the structural and regulatory genes have been cloned (Dooner et al. 1991). Genetic and molecular studies of a wide spectrum of mutants have revealed that the distribution and level of pigment accumulation accurately reflect activity of these genes (Coe et al. 1988). In maize, several genes involved in anthocyanin biosynthesis have been identified. Among these, regulatory genes encoding a suite of transcription factors as well as structural genes encoding biosynthetic enzymes have been extensively characterized at the molecular level (Dooner et al. 1991). From these data, detailed knowledge about the regulation, mode of interaction, and function of maize anthocyanin genes has been assembled.
One of the structural genes, colorless2 (c2), encodes chalcone synthase, the enzyme responsible for the first dedicated step in the pathway. In combination with appropriate regulatory alleles, a normal C2 allele leads to pigment production in many parts of the plant, including the pericarp, the aleurone layer of the endosperm, tassels, and vegetative organs such as ear husks and leaf sheaths. A colorless mutant, initially called Inhibitor diffuse (Idf), was isolated from Peruvian lines as a dominant inhibitor of pericarp pigmentation (Brink and Greenblatt 1954). Later, when the mutation was mapped to the c2 locus, the allele became known as C2-Idf (Brink and Greenblatt 1954). When heterozygous with a normal C2 allele, C2-Idf reduces pigmentation not only in the pericarp, but also in the aleurone. At the enzyme level, the effect on aleurone pigmentation is due to a lack of chalcone synthase enzyme activity (Dooner 1983). At the RNA level, c2-homologous RNA is not detectable in either tassels or aleurone of C2-Idf homozygotes (Franken et al. 1991).
Unlike cases of paramutation (for reviews see Chandler and Stam 2004; Della Vedova and Cone 2004), C2-Idf inhibition is not meiotically heritable. These observations suggest that the absence of enzyme activity in C2-Idf/C2 heterozygotes results from a reduction of C2 mRNA accumulation. In this work, we investigated the mechanism of inhibition of the C2 allele by C2-Idf.
Similar semidominant types of mutations in chalcone synthase genes have been described and analyzed in Antirrhinum majus L. (niv-525 allele) and soybean (chalcone synthase allele I) (Coen and Carpenter 1988; Todd and Vodkin 1996; Tuteja et al. 2004). In these inhibitory mutants, the dominant negative effect likely results from either gene duplication events of the chalcone synthase gene (as shown in the case of soybean; Tuteja et al. 2004) or the production of antisense transcripts (as proposed for the niv-525 allele of A. majus L.; Coen and Carpenter 1988). The inhibitory effects of these mutants were mimicked phenotypically in transgenic petunia lines; plants carrying multiple insertion copies of the chalcone synthase gene showed reduction of normal gene activity, caused by a cosuppression effect that involves RNA silencing (Napoli et al. 1990; van der Krol et al. 1990; Jorgensen et al. 1996; Metzlaff et al. 1997).
RNA silencing refers to a homology-dependent type of gene silencing that employs RNA to mediate the targeted degradation of homologous transcripts. The process is triggered by the production of aberrant RNA, which is usually at least partially double stranded. Double-stranded RNA is recognized by an RNase-III like enzyme, referred to as Dicer (Bernstein et al. 2001), and cleaved into small double-stranded RNAs of 21–26 nt known as small interfering RNAs (siRNAs) (Hamilton and Baulcombe 1999). These siRNAs are incorporated into a ribonucleoprotein complex known as the RNA-induced silencing complex (RISC), which targets homologous transcripts, catalyzing their degradation (Hammond et al. 2001). Thus, any transcript homologous to the aberrant RNA is destroyed, essentially silencing the expression of the cognate gene. The RNA silencing machinery can also participate in chromatin modifications, whereby siRNAs may recognize homologous DNA loci and induce remodeling of the surrounding chromatin into a more restrictive state, effectively silencing transcription from that locus (Pal-Bhadra et al. 2002; Volpe et al. 2002; Cao et al. 2003; Chan et al. 2004; Verdel et al. 2004).
RNA silencing has likely evolved as a defense mechanism against invasive nucleic acids (Herbert 2004). For example, in many plant viruses with a single-stranded RNA genome, replication involves a double-stranded RNA intermediate produced by a virally encoded RNA-dependent RNA polymerase (Baulcombe 1996). This double-stranded transcript is recognized as aberrant by the host plant and triggers RNA silencing. The virus infection is thus controlled by degradation of its RNA, preventing further cycles of replication (Al-Kaff et al. 1998). However, as a counter defense, many plant viruses carry genes that encode proteins capable of suppressing the plant's RNA-silencing machinery. A number of such suppressors of RNA silencing have been identified, but the mechanism by which suppression is achieved remains unclear for most (Ahlquist 2002). The two best-described viral suppression proteins are P1/HC-Pro from potyviruses and p19 from tombusviruses. P1/HC-Pro appears to prevent Dicer cleavage of the aberrant precursor RNA while p19 binds siRNAs and thus prevents them from acting as guides for degradation of homologous transcripts (Dunoyer et al. 2004; Lakatos et al. 2004).
A conserved feature of RNA silencing is that it is triggered by double-stranded RNA molecules, which, in some cases, originate from transcription of repeated DNA segments (Muskens et al. 2000; Wassenegger 2000). Two recently published descriptions of RNA silencing in rice and soybean involve endogenous alleles that are composed of multiple genes arranged in inverted repeat orientations (Kusaba et al. 2003; Tuteja et al. 2004). In this study, we addressed the basis of silencing by C2-Idf in maize and conclude that it involves an RNA-based silencing mechanism.
MATERIALS AND METHODS
Genetic stocks:
Genetic and molecular analyses of the C2-Idf mutant line were carried out in W22 or Mo17 inbred backgrounds carrying R-scm2 and C1 or in the original genetic C2-Idf background (A1, C1, R1, P-wr/W22). For comparison, a color-converted W22 carrying A1, A2, C1, C2, and R1 (Wienand et al. 1986, line C) was used; in the present study, this normal c2 allele is designated C2-W22. For analysis of husk RNA levels, C2 and C2-Idf in a W22 inbred background were backcrossed three times to a stock, which is homozygous for R-g, C1, B-I, Pl-Rhoades, and P-ww. With C2 homozygous, these plants are deep purple. The negative controls were plants homozygous for R-r; c1; B-I; pl-0; P-ww. The pl-0 mutation prevents any accumulation of anthocyanin in vegetative tissue. With C2 homozygous, these plants are green (P. Cooper and K. Cone, unpublished results). All stocks used in this study were homozygous dominant for white pollen1 (whp), a duplicate of the c2 gene that influences pollen viability. C2-Idf plants have normal pollen.
Probes:
c2-specific probes used in DNA and/or RNA analysis were generated from restriction fragments or PCR-amplified products. All PCR products were cloned into pCR-TOPO (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Two probes specific for the c2 promoter were produced. A 388-bp product (−452 to −65; C2-P1) was amplified by PCR with the primers C2PromF (5′-AATTTACCACGGACGACGGAGACGACG-3′) and C2PromR (5′-TCAGTTGGACGGGCGGATGG-3′) and a 753-bp HinfI fragment (−1427 to −675) was subcloned from a genomic c2 clone (C2-P2; Wienand et al. 1986). A probe specific for the 5′-UTR was made by PCR amplification of a 303-bp fragment with forward primer C2 gs 5′ F (5′-GGCTTCCGTCCTCTCCCACCACAG-3′) and reverse primer C2 gs 5′ R (5′-CGCGCTGGGCCTTCCTCACCTCCTCC-3′). Two probes specific for the c2 intron were made: a 1294-bp AvaI fragment (Wienand et al. 1986) and an 884-bp PCR product amplified with forward primer C2st2F (5′-CTCGCGCCATGCACAAAGAC-3′) and reverse primer C2st3R (5′-GCGCGCTAGAAGAAGAAGAAGAGGT-3′). A 164-bp probe specific for the 3′-UTR of C2 was amplified by PCR with the primers C23′ F (5′-CTCCACAGCGTCCCCATCA-3′) and C23′ R (5′-ACACACGACAATTATAGCAGAGA-3′).
Genomic DNA isolation and Southern hybridization:
Genomic DNA was isolated from maize leaves as described (Cone et al. 1986). Ten micrograms of genomic DNA was incubated with 10 units of each of the appropriate restriction endonuclease(s) for 4 hr at the suggested temperature and was subjected to agarose gel electrophoresis and Southern blotting. Hybridizations were performed with randomly labeled radioactive probes under stringent conditions according to Sambrook and Russell (2001).
λ-cloning:
To obtain full-length C2-Idf clones, two independent λ-libraries were generated. To clone a longer sequence of the C2-W22 allele, one λ-library was generated. To produce these libraries, DNA was isolated from young seedlings from either C2 or C2-Idf mutant backgrounds as previously described (Cone et al. 1986; Cocciolone and Cone 1993). Genomic DNA was fragmented by partial digestion with the restriction endonuclease MboI as described previously (Sambrook and Russell 2001). To select for fragments in the range of 9–23 kb, the DNA was fractionated on a 10–40% sucrose gradient or on a 5–20% sodium acetate gradient and subjected to centrifugation at 23,500 rpm for 20 hr at 20°. Fractions in the correct size range were selected as previously described (Sambrook and Russell 2001). DNAs were ligated either by using the Lambda Fix II/XhoI partial fill-in vector kit (Stratagene, La Jolla, CA) or into the EMBL4 λ-vector (Frischauf et al. 1983). For packaging, either the Gigapack III gold packaging extract (Stratagene) system or self-made packaging extracts were used. Phage were plated in top agar either with XL1-Blue MRA (P2) or with K803 Escherichia coli cells. Plaque lifts and hybridization to detect positive clones were performed as previously described (Cocciolone and Cone 1993; Sambrook and Russell 2001), using either the C2-P1 or the C2-P2 probe. DNA was isolated from phage of interest using a QIAGEN (Valencia, CA) Lambda maxi kit according to the manufacturer's instructions and was subcloned using standard procedures. To sequence large clones, the Locus Pocus subcloning system (Novagen, Schwalbach, Germany) was used with clones of the EMBL4 λ-library according to the manufacturer's instructions.
