Abstract
In this study a well-characterized pathological mutation at nucleotide position 3243 of human mitochondrial DNA was introduced into human ρ0 teratocarcinoma (NT2) cells. In cloned and mixed populations of NT2 cells heteroplasmic for the mutation, mitotic segregation toward increasing levels of mutant mitochondrial DNA always occurred. Rapid segregation was frequently followed by complete loss of mitochondrial DNA. These findings support the idea that pathological mitochondrial DNA mutations are particularly deleterious in specific cell types, which can explain some of the tissue-specific aspects of mitochondrial DNA diseases. Moreover, these findings suggest that mitochondrial DNA depletion may be an important and widespread feature of mitochondrial DNA disease.
MITOCHONDRIAL DNA (mtDNA) mutations are now a well-recognized cause of human disease. Pathological mutations often coexist with apparently wild-type mtDNA, a situation termed heteroplasmy (Holt et al. 1988, 1990; Shoffner et al. 1990). The mutation at nucleotide position (np) 3243 of human mtDNA was one of the first pathological mutations to be characterized, often associated with the mitochondrial disease, mitochondrial encephalo(myo)pathy, lactic acidosis, and stroke-like episodes, or MELAS (Goto et al. 1990). In other patients, the same mutation is also linked to diabetes and deafness (MIDD) (van den Ouweland et al. 1992). The A-to-G transition at np 3243 is located in the transfer RNA gene that decodes leucine UUR codons (Goto et al. 1990) and also forms part of the binding site for a transcription termination factor (Christianson and Clayton 1988). The mutation adversely affects the steady-state level, aminoacylation, and extent of wobble base modification of tRNALeuUUR (Chomyn et al. 1992; King et al. 1992; Yasukawa et al. 2001).
Biased segregation or maintenance of A3243G and other mutant mtDNAs has been reported both in vitro (Hayashi et al. 1992; Yoneda et al. 1992; Bourgeron et al. 1993; Dunbar et al. 1995; Holt et al. 1997; Spelbrink et al. 1997; Vergani et al. 1999) and in vivo (Poulton et al. 1995; Blok et al. 1997; Weber et al. 1997; Rahman et al. 2001). However, the direction of segregation of mutant mtDNAs can vary according to cellular background or other factors (Bourgeron et al. 1993; Dunbar et al. 1995) and in some cases is not observed (Matthews et al. 1995; Shoubridge 1995). Nuclear effects on mtDNA segregation have also been reported in mice in the case of a nonpathological mtDNA haplotype (Battersby et al. 2003).
In vitro studies of pathological mtDNA mutations have exploited cell lines that lack mtDNA. These so-called ρ0 cells are fused with cytoplasts from donor cells containing mtDNA mutations. The resultant cybrids manifest the phenotypic consequences of the mutations in control cell backgrounds (Chomyn et al. 1991). These studies have, in the main, utilized cell lines unrelated to the tissues most affected by mitochondrial disease, such as osteosarcoma and cervical or lung carcinomas. The tissues most commonly affected by pathological mtDNA mutations are, conversely, brain, muscle, cochlea, and pancreatic β-cells.
In an attempt to overcome this methodological problem, we selected, as a recipient for cybridization, a pluripotent teratocarcinoma cell line from which a number of differentiated cell types can be derived (Chadalavada et al. 2005), including cells displaying neuronal properties (Younkin et al. 1993). We depleted (undifferentiated) NT2 cells of their endogenous mtDNA by prolonged treatment with dideoxycytidine (ddC), thus creating the desired ρ0 line. The same rationale lay behind a study of two mutations associated with Leber's hereditary optic neuropathy in the NT2 cell background (Wong et al. 2002).
When mitochondria derived from a MELAS patient with a mixture of wild-type and A3243G mutant mtDNA were introduced into ρ0 NT2 cells, segregation toward increasing levels of mutant mtDNA invariably occurred. Moreover, complete loss of mtDNA was observed in some cybrids. These findings support the idea that the cellular background is a determining factor in both the phenotypic expression of mtDNA mutations such as A3243G and their replicative dynamics.