Fosmid cloning:
The CopyControl fosmid library production kit (Epicentre, Madison, WI) was employed to obtain larger-size clones than was possible with λ-cloning. DNA was isolated by cesium chloride centrifugation. Library production was performed using the manufacturer's instructions. Colonies were screened using the protocol for screening bacterial colonies as described by Sambrook and Russell (2001). Filters were hybridized as for plaque lifts, except that the 3′-UTR of c2 was used as a probe. For positive colonies, the plasmids were induced to high copy number using the manufacturer's protocol. DNA was isolated for restriction enzyme mapping and sequencing using a QIAGEN plasmid maxi kit, following the manufacturer's instructions. To expedite sequencing of the large inserts, an in vitro transposition system was employed. The GeneJumper primer insertion kit for sequencing (Invitrogen) was used to introduce bacteriophage Mu into random positions in the cloned C2-Idf-containing DNA.
To prepare DNA for sequencing, the R.E.A.L. Prep 96 kit (QIAGEN) was used with a number of modifications of the manufacturer's instructions. Precultures were grown in 1 ml 2XYT plus 10 μg/ml kanamycin for 16 hr at 37° with shaking at 175 rpm. A 200-μl aliquot of the precultures was added to 2.5 ml of fresh 2XYT containing 25 μg/ml chloramphenicol and 2X CopyControl induction solution (Epicentre, Madison, WI); these cultures were grown for 16 hr at 37° with shaking at 175 rpm. The cells were harvested by centrifugation for 10 min at 1000 × g. Resuspension, lysis, precipitation of cellular debris, and lysate clearing were performed as per manufacturer's instructions. To precipitate the DNA, 0.7 volume of room temperature isopropanol and 2 μg glycogen were added to each well. Samples were centrifuged at 2254 × g for 60 min to pellet the DNA. Ethanol wash and redissolving DNA were performed as per manufacturer's instructions. To complete the sequencing of C2-Idf-I/II a 6.1-kb PstI fragment, containing sequence between the inverted promoters, was selected by gel purification and subcloned.
DNA sequencing:
DNA sequencing reactions were performed either by the University of Missouri DNA Core facility or by the Wienand laboratory with Applied Biosystems (Foster City, CA) 377 automated DNA sequencers using Applied Biosystems Prism BigDye terminator cycle sequencing chemistry). Computer-assisted sequence assembly was performed using SeqMan (DNAStar, Madison, WI) and the programs provided by the ExPASy proteomics server (http://www.expasy.ch/). Alignments were calculated with the CLUSTAL X 1.83 program (Thompson et al. 1997) and manually edited using GeneDoc 2.6 (Nicholas et al. 1997).
RNA blot analysis:
Total RNA was isolated from husks or leaf sheaths just after silk emergence, using Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA was stored at −20° in 100% formamide at a concentration of 500 ng/μl. Total RNA was fractionated on formaldehyde gels as described (Sambrook and Russell 2001), except that blots were stained with 0.04% methylene blue, 0.5 m sodium acetate for 5 min. The blots were rinsed with deionized water and then destained in 0.2× SSC, 1% SDS. RNA was blotted to Magnagraph nylon transfer membrane (Osmonics, Minnetonka, MN) and hybridized as previously described (Cone et al. 1986). Blots were hybridized with a probe specific for the 3′-untranslated region of c2. RNA blots were also hybridized with a maize actin probe (Shah et al. 1983) as a loading control. Signal was detected by exposure to a Fuji Bas-III S imaging plate followed by detection by a Fuji Bas-1000 phosphorimager. c2 RNA levels were normalized to actin by subtracting the background signal from both c2 and actin and then dividing the c2 signal by the actin signal.
Slot blots:
For use in nuclear run-on transcription the following probes were linearized: a 1.5-kb c2 cDNA, a 2.5-kb maize actin cDNA, a 200-bp fragment from the c2 promoter, and empty pCR-TOPO as negative control. For the slot blots, 20 μg of each linearized plasmid was denatured in 0.1 n NaOH. The total volume was increased to 1 ml with 6× SSC and 250 μl of each sample was added to a slot of a Bio-Dot SF microfiltration apparatus (Bio-Rad, Hercules, CA) containing nitrocellulose hydrated with TE and equilibrated with 2× SSC. The membrane was dried and baked under vacuum at 80° for 90 min.
Nuclear run-on transcription:
Nuclei were isolated from husks just after silk emergence as described previously (Cone et al. 1993a; Hoekenga et al. 2000). Nuclear run-on transcription was performed using ∼5 × 106 nuclei per reaction. Counts per minute (cpm) were determined as described previously (Cone et al. 1986) and labeled RNA (3–5 × 106 cpm) was added to slot blots in hybridization solution. Hybridization was carried out for 2–4 days at 42°. Following hybridization, membranes were washed with 2× SSC for 15 min at room temperature followed by four washes, each one for 15 min in 0.1× SSC, 0.1% SDS at 50°. Signal was detected by exposure to a Fuji Bas-III S imaging plate followed by detection by a Fuji Bas-1000 phosphorimager. Transcription levels were normalized to actin by subtracting empty vector signal from both c2 and actin and then dividing the c2 signal by the actin signal.
Viral inoculation methods:
Inoculum for Maize necrotic streak virus (MNeSV) (Louie et al. 2000) was prepared by grinding a small amount of virus-infected leaf material in 3 volumes of 0.1 m potassium phosphate, pH 7, and collecting the supernatant from a 15,000 × g centrifugation. Maize seeds were inoculated with MNeSV by vascular puncture inoculation of embryos (Louie 1995). Briefly, seeds were soaked (2.5 hr) in water at 28° and then arranged on four layers of wet paper towels in a petri dish. Virus extract, 3–5 μl, was placed on the embryo and five minuten pins were pushed through the inoculum into the vascular tissue on each side of the embryo. After 2 days at 28°, the seeds were planted and transferred to a greenhouse without supplemental light. When the plants were mature, virally infected streaks were excised for RNA extraction. Similar tissues were processed as controls from uninfected plants.
Inoculum for Maize dwarf mosaic virus-A (MDMV-A) infection was prepared by grinding a small amount of virus-infected leaf tissue with roughly 4 volumes of 10 mm phosphate buffer (pH 7.5) and a small amount of carborundum. The slurry was manually applied to maize plants at the three-leaf stage by rubbing the slurry onto the top two leaves. The plants were then rinsed with water. Inoculation was judged to be successful if visible symptoms of infection—chlorotic mosaic and streaks on leaves and leaf sheaths—were apparent after 7–10 days. Plants not showing symptoms of infection by 14 days after inoculation were removed from the experiment. Uninfected control plants were kept separate from infected plants and any noninoculated plants showing symptoms of infection at any time were removed from the experiment. In the field, the uninfected and infected plants were in separate locations and the same criteria were used to select plants to be analyzed. MDMV-A was propagated by sequentially infecting young plants from previously infected plants or by inoculation with infected tissue that had been stored at −80°. At maturity, husks were harvested for RNA isolation.
siRNA detection:
RNA was isolated with Trizol (Invitrogen) and the final pellet was resuspended in 2.5 ml water in a 15-ml Corex tube. High-molecular-weight RNA was precipitated by adding 0.5 ml 5 m NaCl and 2 ml 20% PEG-8000, 30 mm MgCl2 and incubating overnight at 0° (on ice in a 4° refrigerator). Following precipitation, the RNA was subjected to centrifugation at 4° in a Beckman JS13.1 rotor at 10,000 × g for 30 min. The supernatant, containing RNA ∼200 nt and smaller, was transferred to a 30-ml Corex tube and extracted with 100:100:1 phenol:chloroform:isoamyl alcohol followed by centrifugation at 4° in a JS13.1 rotor for 10 min at 10,000 × g. The aqueous phase was transferred to a new 30-ml Corex tube and the RNA was precipitated by adding 0.5 ml 3 m sodium acetate and 15 ml absolute ethanol and storing overnight at −20°. RNA was pelleted at 4° in a JS13.1 rotor at 10,000 × g for 1 hr. The pellet was suspended in 250 μl of water and the total amount of RNA was determined spectrophotometrically. The samples were transferred to a 1.5-ml microcentrifuge tube and the RNA was precipitated by adding 10 μl 3 m sodium acetate and 330 μl ethanol and storing at −20° overnight. The RNA was pelleted by centrifugation in a microcentrifuge at maximum speed for 1 hr and the pellet was resuspended in water to a concentration of 2.5–5 μg/μl. The samples were mixed with an equal volume of deionized formamide and denatured by heating at 95° for 5 min. An aliquot of 25–50 μg of RNA was fractionated on 20% polyacrylamide, 8 m urea denaturing gels. The acrylamide gels were blotted to Hybond N+ nylon membranes (Amersham Biosciences, Piscataway, NJ), using a Trans-blot cell (Bio-Rad) at 22 V overnight. Prehybridization and hybridization were performed as described previously (Cone et al. 1986) except that ∼6 × 106 cpm of probe were added to each blot. Blots were washed twice in 1× SSC, 0.1% SDS for 20 min at 50° followed by two washes in 0.5× SSC, 0.1% SDS for 1 hr at 50°. Signal was detected by autoradiography.
RESULTS
Phenotypic characterization of the C2-Idf mutation:
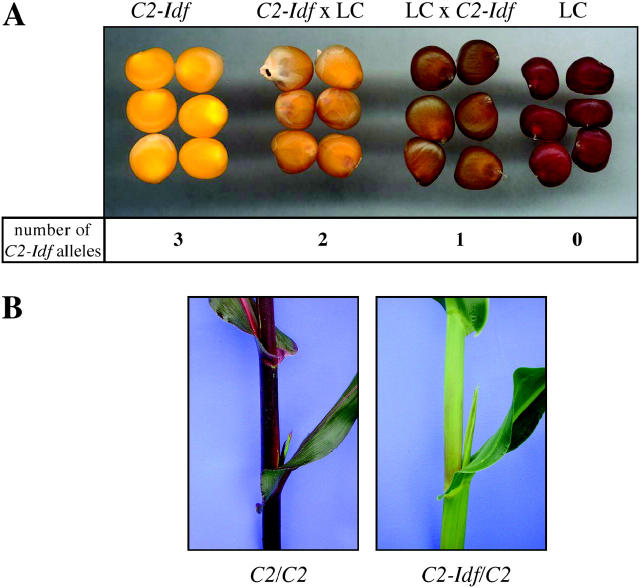
The inhibitory effect of the C2-Idf allele is clearly visible as a reduction of aleurone pigmentation in kernels of reciprocal crosses between C2-Idf and a normal line (Figure 1A). Kernels of a color-converted W22 line (LC) are dark red whereas homozygous C2-Idf mutant kernels are colorless. Due to the triploid nature of the aleurone, either one or two C2-Idf alleles are present in kernels of the reciprocal crosses. An increase in dosage of C2-Idf alleles resulted in a dosage-dependent decrease in aleurone pigmentation. However, even a single dose of C2-Idf produces a less pigmented aleurone phenotype than does a single dose of a recessive loss-of-function allele (Dooner 1983). The phenotype of the C2-Idf mutant allele in vegetative tissues is shown in Figure 1B. Plants homozygous for a functional C2 allele produce anthocyanin pigments in leaf sheaths and husks. A single copy of the dominant C2-Idf allele nearly eliminates this pigmentation. C2-Idf/C2-Idf homozygotes are completely lacking anthocyanin.
Figure 1.—
C2-Idf phenotypes. (A) Phenotypes of maize kernels from the mutant line C2-Idf, a normal line (LC), and reciprocal crosses of both lines. The corresponding number of C2-Idf alleles in the triploid aleurone is given. (B) Vegetative phenotype of C2/C2 and C2-Idf/C2 plants.
The c2 locus is substantially rearranged in the C2-Idf mutation:
As an explanation of the dominant inhibitory effect of the C2-Idf mutation, an epigenetic silencing mechanism was postulated as a working hypothesis. Such effects might result from gene duplication events or rearrangements of the c2 locus. To address this hypothesis, the C2-Idf locus was investigated by detailed Southern and sequence analyses.
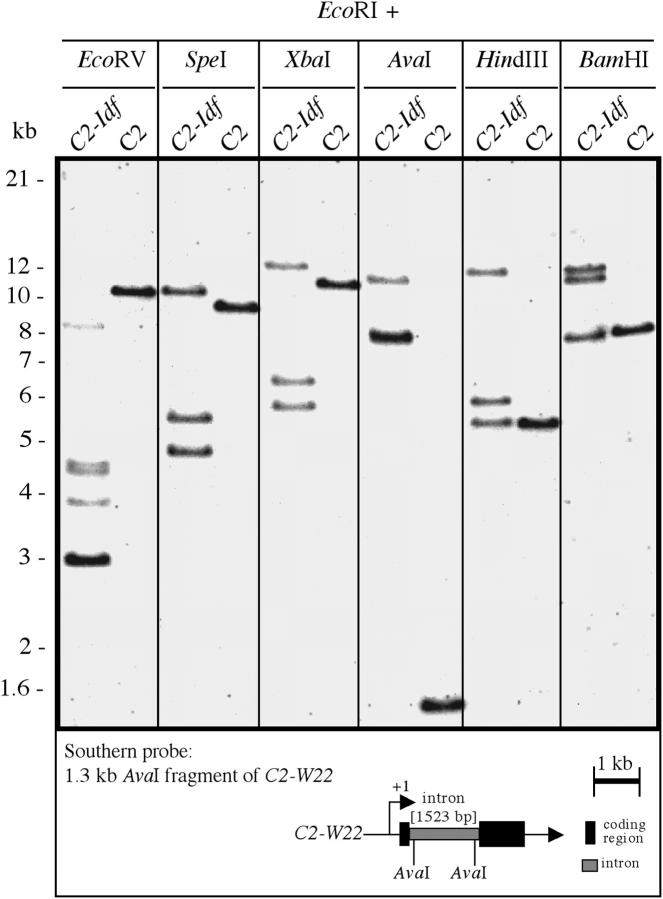
To examine the c2 locus arrangement in C2-Idf, Southern analyses with DNA probes derived from the c2 intron (Figure 2) and the c2 promoter (data not shown) were performed. The c2 gene of line LC consists of two exons and an intron of 1523 bp (Figure 2; Wienand et al. 1986). Hybridizations with the c2-intron probe revealed a higher number of c2 hybridizing fragments in the C2-Idf line as compared to LC. Restriction analyses with different endonucleases in most cases revealed three fragments in C2-Idf DNA and only one fragment in LC DNA (Figure 2). In some cases, only two c2 homologous fragments were detectable (e.g., Figure 2, EcoRI + AvaI); however, one of these fragments showed a stronger hybridization signal, suggesting comigration of two of the three putative c2-homologous fragments. These data suggested that three different c2 homologous regions (or c2 gene copies) are present in the C2-Idf allele.
Figure 2.—
Analysis of C2-Idf allelic structure. Southern blot of genomic DNA from homozygous C2-Idf and C2 (LC) leaves, respectively, is shown. DNA samples were digested with the restriction enzyme EcoRI and with a second enzyme as indicated. DNA was fractionated on an agarose gel, blotted to a nylon membrane, and hybridized with an intron-specific probe that was generated by AvaI restriction of a genomic C2 clone from LC (Wienand et al. 1986). The schematic structure of this normal C2-W22 gene is given (solid areas, exons; shaded area, intron; +1, transcription start). Sizes of fragments in kilobases are indicated at left.
Cloning and genomic structure of the C2-Idf allele:
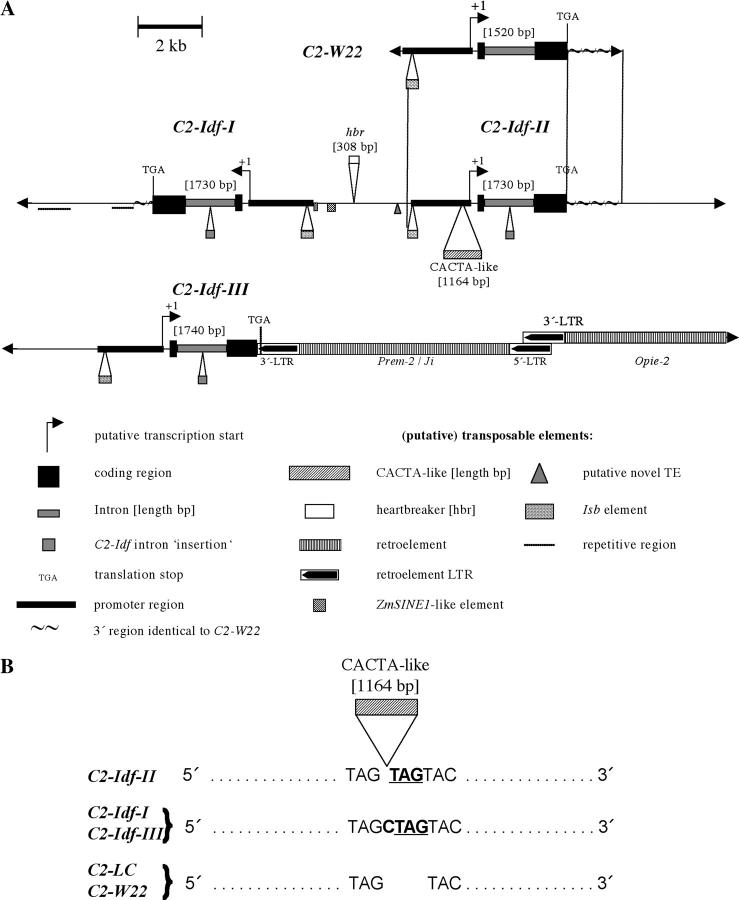
Approximately 46 kb of the genomic C2-Idf locus were cloned and sequenced. Four independent c2-hybridizing clones were identified during screening of two lambda λ-phage libraries. Moreover, three longer genomic DNA clones were identified during screening of a fosmid C2-Idf library. A segment of 2.1 kb not covered by the cloned DNA fragments was amplified by PCR. For comparison, 7.2 kb of the C2-W22 allele, a functional c2 allele, were cloned from a λ genomic DNA library and sequenced (Figure 3A). As expected, the corresponding parts of this sequence were almost identical to the previously published 3.8 kb of C2 from line C (Franken et al. 1991).