MATERIALS AND METHODS
All cell lines were routinely maintained in Dulbecco's modified Eagle's medium (DMEM) containing 25 mm glucose and 110 μg/ml sodium pyruvate, supplemented with 10% fetal bovine serum (FBS). NT2 teratocarcinoma ρ0 cells were, in addition, supplemented with 50 μg/ml uridine. NT2 cells were denuded of their endogenous mtDNA by treatment with 50 μm ddC for 21 days in medium containing 20% FBS, 80% DMEM, and 50 μg/ml uridine. The absence of mtDNA was confirmed by Southern blotting and PCR (data not shown). A G418-resistant NT2 ρ0 cell line was generated by transfection of the cells with pcDNA3.1neo (Invitrogen, San Diego) using lipofectamine (Invitrogen). After transfection, a stably G418-resistant clonal cell line was selected and maintained by culturing in the presence of 300 μg/ml G418 (Invitrogen).
Mitochondrial DNA carrying the A3243G mutation was introduced into NT2 ρ0 cells from 143B osteosarcoma cybrids, A549 lung carcinoma cybrids, or a mixed myoblast/fibroblast culture, derived from a single patient by cell-cytoplast fusion, as previously described (King and Attardi 1989; Dunbar et al. 1995). Briefly, cytoplasts were generated by inverting 35-mm tissue culture plates, 50–90% confluent, in 95% DMEM, 5% FBS (nondialyzed) with 10 μg/ml cytochalasin B (Calbiochem, La Jolla, CA) and centrifuging at 7000 × gmax for 20 min. The resultant cytoplast lawn was incubated for 3 hr at 37° with ∼8 × 105 ρ0 cells. Cell-cytoplast fusion was induced by the addition of 50% (w/v) PEG 1500, 45% DMEM, and 5% DMSO. After 1 min, the cells were washed twice in 90% DMEM, 10% DMSO, and three times in DMEM alone and then incubated overnight in 90% DMEM and 10% FBS without uridine supplementation. Putative transformant cells were replated on 90-mm dishes in 90% DMEM and 10% FBS without supplemental uridine and individual colonies were picked ∼14 days later. Where the nuclear recipient carried a neomycin resistance gene, G418 was included in the growth medium at a concentration of 300 μg/ml for a period of ∼1 month. NT2 cybrids carrying mtDNA molecules with the A3243G mutation were designated NT2.3243, even where cells did not maintain A3243G mtDNA molecules.
DNA extraction from cultured cells was as described by Laird et al. (1991). DNA samples were digested with PvuII, ApaI, or AflII according to the manufacturer's instructions (New England Biolabs, Beverly, MA). PvuII-digested samples were electrophoresed at 1.3 V/cm for 16–20 hr in a 0.8% agarose gel. The relative amounts of mitochondrial and nuclear DNA were determined either by probing Southern blots simultaneously with labeled, cloned fragments of mtDNA and 18S ribosomal DNA or by multiplex PCR of mtDNA and nuclear DNA, as described previously (El Meziane et al. 1998). Southern blotting and hybridization were as described in Sambrook et al. (1990). Quantification of the relative amounts of mutant and wild-type mtDNA was determined using a Packard (Meriden, CT) cyclone phosphorimager.
The proportion of A3243G and G12300A mtDNA was estimated by separating amplified DNA fragments digested with ApaI or AflII, respectively, on 6% nondenaturing polyacrylamide gels. Regions of mtDNA spanning the 3243 and 12,300 regions were amplified by 21–24 cycles of 95° for 30 sec, 60° for 30 sec, and 72° for 45 sec. Reactions comprised 100 nm of each primer (see below), 1× Taq buffer (Promega, Madison, WI), 16 μm dNTPs, 1 unit Taq polymerase (Promega), and 1 μl template DNA in 50 μl reactions. Prior to the last cycle, 1 μl of [32P]dCTP (Amersham, Buckinghamshire, UK) was added and the reactions were incubated at 95° for 3 min, 60° for 1 min, and 72° for 10 min. Primers for the nt 2968–3573 region, including np 3243, were 5′-TCAACAATAGGGTTTACGACCTCG-3′ and 5′-AGGGGGGTTCATAG TAGAAGAGCG-3′. Those for the np 12,300 region (nt 11,992–12,898) were 5′-TTACCACAACACAATG GGGCTC-3′ and 5′-TGCTAAGGCGAGGATGAAACC-3′. Intact-cell oxygen consumption rates were determined in a Clark-type oxygen electrode with 500–1000 μl of 5 × 106 cells/ml in RPMI 1640 medium without glucose (Invitrogen) (Chomyn et al. 1991).