Figure 3.—
Schematic comparison between the C2 and the C2-Idf allelic structures. (A) Genomic regions of the C2-Idf allele were cloned from corresponding λ- and fosmid-libraries, respectively. Sequences were assembled and analyzed with the help of bioinformatic software. For comparison, 7.2 kb of the C2 gene from the normal line W22 were cloned and sequenced. The C2-Idf allele consists of three C2 gene copies, C2-Idf-I, C2-Idf-II, and C2-Idf-III. Two of the gene copies, C2-Idf-I and C2-Idf-II, are located in close proximity to each other in a head-to-head orientation on the same contig (23,255 bp). C2-Idf-III was identified on a second contig of 22,981 bp. Both contigs contained no overlapping flanking sequences so that the relative position of C2-Idf-III within the C2-Idf allele is unknown. For comparison, corresponding regions of C2-Idf-II and the normal C2 gene are aligned (vertical lines). Putative positions of the transcription start sites of the C2-Idf gene copies are marked according to the experimentally determined start site of the normal C2 gene (Franken et al. 1991). Major and minor characteristic sequences of the C2-Idf allele are given as boxes or triangles. (B) Insertion of the CACTA-type element in the promoter of C2-Idf-II and footprints of the insertion/excision event in C2-Idf-I and C2-Idf-III. The insertion of the element generated a 3-bp target site duplication (TAG) in C2-Idf-II. The identical footprint of this transposon was found in C2-Idf-I and C2-Idf-III. One additional nucleotide [C] was present in both sequences. No such footprint was found in normal c2 promoters of LC and W22.
Assembly of overlapping C2-Idf sequences generated two contigs of ∼23 kb each. Three different copies of the c2 gene could be identified on these clones (Figure 3A). The three copies were designated as C2-Idf-I, C2-Idf-II, and C2-Idf-III, respectively. Two copies (C2-Idf-I and C2-Idf-II) were found to be oriented head-to-head with a distance of ∼3.5 kb between the postulated promoter regions (Figure 3A). These regions were defined on the basis of the similarity of their sequences to the corresponding sequenced region upstream of the C2-W22 protein-coding sequence. For the third copy, C2-Idf-III, a position relative to C2-Idf-I and C2-Idf-II could not be determined because no sequence overlaps were present in the two contigs. On the basis of the lengths of noncoding sequences that border the three C2-Idf gene copies on both contigs, the minimal distance between the C2-Idf-III gene copy and the C2-Idf-I and -II gene cluster was calculated to be at least 6.7 kb. However, the fact that C2-Idf has segregated genetically as a stably transmitted single locus for over seven generations of backcrossing to inbreds Mo17 and W22 (our unpublished data) argues that all three C2-Idf gene copies are likely to be located in close proximity to each other. The sizes of the predicted restriction fragments of the sequenced C2-Idf allele were identical to the sizes of fragments detected by Southern analysis (Figure 2). These data demonstrate that the C2-Idf locus indeed consists of three different c2 gene copies.
The genomic structure of the individual C2-Idf gene copies is very similar to that of C2-W22 and contains two exons. The protein-coding regions of C2-Idf-I and C2-Idf-II show only two single-nucleotide exchanges in comparison to C2-W22; neither change would alter the amino acid sequence of a putative C2-Idf chalcone synthase protein. C2-Idf-III is truncated and is missing 77 bp at the 3′ end of the second protein-encoding segment. This truncation is due to an insertion of a PREM-2/Ji-like (SanMiguel et al. 1996; Turcich et al. 1996) retrotransposon of ∼9.2 kb. The 3′-long terminal repeat (LTR) of this element, adjacent to the remaining part of the C2-Idf-III gene copy, introduces a new stop codon, such that translation of this gene copy would result in a C2 protein shortened by 21 amino acids and containing four changed amino acids at its C-terminal end (Figure 3A). Because the C terminus is highly conserved in chalcone synthases (Niesbach-Klösgen et al. 1987), it is very likely that this 3′ truncation would have a negative effect on a putative C2-Idf-III protein. However, theoretically, translation from the C2-Idf-I and C2-Idf-II copies could produce active, functional C2 proteins.
At its 5′-LTR, the PREM-2/Ji-like element is flanked by an Opie-like retrotransposable element (Meyers et al. 2001). A 5.5-kb portion of this Opie-2-like element was sequenced without reaching its putative 5′-LTR. The exact border between both elements could not be determined by sequence similarity analysis. Both elements are members of retroelement families that are highly abundant in maize (Meyers et al. 2001).
The intron lengths of the three C2-Idf gene copies were 1730 bp for C2-Idf-I and C2-Idf-II and 1740 bp for C2-Idf-III, respectively. All three introns were highly similar to each other (98–99% identity). The C2-Idf introns differ from the 1520-bp C2-W22 intron by the presence of several small insertions/deletions (indels) and insertion of a 239-bp sequence stretch that is specific for C2-Idf (Figure 3A, C2-Idf intron “insertion”). This insertion has no significant homology to known sequences.
The promoter sequences of the C2-Idf copies are identical over a sequence range between −1500 and −1 relative to the predicted transcription start with the exception of two small indels in C2-Idf-I and an insertion of a 1163-bp transposable element in C2-Idf-II. This element belongs to the CACTA-type family and is inserted at position −211 bp relative to the putative start of transcription. A 3-bp target site duplication (TAG), which is typical for the CACTA family elements, is present at the site of insertion of the element (Figure 3B). A footprint of this transposable element (TAGCTAG) is found at identical positions in C2-Idf-I and C2-Idf-III (Figure 3B). On the basis of this finding, it seems likely that the CACTA-element was already present before the first duplication event of the c2 gene in the C2-Idf allele. This conclusion is further supported by sequence analysis of the regions 3′ of the C2-Idf-I and II genes. The 3′ region following the C2-Idf-II protein-coding sequence is almost identical to the corresponding region of C2-W22 (∼1.3 kb; Figure 3A). In contrast, the sequence identity between the 3′ regions of C2-Idf-I and C2-W22 spans only 556 bp. Further downstream in C2-Idf-I are clusters of repetitive sequences that probably originated from the insertion/activity of additional mobile elements (Figure 3A). The C2-Idf-II copy seems to be the progenitor in this cluster and the C2-Idf-I and C2-Idf-III copies most likely duplicated from this copy.
A comparison between the promoter regions of C2-Idf-I and C2-W22 revealed overall identity of 98% with two deletions of a CGCGC motif at −105 and −147, respectively, one insertion of GCTA at −209, and four single-nucleotide exchanges. Each promoter of the C2-Idf copies and of the C2-W22 version contained a small defective CACTA-like Isb transposable element at identical positions but with some minor sequence variations in the element (Figure 3A). Isb was previously identified to be of ancient origin and is present in a number of different anthocyanin genes (Techen et al. 1999). The high degree of similarity between the C2-W22 promoter and the promoters of C2-Idf copies extends close to the respective Isb elements (Figure 3A, promoter region).
Several transposable element-like sequences were identified in the intergenic region between C2-Idf-I and C2-Idf-II. Two imperfect direct repeats of ∼100 bp each were identified at −2092 and −2603 relative to the putative transcription start of C2-Idf-I (+1 C2-Idf-I; Figure 3A). Both repeats have significant similarity to a part of the 300-bp ZmSINE1 element (Yao et al. 2002; GenBank accession AF434193). Additionally, a 308-bp heartbreaker element (MITE-like) (Hbr, Zhang et al. 2000) was identified at position −3304 (relative to +1 of C2-Idf-I; Figure 3A). The 3-bp target site duplication of this element (TTT) is identical to the previously described target site duplication of Hbr22 (Zhang et al. 2000). Another putative transposable element (TE) was identified by the presence of two nearly identical 56-bp terminal inverted repeats (putative TIRs) at positions −4946 and −5046, relative to +1 of C2-Idf-I and separated by 46 bp (putative novel TE; Figure 3A). Similar TIR-like sequences, enclosing sequences of variable lengths, were also found in other genomic sequences of Zea mays subsp. mays and subsp. parviglumis (data not shown). Hence, these sequences might be parts of a novel genetic element.
C2-Idf promoter analysis:
The presence of multiple c2 gene copies in the C2-Idf allele prompted us to ask which ones might be capable of driving expression. In plants, transcriptionally active genes have loosely packed chromatin, which is associated with low levels of cytosine methylation on DNA. Conversely, transcriptionally silent genes have more tightly packed chromatin organization and higher levels of cytosine methylation (Jenuwein and Allis 2001; Jackson et al. 2002; Soppe et al. 2002). To assess the expression potential of the C2-Idf genes, we used methylation-sensitive restriction enzyme digests to compare the methylation status of the C2-Idf gene promoters to that for the wild-type C2-W22 allele. To provide a reference, DNA was first digested with the non-methylation-sensitive enzyme, NdeI. Probing with the c2 promoter probe yielded a single hybridizing band of 2.1 kb in C2-W22, which is the predicted fragment based on sequence data (Figure 4). In C2-Idf, there were two hybridizing bands; on the basis of sequence data, these represent all three copies, with C2-Idf-I and C2-Idf-III producing 2.1-kb fragments and C2-Idf-II producing a larger 3.2-kb fragment due to the CACTA element insertion. When digested subsequently with a methylation-sensitive enzyme, these fragments should be cleaved into smaller products if the restriction sites are unmethylated. If the sites are methylated, the larger fragments will remain uncut.