RESULTS AND DISCUSSION
NT2.3243 cybrids segregate to mutant in bulk culture and display mtDNA loss:
In trial fusions between NT2 ρ0 cells and cytoplasts from a heteroplasmic donor cell line containing 74% A3243G mutant mtDNA, we observed that cloning efficiency of the resultant cybrids was extremely low (data not shown). We therefore studied the behavior of cybrids initially in bulk culture, following fusion. The presence of mtDNA, by definition deriving from the donor cytoplasts, was verified by Southern blotting, and the relative amount of A3243G mutant mtDNA was quantified using ApaI restriction-site gain. Cybrids were G418 resistant (like the NT2 ρ0 cell recipient) and nuclear microsatellite (Mariotti et al. 1994) or SNP markers (Loktionov et al. 2001) were always of the recipient (NT2) rather than the donor type, confirming that the cells surviving the fusion procedure were true cybrids.
In five independent experiments, the level of A3243G mutant mtDNA was always higher than that in the donor cells by the time it could first be measured in the bulk culture of cybrids, 15–20 days after fusion (Figure 1 and other data not shown). In all five cases, it continued to increase, typically reaching 85–95% of total mtDNA before a rapid loss of mtDNA was observed. Mitochondrial DNA then became undetectable in the cells between 45 and 80 days after fusion.
Figure 1.—
Increase in proportion of mutant (A3243G) mtDNA and subsequent loss of mtDNA in pooled populations of NT2 cybrids. Solid symbols are percentages of A3243G mtDNA; open symbols are mtDNA nuclear DNA. Squares and circles are two populations of cybrid cells from independent fusion experiments. X indicates the proportion of A3243G mtDNA of the mitochondrial donor cytoplasts (t = 0). DNA was harvested for the first time 17 days after cell-cytoplast fusion; analysis at earlier time points was inappropriate both because cell numbers were low and because unenucleated osteosarcoma cells survived for up to 14 days in G418. The proportion of A3243G mtDNA was based on the mean of three measurements with a standard deviation of <2%, determined by last-cycle PCR analysis. The ratio of mitochondrial to nuclear DNA was derived from densitometry of multiplex PCR samples also done in triplicate (standard deviation <6%).
To test whether growth conditions influence the outcome of such experiments, we plated cells from one of the same fusions as shown in Figure 1 at a fourfold higher initial density. Although this culture once again exhibited a progressive increase in the proportion of A3243G mutant mtDNA, it showed two clear differences from cybrid cultures plated initially at lower density, including cells from the same fusion (Figure 2). First, the proportion of mutant mtDNA was considerably lower than that in cells plated at low density at the first time point when it could be measured: 57% A3243G compared to 78% A3243G (Figure 2). Second, there was no loss of mtDNA from the cells, which maintained a normal copy number even 180 days after fusion. This suggests that the rate and outcome of mitotic segregation in this cell background depends upon a complex interplay of intercellular and intracellular signals.
Figure 2.—
Maintenance or loss of mtDNA in NT2 cybrids dependent on the extent of dilution after fusion. NT2 ρ0 cells and osteosarcoma cytoplasts carrying 74% A3243G mtDNA, n = 9, standard deviation = 2.37 (X) were fused and 16 hr later passaged and fractions of eight-tenths, one-tenth, and one-fortieth were maintained as separate populations. The proportion of A3243G mtDNA rose to 95% in the case of the 1 in 40 dilution (solid circles); thereafter, mtDNA was undetectable by PCR. In contrast, cells of the 1 in 10 dilution (open triangles) mtDNA continued to be detected up to 180 days, after which the experiment was terminated. The cell line comprising 80% of the original fusion also maintained mtDNA over a long term (data not shown).
NT2.3243 cybrid clones segregate to mutant and display mtDNA loss:
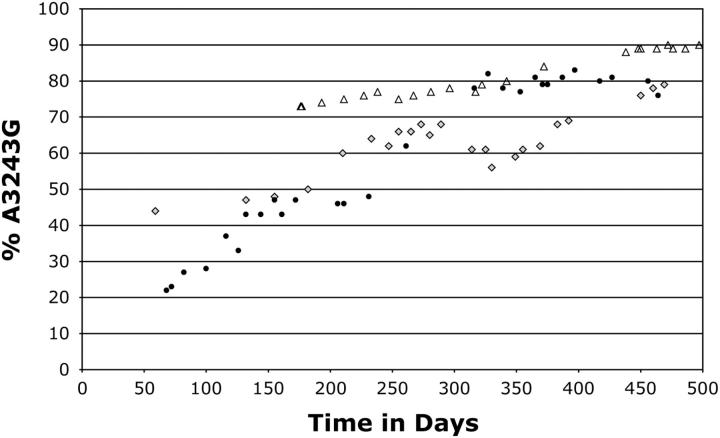
A small number of cybrid clones were obtained from fusions of NT2 ρ0 cells with cytoplasts heteroplasmic for A3243G. Mostly, these exhibited either of two behaviors broadly similar to those of the bulk cultures. Of 22 such clones, 17 thus showed higher levels of mutant mtDNA than the mitochondrial donors did and eventually underwent complete loss of mtDNA. The remaining clones, however, had lower levels of mutant mtDNA (≤50%, 2 of them being 0%) and did not lose mtDNA. Those that were heteroplasmic showed a slow increase in the amount of mutant mtDNA over many months, but eventually stabilized at ∼80–90% heteroplasmy (Figure 3).
Figure 3.—
Rare NT2.3243 clones that maintained mtDNA displayed biased segregation to mutant mtDNA. Two of three heteroplasmic clonal cell lines that had a normal mtDNA copy number were maintained in continuous culture for ∼500 days after fusion: NT2.3243a (shaded diamonds) and NT2.3243b (solid circles). A subclone of the NT2.3243a cell line, denoted NT2.3243a2, was also monitored (open triangles). These cybrids were products of a fusion between a mixed fibroblast/myoblast line with 45% A3243G mtDNA (n = 5, standard deviation ±3.4%) and NT2 ρ0 cells; in another fusion experiment, the same cells yielded cybrids, all of which lost mtDNA, as was the case with fusions derived from osteosarcoma cytoplasts containing 74% A3243G mtDNA.
Since wild-type mtDNA was maintained in the NT2 cell background, loss of mtDNA cannot be a general property of this particular recipient cell line. To test whether it applied also to mtDNAs carrying other pathological mutations, we carried out parallel cybrid fusions with donor cytoplasts carrying the A1555G mutation (Prezant et al. 1993). A total of 12 cybrid colonies selected at random were all homoplasmic for A1555G and maintained normal levels of mtDNA (data not shown). Therefore, systematic loss of mtDNA in the NT2 cell background is specific to A3243G and is not shared with another pathological mutation or with wild-type mtDNA that is otherwise identical. Notwithstanding, other heteroplasmic pathological mtDNA variants might also precipitate mtDNA loss in the NT2 cell background. Finally, it must be borne in mind that these experiments have studied the behavior of mtDNA from only one A3243G patient. Although internally controlled by wild-type mtDNA from the same patient, it cannot be excluded that other polymorphisms present in the mtDNA influence the behavior of A3243G mutant mtDNA. If this applies in vivo, the phenotypic outcome may thus also depend on the mtDNA haplotype.