Figure 4.—
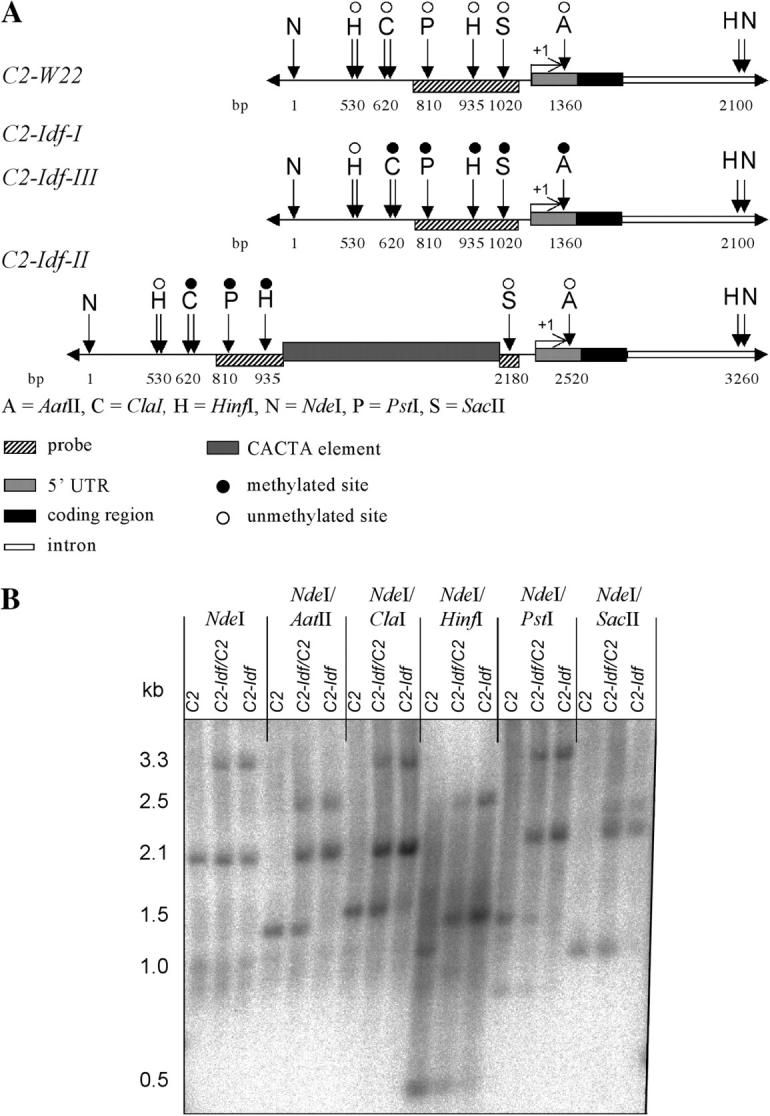
Promoter methylation. DNA from C2-W22, C2-Idf, and C2-Idf/C2-W22 heterozygotes was digested with the methylation-insensitive enzyme NdeI. Samples were then digested with methylation-sensitive enzymes. (A) A diagram of restriction sites assayed. Open circles represent unmethylated sites and solid circles represent methylated sites. Because the 3′-most HinfI site is located very close to the 3′ NdeI site, its methylation status could not be determined by Southern analysis. (B) Representative Southern blot probed with the c2 promoter.
In the normal allele, all of the sites tested are unmethylated, which is the expectation in a transcriptionally active allele (Figure 4). The C2-Idf allele has a more complex pattern of methylation (Figure 4). In C2-Idf-I and C2-Idf-III, the sites in the promoter nearest to the putative transcription start site (ClaI, PstI, HinfI, SacI, and AatII) are methylated, which is typical of a transcriptionally silent allele. In C2-Idf-II, which has a CACTA element inserted in the promoter region, sites upstream of the insertion are methylated, as in C2-Idf-I and C2-Idf-II; however, the two sites closest to the start of transcription (SacI and AatII) are unmethylated, as would be expected if this gene were transcriptionally active. This unmethylated region appears to be localized, because Southern analysis with a c2-intron probe (Figure 2) showed that the intron regions of all three C2-Idf gene copies are subject to methylation; in an EcoRI/AvaI digest, none of the three expected C2-Idf fragments (C2-Idf-I and II, 1494 bp; C2-Idf-III, 1505 bp) could be detected, whereas the expected fragment from the wild-type allele was detected in LC DNA (1295 bp).
In some cases of silencing, the methylation pattern of one allele can be transferred to a second, homologous allele (Luff et al. 1999; Walker and Panavas 2001). To ask whether the promoter methylation pattern in C2-Idf is transferred to the normal allele, methylation-sensitive restriction enzyme analysis was carried out in heterozygotes (Figure 4). In these plants, restriction fragments characteristic of both alleles are seen, indicating that trans-methylation of the wild-type C2 allele did not occur. This result fits with the genetic behavior of C2-Idf, in that C2-Idf does not heritably alter the expression of the normal allele.
RNA blot analysis:
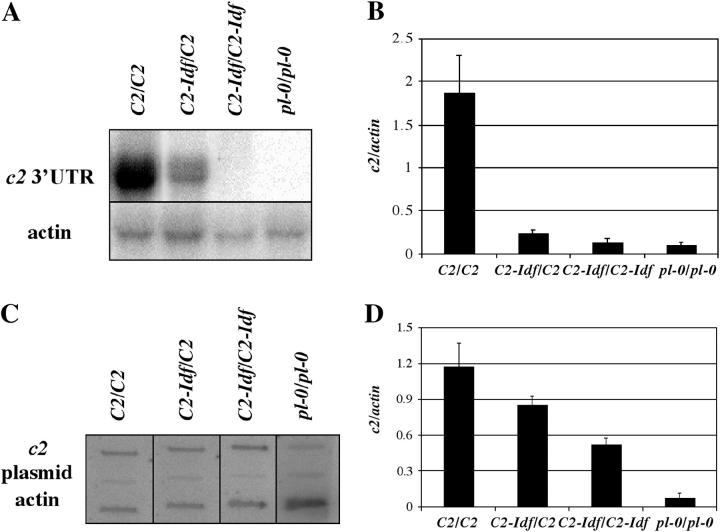
To investigate the nature of the C2 silencing by C2-Idf, steady-state RNA levels were determined. RNA was isolated from husks of C2-Idf/C2 and C2/C2, blotted, and hybridized with a probe derived from the 3′-untranslated region of c2 and an actin cDNA (Shah et al. 1983) as a loading control (Figure 5A). In husks from C2-Idf/C2 plants, c2-homologous transcript accumulated at ∼20% of the C2/C2 level (Figure 5B). In homozygous C2-Idf/C2-Idf plants, RNA levels were nearly as low as in the negative control plants (pl-0/pl-0). These data confirm that C2-Idf inhibition occurs at the RNA level.
Figure 5.—
RNA levels in C2-Idf mutants. (A) Representative RNA blot probed with the C2 3′-UTR. The blot was stripped and rehybridized with an actin probe as a loading control. (B) Relative steady-state c2 transcript levels normalized against actin (n = 6–8; error bars are the standard errors of the means). (C) Representative slot blots probed with radioactively labeled nuclear RNA. (D) Relative transcription rate normalized against actin (n = 6; error bars represent the standard errors of the means).
Nuclear run-on transcription:
To determine whether C2-Idf is transcribed, we performed nuclear run-on transcription on nuclei isolated from husks. In run-on transcription, nascent nuclear transcripts are radioactively labeled and used as hybridization probes on DNA slot blots. This experiment allows measurement of the amount of transcription from genes of interest (C2 and C2-Idf) relative to negative (empty vector) and positive (actin) controls. If C2-Idf is transcribed and has no influence on C2 transcription, then the signal from C2-Idf/C2 heterozygotes should be >50% that of C2/C2. Alternatively, if C2-Idf represses transcription of the C2 allele, then signal from C2-Idf/C2 heterozygotes should reflect the levels of steady-state transcript accumulation.
Run-on transcription revealed that the C2-Idf locus is transcribed (Figure 5, C and D). In C2-Idf/C2-Idf homozygotes, the level of transcription was 44% of the C2/C2 level and significantly higher than the level of the negative control (pl-0/pl-0), indicating that the C2-Idf allele produces c2-homologous transcripts, albeit at a lower level than C2. In C2-Idf/C2 heterozygotes, c2 transcription was 73% that of C2/C2, indicating that C2-Idf does not transcriptionally silence the normal allele.
We should note that for this assay, the DNA used on the slot blots was a C2 cDNA clone. Transcripts hybridizing to the cDNA could come from either the c2 gene or a duplicate gene called white pollen1 (whp). The whp gene is 94% identical to c2 within the coding region (Franken et al. 1991) and is expressed at low levels in husks. If whp transcription is unaffected by the C2-Idf genotype, then in each sample the same fraction of the signal is expected to represent whp transcription. In fact, the low level of transcription observed in the pl-0/pl-0 negative control is probably due to whp. Two lines of evidence suggest that C2-Idf does not affect whp expression. First, plants that lack chalcone synthase activity in pollen are self-sterile due to defective pollen. If C2-Idf silences whp1, then C2-Idf homozygotes would be self-sterile; this is not the case. Second, in a homozygous intensifier1 (in1) mutant background, whp1 is expressed in the kernel aleurone, resulting in a colored phenotype in a recessive loss-of-function c2 mutant. Plants homozygous for the in1 mutant and C2-Idf produce kernels with a colored aleurone, indicating that C2-Idf does not affect whp expression (our unpublished data). Thus, the presence of whp signal does not alter the conclusions from this experiment.
In some cases of silencing, ectopic transcription of promoter sequences leads to production of an aberrant RNA that triggers transcriptional silencing of the gene through chromatin modifications to the promoter (Mette et al. 1999). To test whether the C2-Idf promoter is transcribed, we hybridized labeled nuclear transcripts to a c2 promoter DNA fragment. There was no signal in any genotype (data not shown), indicating that the promoters are not transcribed at a detectable level.
Viral suppression of silencing by MNeSV:
A number of viruses encoding proteins that are known to effectively inhibit RNA silencing are common maize pathogens. MNeSV has been tentatively identified as the first monocot-infecting tombusvirus. It encodes a protein highly homologous to the p19 protein from dicot-infecting tombusviruses, such as Tomato bushy stunt virus (TBSV) and Carnation Italian ringspot virus (CIRV) (Louie et al. 2000). The p19 protein acts as a suppressor of RNA silencing and for TBSV and CIRV has been shown to bind siRNAs (Vargason et al. 2003; Ye et al. 2003), presumably inhibiting their incorporation into the RISC and curtailing downstream roles in silencing (Lakatos et al. 2004).