Loss of mtDNA in NT2.3243 cybrids is independent of respiratory phenotype:
Respiration in NT2.3243 cybrids showed a classical threshold effect (Figure 4A). Oxygen consumption of cybrids with 69% or even 80% A3243G mutant mtDNA was indistinguishable from that of controls with 0% mutant mtDNA, whereas a cybrid bearing 84% mutant mtDNA showed a much lower rate of oxygen consumption. This suggests that above a certain heteroplasmic threshold, the presence of A3243G mutant mtDNA is of little metabolic benefit to NT2 cells and might even be harmful or burdensome, providing a selective mechanism favoring its loss.
Figure 4.—
High levels of A3243G mtDNA are associated with impaired respiratory capacity in the NT2 cell background. (A) Oxygen consumption was measured in NT2.3243 cybrids with 0, 69, 80, and 84% A3243G mtDNA; the results suggest that only with >80% A3243G mtDNA is there a measurable effect on respiration. Four replicate measurements of percentage of heteroplasmy levels gave values of 80, standard deviations ±1.4 and 84 and standard deviation ±1.7 for the two most similar cybrids, suggesting that their different respiratory phenotypes are meaningful. (B) Ninety-nine percent A3243G is associated with a marked decrease in oxygen consumption, which is rescued by a second mutation in tRNALeuCUN at np 12,300.
To test this hypothesis, we transferred mtDNA from a donor lung-carcinoma cell line containing >99% A3243G mutant mtDNA to NT2 ρ0 cells. Once again, a majority of cybrid clones (14/22) showed loss of mtDNA, but mtDNA appeared to be stably maintained in the remaining cybrids, even though they showed very low rates of respiration (Figure 4B). Conversely, when the donor mitochondria used were also heteroplasmic for the G12300A suppressor mutation (El Meziane et al. 1998), as well as >99% mutant for A3243G, a majority of cybrid clones isolated (25/29) still lost mtDNA, despite the fact that the presence of the suppressor improved respiration. This demonstrates clearly that mtDNA loss or retention is independent of respiratory phenotype.
The fact that some NT2 cybrid clones with >99% A3243G mutant mtDNA were able to maintain mtDNA suggests that the level of mutant mtDNA is also not an absolute determinant of loss or retention of mtDNA.
Intercellular selection can affect the direction and endpoint of segregation in NT2.3243 cybrids:
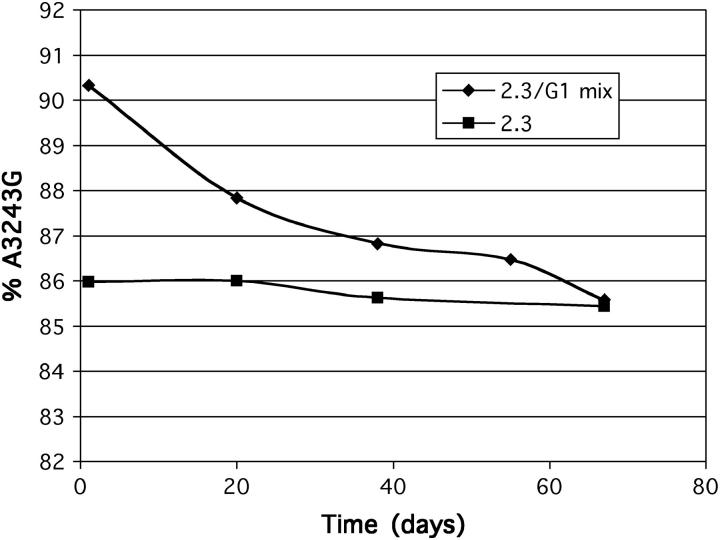
On the basis of the above experiments, the retention or loss of mtDNA did not appear to be influenced by respiratory selection. However, this does not exclude intercellular selection from influencing the change over time in the proportion of mutant vs. wild-type mtDNA in those cybrids that retained mtDNA. To test this, we co-cultured a cybrid clone with >99% mutant mtDNA, showing a very low rate of respiration, with one that had stabilized at 86% mutant mtDNA and exhibited only a partial respiratory impairment. The initial mixture contained ∼50% of each clone of cells. Over a period of 8 weeks of continuous culture, the mixed culture showed a progressive reduction in the average level of A3243G mutant mtDNA until reaching 86% (Figure 5). This implies that cells with 86% A3243G mtDNA have a growth advantage over those with 99% mutant mtDNA, especially given that a parallel culture of the 86% mutant cybrid retained a constant proportion of mutant mtDNA throughout the experiment. The outcome of the experiment indicates that intercellular competition based on respiratory competence or on some other growth-regulatory parameter can influence the direction as well as the endpoint of segregation in NT2 cybrids. The tendency for the proportion of A3243G mutant mtDNA to increase in bulk culture or in individual cybrid clones appears to be an inherent property of the mutation and cell background, since it occurred across a range of heteroplasmy levels where respiration was unaffected. However, eventual stabilization of the level of heteroplasmy in the 80–90% range appears to be a trade-off between this natural tendency toward higher levels of mutant mtDNA, counteracted by the selective disadvantage to growth of exceeding this threshold level.