To assess whether infection with this virus relieved C2-Idf inhibition of the C2 allele, C2-Idf mutant and C2 control plants were infected with MNeSV. If C2-Idf inhibits the functional C2 allele by RNA silencing, then infection of C2-Idf/C2 heterozygotes with this virus should result in a higher level of steady-state transcript and a more pigmented phenotype. Infected C2-Idf/C2 heterozygotes exhibited anthocyanin accumulation that corresponded to the MNeSV-infected lesions on the leaf sheath (Figure 6A). There was no anthocyanin accumulation in infected pl-0/pl-0 plants, nor did we observe anthocyanin accumulation in C2-Idf/C2 plants infected with the unrelated Maize chlorotic mottle virus (MCMV), a machlomovirus (data not shown). The latter result indicates that viral infection per se does not activate the anthocyanin pathway and that MCMV does not suppress C2-Idf silencing.
Figure 6.—
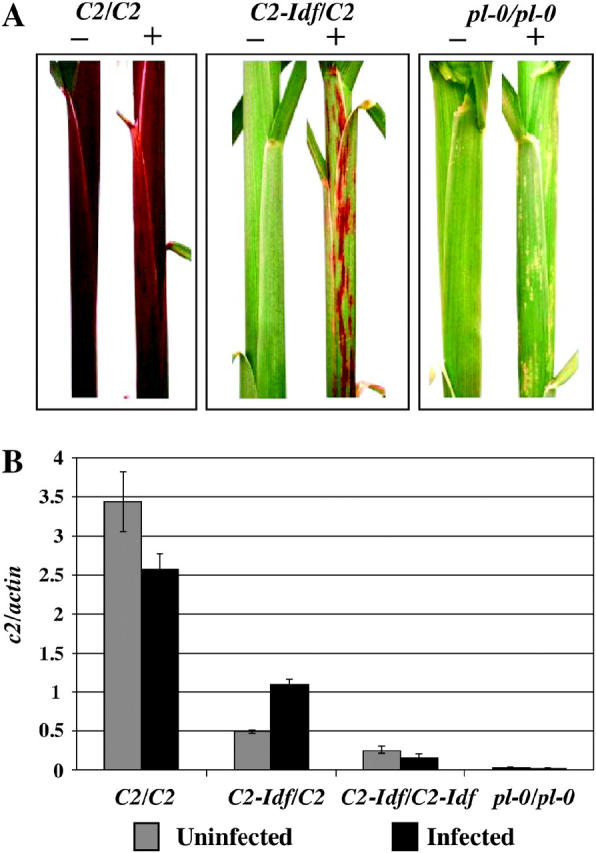
The effect of infection with MNeSV on C2-Idf silencing. (A) Phenotypes of MNeSV infected (+) and uninfected plants (−). (B) Relative steady-state c2 transcript levels normalized against actin (n = 3; error bars are the standard errors of the means).
Total RNA was extracted from MNeSV-infected streaks and from uninfected control tissue and subsequently analyzed by RNA blot hybridization with a probe derived from c2. MNeSV-infected C2-Idf/C2 heterozygotes accumulated twice as much c2 mRNA as uninfected plants (Figure 6B). In contrast, infected C2/C2 and C2-Idf/C2-Idf plants showed a slight decrease in c2 mRNA, relative to controls. Infection of pl-0/pl-0 plants did not alter c2 mRNA levels. Together, the pigment phenotypes and the RNA levels indicate that infection with MNeSV relieves C2-Idf inhibition of C2.
Viral suppression of silencing by MDMV-A:
As a second test for viral suppression of RNA silencing, we infected homozygous C2-Idf mutant and C2 control plants with MDMV-A, a potyvirus, which produces the P1/HC-Pro polyprotein. P1/HC-Pro has been shown to act as an effective inhibitor of RNA silencing in other systems (Anandalakshmi et al. 1998; Kasschau and Carrington 1998). In infected C2-Idf/C2 heterozygotes, suppression of the typical colorless or faintly colored phenotype first became evident ∼4 weeks after inoculation as dark red or purple streaks along the sheaths in a pattern characteristic of the viral infection pattern (Figure 7A). By anthesis, strong purple pigmentation was visible on leaf sheaths, husks, some adult leaves, and tassel glumes.
Figure 7.—
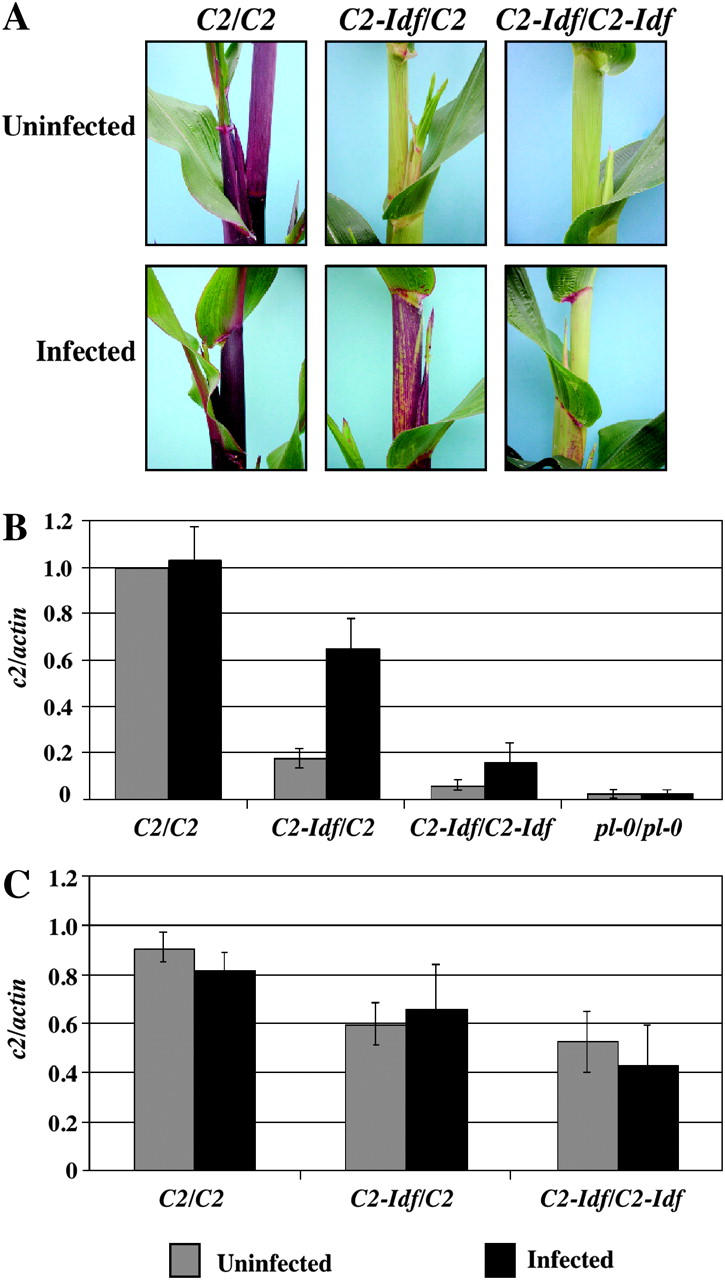
The effect of infection with MDMV-A on C2-Idf silencing. (A) Phenotypes of uninfected control plants (top) and MDMV-A-infected plants (bottom). (B) Steady-state c2 transcript levels normalized against actin (n = 6–8; error bars are the standard errors of the means). (C) Relative c2 transcription rate was measured by run-on transcription assays of infected plants and uninfected controls (n = 3, error bars are the standard errors of the means).
Infected C2-Idf/C2 heterozygotes accumulated more than three times the amount of steady-state c2 mRNA in husks than did uninfected heterozygotes (Figure 7B). The mRNA level in the infected C2-Idf/C2 plants was ∼65% of the mRNA level in C2/C2 husks. A slight but lesser increase of c2 mRNA level was also observed between infected and uninfected C2-Idf/C2-Idf homozygotes. This coincides with the slight phenotypic suppression of silencing in the infected C2-Idf/C2-Idf homozygote plants. Neither C2/C2 nor pl-0/pl-0 homozygotes exhibited a significant difference in transcript accumulation upon MDMV-A infection.
The increase in steady-state transcript levels in MDMV-A-infected plants carrying C2-Idf could be explained either by an increase in transcription or by a decrease in mRNA degradation. To distinguish between these possibilities, we used run-on transcription assays to measure the amount of transcription in husks of infected and uninfected plants. As shown in Figure 7C, MDMV-A infection did not significantly alter transcription. These results indicate that the increase in steady-state mRNA levels in MDMV-A-infected plants containing C2-Idf alleles is likely the result of reduced transcript degradation.