Figure 5.—
Intercellular competition favors cells with 86% A3243G mtDNA over cells with 99% A3243G mtDNA. A clonal teratocarcinoma line (2.3, ▪) was cultured for 70 days alone or co-cultured with a sister line carrying 99% A3243G mtDNA (2.3/G1 mix, ♦). The cultures were sampled at intervals to determine the proportion of mutant mtDNA (percentage of A3243G). By 70 days, both cultures had a similar level of A3243G mtDNA.
Mitotic segregation in NT2.3243 cybrids:
The overall tendency for mutant mtDNA levels to increase in bulk culture of NT2.3243 cybrids, even after cloning, might mask the fact that random mitotic segregation can shift the proportion of mutant mtDNA in either direction at any given cell division. To measure the extent of mitotic segregation in NT2.3243 cybrids, we analyzed the range of heteroplasmy values in subclones obtained by plating at limiting dilution and derived from a specific cybrid clone maintained for 35 weeks after cybrid formation. The rate and extent of segregation depends on the effective number of segregating units. For instance, a cell with 50% mutant mtDNA transmitted as a small number of segregating units will give rise to cells with the full range of mutant and wild-type mtDNAs after a few cell divisions, whereas much higher numbers of a segregating unit (103 or greater) will tend to cluster around the original value, even after 100 or more cell divisions.
To exclude any effect of respiratory selection, we chose a cybrid clone exhibiting a relatively low average level of heteroplasmy for A3243G (43%). Ignoring any small additional shift toward higher levels of average heteroplasmy during the time of the experiment, we compared the results with the distributions of heteroplasmy levels predicted to have arisen from random, mitotic segregation, using mathematical models based on 1000, 200, or <200 segregating units. The observed range of heteroplasmy levels was narrower than predicted even on the basis of 1000 segregating units (Figure 6). This finding, coupled with the knowledge that mtDNA is arranged in multigenomic structures, or nucleoids, of approximately five copies (Iborra et al. 2004; Legros et al. 2004), which display limited movement (Garrido et al. 2003), suggests that heteroplasmy for A3243G mtDNA in NT2 cybrids is not maintained simply by stochastic processes, but is subject to mechanistic constraints connected with mtDNA replication or partition, such as faithful duplication of nucleoids (Jacobs et al. 2000).
Figure 6.—
The unit of segregation in NT2 cybrids is >1000. A cybrid cell line with a mean level of 43% A3243G mtDNA (generated from fusion with cytoplasts carrying 45% A3243G mtDNA) was serially diluted and mtDNA of 24 subclones was genotyped 27 weeks after fusion. The observed results were grouped in intervals of 10% (shaded bars) and compared to the predicted distribution for random partitioning of independently segregating units of 200 (open bars) and 1000 (solid bars) (see Lehtinen et al. 2000 for details of the method of statistical analysis).
Relevance of the findings to mitochondrial disease:
The clinical phenotypes associated with the A3243G mutation are diverse, and the progression of the disease is also unpredictable. Since the mutation is always heteroplasmic, the patterns of evolution of heteroplasmy in a given patient, both during development and in adulthood, may have a crucial bearing on the clinical phenotype. However, it has thus far been difficult to study this parameter in the relevant tissues in vivo, especially in the central nervous system and sensory epithelia (as well as pancreatic β-cells).