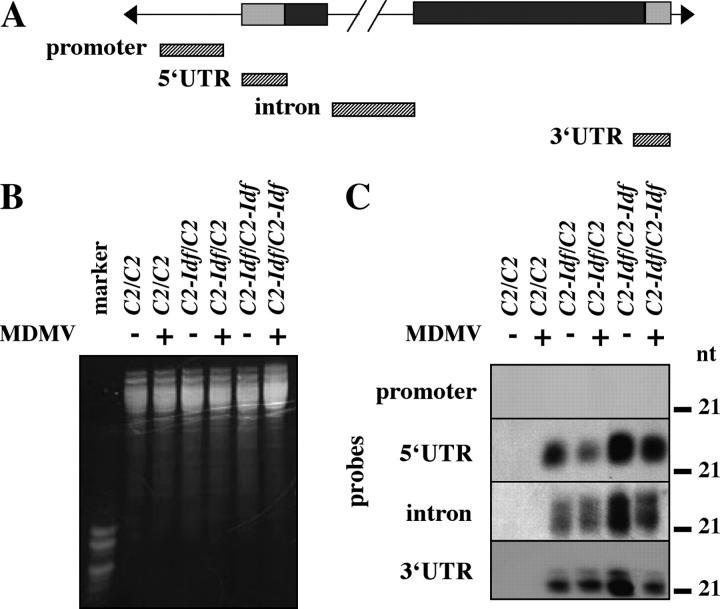
Detection of small interfering RNAs:
Accumulation of siRNAs, processed from double-stranded RNAs by Dicer, is a hallmark of RNA silencing. To determine whether c2-homologous siRNAs are present in plants carrying a C2-Idf allele, we analyzed RNA from husks of C2/C2, C2-Idf/C2, and C2-Idf/C2-Idf plants on RNA blots using probes from different parts of the c2 gene (Figure 8A). The siRNAs were not detected in normal C2/C2 with any of the probes or in any genotype with the promoter probe. However, all three probes derived from the transcribed regions of the c2 gene detected siRNAs in husks of plants carrying C2-Idf (Figure 8C). Two classes of siRNAs were present. Multiple size classes of siRNAs have been described previously and may play different roles in silencing (Hamilton et al. 2002).
Figure 8.—
siRNA analysis from plants infected with MDMV-A. (A) Diagram of the c2 gene showing location of hybridization probes (hatched boxes). Shaded boxes represent untranslated regions; solid boxes represent protein-encoding regions. (B) Ethidium bromide-stained image of RNA gel to show comparable loading in all lanes. (C) RNA blots hybridized with probes from A. RNA was isolated from MDMV-A-infected plants (+) and uninfected controls (−). Position of 21-nt size standard in gel is indicated to the left of each blot.
We also assayed for the presence of siRNAs in MDMV-A-infected plants. The three probes from the transcribed region of c2 detected siRNAs in husks from both infected and uninfected C2-Idf plants (heterozygotes and homozygotes); however, while this assay is not strictly quantitative, in most cases the siRNA levels appeared to be lower in the infected plants. This reduction is consistent with current thinking about how P1/HC-Pro suppresses RNA silencing. The viral protein is thought to interfere with either production of the aberrant double-stranded precursor or processing of siRNAs (Dunoyer et al. 2004); either possibility would be consistent with reduced levels of siRNAs in infected plants.
DISCUSSION
C2-Idf inhibits expression of normal C2 alleles to produce a colorless phenotype that is distinct from the deeply pigmented phenotype of the normal plants. The results of our experiments indicate that this phenotypic inhibition occurs by RNA silencing. The sequence analysis of the C2-Idf allele revealed that the structural difference between the mutant and a normal functional allele is the presence of three nearly full-length copies of the c2 gene in C2-Idf. The C2-Idf allele is transcribed, as assessed by nuclear run-on transcription assays, but there is little accumulation of full-length transcripts. Instead, plants carrying C2-Idf produce siRNAs with homology to the transcribed portions of the c2 gene. Furthermore, infection of C2-Idf/C2 heterozygotes with either MNeSV or MDMV-A partially relieves silencing, leading to production of more pigment and higher levels of steady-state c2 mRNA than in uninfected plants. In the case of MDMV-A infection, the increase in c2 mRNA accumulation is not due to higher levels of transcription, but instead is correlated with lower levels of siRNA accumulation.
Two previously described cases in which a dominant mutant induces RNA silencing of an endogenous gene bear similarities to C2-Idf silencing. In the first case, a rice mutant, Low glutelin content1 (Lgc1), contains a deletion of sequences between two tail-to-tail-inverted repeat gene segments. The deletion leads to read-through transcription from one gene into the inverted repeat, producing a double-stranded RNA capable of inducing RNA silencing. When Lgc1 is crossed to wild type, gene expression from the wild-type allele is reduced and the reduction is correlated with production of siRNAs (Kusaba et al. 2003). In soybean, the Inhibitor mutation, which is a dominant negative variant of the chalcone synthase (CHS) locus, contains multiple CHS gene copies, some arranged as inverted repeats (Tuteja et al. 2004). When Inhibitor is crossed to wild type, expression of the wild-type gene is silenced. Silencing is accompanied by reduction of CHS mRNA and production of CHS-homologous siRNAs. Furthermore, infection with viruses carrying suppressors of silencing partially restores CHS expression in heterozygotes (Senda et al. 2004).
The C2-Idf allele is also composed of multiple c2 gene copies. C2-Idf-I and C2-Idf-II are arranged as a head-to-head inverted repeat and a novel CACTA transposable element is found in the promoter of C2-Idf-II. C2-Idf-III is truncated in the second exon by a Ji/Prem-2 retrotransposon. The presence of these transposable elements suggests a possible sequence of events that led to the present structure of the C2-Idf allele. First, a CACTA element inserted into the ancestral c2 gene and then this gene was duplicated. This event was followed by excision of the CACTA from one c2 copy followed by a second duplication of this gene, leading to production of C2-Idf-I and C2-Idf-III with identical transposon footprints. The CACTA element remains in the C2-Idf-II gene copy. It is not clear which duplication event coincided with production of the head-to-head arrangement of C2-Idf-I and C2-Idf-II. Later, C2-Idf-III was modified by retroelement insertion or by a deletion event to abut the 3′ end of the gene to a retroelement. It is also possible that transposition events of the CACTA element might have occurred independently after duplication.
Although there are three c2 gene copies in the C2-Idf allele, it is not clear which gene or genes are transcribed. The results of run-on transcription assays, which showed that transcription in C2-Idf homozygotes is ∼40% of that in C2 homozygotes, indicate that it is unlikely that all three genes are each transcribed at normal levels to produce RNA homologous to the c2 coding region. Methylation analysis showed that the only gene with an unmethylated promoter is C2-Idf-II, the copy carrying a CACTA element. However, the influence of the CACTA insertion at position −211 is difficult to predict, as insertions of CACTA elements in the promoter regions of c2 and other genes can have either positive or negative effects. For example, in the c2-m1-130 allele, insertion of an En/Spm CACTA-like element at position −94 results in a complete loss of C2 expression (Wienand et al. 1986). In contrast, CACTA-like Doppia elements located upstream of coding sequences in some alleles of the anthocyanin regulatory genes r1 (Walker et al. 1995; May and Dellaporta 1998; Bercury et al. 2001) and pl1 (Cone et al. 1993b) appear to be essential for proper promoter activity.
The nature of the RNA that triggers RNA silencing in C2-Idf is not known. Unlike the silencing associated with the soybean CHS genes or the rice Lgc1 gene, the C2-Idf genes are not arranged in the type of tail-to-tail inverted repeat that would lead to read-through production of an antisense transcript. However, in C2-Idf, one possible source of triggering RNA could be transcription from the Prem-2/Ji retroelement inserted at the end of the second exon in C2-Idf-III. The orientation of this element is such that transcription from its 3′ LTR could produce an antisense c2 transcript that could associate with RNA produced from the transcribed C2-Idf-II gene to induce silencing.
In plants carrying the C2-Idf mutation, two species of siRNAs are present—one of 21–22 nt and a second of 24–25 nt. These two size classes of siRNAs play different roles in silencing (Hamilton et al. 2002). The short siRNAs, which are typically produced from silenced transgenes, are required for mRNA degradation. In contrast, the long siRNAs, produced from silenced transgenes and from retroelements, result in modification of the chromatin in sequences with homology to degraded transcripts; this results in silencing of homologous loci (Aufsatz et al. 2002; Volpe et al. 2002, 2003; Cao et al. 2003). The presence of these RNAs raises the possibility that one or more gene copies in C2-Idf are transcriptionally silenced to some degree. However, the available evidence indicates that the effect of C2-Idf on normal C2 is post-transcriptional and that this is the process suppressed by viral infection. The respective roles of the two classes of siRNAs in the observed silencing for C2-Idf are currently unknown.
The finding that C2-Idf inhibition of C2 involves an RNA silencing mechanism and that this silencing is suppressed by some maize viruses establishes a phenotypic system in which silencing mechanisms can be easily studied. This reporter system should allow the detection of mutations defective in silencing functions and will facilitate the study of silencing suppression by maize viruses.
Acknowledgments
We thank Peter A. Peterson (Iowa State University) and the Maize Genetics Cooperation stock center (University of Illinois) for seed material, Barbara Sonderman (University of Missouri) for technical assistance, and Jim Schoelz (University of Missouri) for productive discussions. This research was supported by Research Incentive Funds to K.C.C. and J.B. C.D.V. was supported by a predoctoral fellowship from the National Institutes of Health (5T32GM08396-13).
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AY728478 [c2 gene chalcone synthase (wild type) C2-W22], AY728476 (Zea mays L. C2-Idf allele; gene copies C2-Idf-I and C2-Idf-II), and AY728477 (Zea mays L. C2-Idf allele; gene copy C2-Idf-III).
References
- Ahlquist, P., 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296: 1270–1273. [DOI] [PubMed] [Google Scholar]
- Al-Kaff, N. S., S. N. Covey, M. M. Kreike, A. M. Page, R. Pinder et al., 1998. Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279: 2113. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi, R., G. Pruss, X. Ge, R. Marathe, A. Mallory et al., 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95: 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz, W., M. F. Mette, J. van der Winden, A. J. M. Matzke and M. A. Matzke, 2002. RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 16499–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. C., 1996. RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol. Biol. 32: 79–88. [DOI] [PubMed] [Google Scholar]
- Bercury, S. D., T. Panavas, K. Irenze and E. L. Walker, 2001. Molecular analysis of the Doppia transposable element of maize. Plant Mol. Biol. 47: 341–351. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., A. Caudy, S. M. Hammond and G. J. Hannon, 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Brink, R. A., and I. M. Greenblatt, 1954. Diffuse, a pattern gene in maize. J. Hered. 45: 47–50. [Google Scholar]
- Cao, X., W. Aufsatz, D. Zilberman, M. F. Mette, M. S. Huang et al., 2003. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13: 2212–2217. [DOI] [PubMed] [Google Scholar]
- Chan, S. W.-L., D. Zilberman, Z. Xie, L. K. Johansen, J. C. Carrington et al., 2004. RNA silencing genes control de novo DNA methylation. Science 303: 1336. [DOI] [PubMed] [Google Scholar]
- Chandler, V. L., and M. Stam, 2004. Chromatin conversations: mechanisms and implications of paramutation. Nat. Rev. Genet. 5: 532–544. [DOI] [PubMed] [Google Scholar]
- Cocciolone, S. M., and K. C. Cone, 1993. Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135: 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, E. H., M. G. Neuffer and D. A. Hoisington, 1988 The genetics of corn, pp. 81–258 in Corn and Corn Improvement, Ed. 3, edited by J. W. Dudley. American Society of Agronomy, Crop Science Society of America, and Soil Science Society of America, Madison, WI.
- Coen, E. S., and R. Carpenter, 1988. A semi-dominant allele, niv-525, acts in trans to inhibit expression of its wild-type homolog in Antirrhinum majus. EMBO J. 7: 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K. C., F. A. Burr and B. Burr, 1986. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc. Natl. Acad. Sci. USA 83: 9631–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K. C., S. M. Cocciolone, F. A. Burr and B. Burr, 1993. a Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5: 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K. C., S. M. Cocciolone, C. A. Moehlenkamp, T. Weber, B. J. Drummond et al., 1993. b Role of the regulatory gene pl in the photocontrol of maize anthocyanin pigmentation. Plant Cell 5: 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Vedova, C. B., and K. C. Cone, 2004. Paramutation: the chromatin connection. Plant Cell 16: 1358–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., 1983. Coordinate genetic regulation of flavonoid biosynthetic enzymes in maize. Mol. Gen. Genet. 189: 136–141. [Google Scholar]
- Dooner, H. K., T. P. Robbins and R. A. Jorgensen, 1991. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25: 173–199. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., C.-H. Lecellier, E. A. Parizotto, C. Himber and O. Voinnet, 2004. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16: 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Franken, P., U. Niesbach-Klosgen, U. Weydemann, L. Marechal-Drouard, H. Saedler et al., 1991. The duplicated chalcone synthase genes C2 and Whp (white pollen) of Zea mays are independently regulated; evidence for translational control of Whp expression by the anthocyanin intensifying gene in. EMBO J. 10: 2605–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf, A. M., H. Lehrach, A. Poutska and N. Murray, 1983. Lambda replacement vectors carrying polylinker sequences. J. Mol. Biol. 170: 827–842. [DOI] [PubMed] [Google Scholar]
- Hamilton, A. J., and D. C. Baulcombe, 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952. [DOI] [PubMed] [Google Scholar]
- Hamilton, A. J., O. Voinnet, L. Chappell and D. C. Baulcombe, 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S. M., S. Boettcher, A. A. Caudy, A. Kobayashi and G. J. Hannon, 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150. [DOI] [PubMed] [Google Scholar]
- Herbert, A., 2004. The four Rs of RNA-directed evolution. Nat. Genet. 36: 19–25. [DOI] [PubMed] [Google Scholar]
- Hoekenga, O. A., M. G. Muszynski and K. C. Cone, 2000. Developmental patterns of chromatin structure and DNA methylation responsible for epigenetic expression of a maize regulatory gene. Genetics 155: 1889–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, J., A. Lindroth, X. Cao and S. E. Jacobsen, 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3-methyltransferase. Nature 416: 554–560. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and C. D. Allis, 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R. A., P. D. Cluster, J. English, Q. Que and C. A. Napoli, 1996. Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single copy vs. complex T-DNA sequences. Plant Mol. Biol. 31: 957–973. [DOI] [PubMed] [Google Scholar]
- Kasschau, K. D., and J. C. Carrington, 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95: 461–470. [DOI] [PubMed] [Google Scholar]
- Kusaba, M., K. Myiahara, S. Iida, H. Fukuoka, T. Takano et al., 2003. Low glutelin content1: a dominant mutation that suppresses the Glutelin multigene family via RNA silencing in rice. Plant Cell 15: 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos, L., G. Szittya, D. Silhavy and J. Burgyan, 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23: 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie, R., 1995. Vascular puncture of maize kernels for the mechanical transmission of Maize white line mosaic virus and other viruses of maize. Phytopathology 85: 139–143. [Google Scholar]
- Louie, R., M. G. Redinbaugh, D. T. Gordon, J. J. Abt and R. J. Anderson, 2000. Maize necrotic streak virus, a new maize virus with similarity to species of the family Tombusviridae. Plant Dis. 84: 1133–1139. [DOI] [PubMed] [Google Scholar]
- Luff, B., L. Pawlowski and J. Bender, 1999. An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell 3: 505–511. [DOI] [PubMed] [Google Scholar]
- May, B. P., and S. L. Dellaporta, 1998. Transposon sequences drive tissue-specific expression of the maize regulatory gene R-s. Plant J. 13: 241–247. [Google Scholar]
- Mette, M. F., J. van der Winden, M. A. Matzke and A. J. M. Matzke, 1999. Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters in trans. EMBO J. 18: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzlaff, M., M. O'Dell, P. D. Cluster and R. B. Flavell, 1997. RNA-mediated RNA degradation and chalcone synthase silencing in petunia. Cell 88: 845–854. [DOI] [PubMed] [Google Scholar]
- Meyers, B. C., S. V. Tingey and M. Morgante, 2001. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11: 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskens, M. W. M., A. P. A. Vissers, J. N. M. Mol and J. M. Kooter, 2000. Role of inverted DNA repeats in transcriptional and post-transcriptional gene silencing. Plant Mol. Biol. 43: 243–260. [DOI] [PubMed] [Google Scholar]
- Napoli, C. A., C. Lemieux and R. A. Jorgensen, 1990. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas, K., H. Nicholas, Jr. and D. Deerfield, II, 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW. NEWS 4: 14. [Google Scholar]
- Niesbach-Klösgen, U., E. Barzen, J. Berhardt, W. Rohde, Z. Schwarz-Sommer et al., 1987. Chalcone synthase genes in plants: a tool to study evolutionary relationships. J. Mol. Evol. 26: 213–225. [Google Scholar]
- Pal-Bhadra, M., U. Bhadra and J. A. Birchler, 2002. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9: 315–327. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- SanMiguel, P. J., A. Tikhonov, Y.-K. Jin, N. Motchoulskaia, D. Zakharov et al., 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768. [DOI] [PubMed] [Google Scholar]
- Senda, M., C. Masuta, S. Ohnishi, K. Goto, A. Kasai et al., 2004. Patterning of virus-infected Glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 16: 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, D. M., R. C. Hightower and R. B. Meagher, 1983. Genes encoding actin in higher plants: intron positions are highly conserved but the coding sequences are not. J. Mol. Appl. Genet. 2: 111–126. [PubMed] [Google Scholar]
- Soppe, W. J., Z. Jasencakova, A. Houben, T. Kakutani, A. Meister et al., 2002. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21: 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Techen, N., L. Borchert, B. E. Scheffler and U. Wienand, 1999. Isolation of two new CACTA transposable elements from anthocyanin genes in maize. Maize Coop. News Lett. 73: 35–36. [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, J. J., and L. O. Vodkin, 1996. Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant Cell 8: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcich, M., A. Bokhari-Rizi, D. Hamilton, C. He, W. Messier et al., 1996. PREM-2, a copia-type retroelement in maize is expressed preferentially in early microspores. Sex. Plant Reprod. 9: 65–74. [Google Scholar]
- Tuteja, J. H., S. J. Clough, W.-C. Chan and L. O. Vodkin, 2004. Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell 16: 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol, A. R., L. A. Mur, M. Beld, J. N. M. Mol and A. R. Stuitje, 1990. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason, J. M., G. Szittya, J. Burgyan and T. M. Tanaka Hall, 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115: 799–811. [DOI] [PubMed] [Google Scholar]
- Verdel, A., J. Songtao, S. Gerber, T. Sugiyama, S. Gygi et al., 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe, T., C. Kidner, I. M. Hall, G. Teng, S. I. S. Grewal et al., 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Volpe, T., V. Schramke, G. L. Hamilton, S. A. White, G. Teng et al., 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11: 137–146. [DOI] [PubMed] [Google Scholar]
- Walker, E. L., and T. Panavas, 2001. Structural features and methylation patterns associated with paramutation at the r1 locus of Zea mays. Genetics 159: 1201–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, E. L., T. P. Robbins, T. E. Bureau, J. Kermicle and S. L. Dellaporta, 1995. Transposon mediated chromosomal rearrangements and gene duplications in the formation of the maize R-r complex. EMBO J. 14: 2350–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M., 2000. RNA-directed DNA methylation. Plant Mol. Biol. 43: 203–220. [DOI] [PubMed] [Google Scholar]
- Wienand, U., U. Weydemann, U. Niesbach-Klosgen, P. A. Peterson and H. Saedler, 1986. Molecular cloning of the c2 locus of Zea mays, the gene coding for chalcone synthase. Mol. Gen. Genet. 203: 202–207. [Google Scholar]
- Yao, H., Q. Zhou, J. Li, H. Smith, M. Yandeau et al., 2002. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc. Natl. Acad. Sci. USA 99: 6157–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, K., L. Malinina and D. J. Patel, 2003. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426: 874–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., J. Arbuckle and S. R. Wessler, 2000. Recent, extensive, and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker into genic regions of maize. Proc. Natl. Acad. Sci. USA 97: 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]