Previous analyses of heteroplasmic evolution in cybrid cell models showed that cellular background can confer systematic segregation toward or away from A3243G mutant mtDNA, but that stable heteroplasmy with minimal mitotic segregation can also be maintained over long periods of time. This study is the first to analyze the behavior of A3243G mutant mtDNA in a cellular background potentially more relevant to the main disease features.
The findings are highly provocative. First, they confirm the tendency of mutant mtDNA to increase with time, a tendency shared with some, but not all, other cell backgrounds studied. In NT2 cells, however, mutant mtDNA invariably increases to >80% in contrast to other cell backgrounds where it can apparently stabilize at much lower levels. Second, the findings indicate two alternate endpoints of this process: either stabilization at a “tolerated” level of heteroplasmy, determined by an equilibrium between further segregation toward mutant mtDNA and phenotypic counterselection at the cellular level, or complete loss of mtDNA, perhaps once a specific threshold level of mutant mtDNA is crossed. Third, the findings show that mtDNA loss is a property not shared with all other pathological mutations; it does not apply, for example, to A1555G.
Complete loss of mtDNA may be considered a drastic outcome of mitotic segregation, pointing toward clinically severe consequences. If it applies in vivo, it may go some way toward explaining why MELAS has such potentially devastating effects on the nervous system, although not in every individual carrying the A3243G mutation. It is therefore important to identify the genetic, epigenetic, or environmental factors that determine which of the two endpoints is reached, as well as the process that underlies “paradoxical” segregation toward higher levels of mutant mtDNA in the first place.
Because these experiments were conducted in tumor-derived, cultured cells, some caution is needed in their interpretation. Although supposedly homogeneous, many tumor-derived lines are karyotypically unstable, even at a microscopic level. Others can exhibit epigenetic silencing or reactivation of many genes. The two observed outcomes of segregation may therefore be due to functional nullizygosity for one or more key genes involved in mtDNA metabolism, nucleoid dynamics, or adaptation to respiratory chain deficiency. The application of genome, transcriptome, and proteome screening to cybrid clones of the two different types could therefore help to identify the relevant pathways and genes.
The level of heteroplasmy for A3243G appears to play some role in the outcome, since all of the cells that experienced mtDNA loss as the outcome were already at high levels of heteroplasmy at the first time point when analyzed, whereas those that reached equilibrium at 80–90% mutant mtDNA generally started out at much lower levels of heteroplasmy. Interestingly, the latter type of cells almost invariably traversed the heteroplasmy levels at which the former began, indicating that the heteroplasmy level itself is not the determining parameter, but rather the time that cells have to adapt to progressively higher levels of mutant mtDNA.
Loss of mtDNA may, indeed, be a relevant disease mechanism in vivo at least in A3243G patients who suffer from diabetes. Pancreatic islet cells from a diabetic patient were reported to lack the high proportion of A3243G mtDNA typical of other affected tissues (Lynn et al. 2003). There was also decreased overall β-cell mass. These observations might be explained on the basis of mtDNA loss, leading to apoptosis of pancreatic cells in which A3243G mutant mtDNA had accumulated above a critical threshold level, as seen in NT2 cell cybrids in this study. The final phenotype in A3243G disease may therefore be a combination of the effects of mitochondrial translational dysfunction and mtDNA loss, influenced by the directional patterns of mitotic segregation in different tissues.
Acknowledgments
We are indebted to Alex Lotonikov of the Dunn Human Nutrition Unit for SNP analysis. The work was supported by the United Kingdom Medical Research Council; the Wellcome Trust; the European Union's integrated project, Rational Treatment Strategies Combating Mitochondrial Oxidative Phosphorylation Disorders (EUMITOCOMBAT), concerted action on Mitochondrial Biogenesis and Disease (mitEURO), and Thematic Network on the Genetics of Deafness (GENDEAF); and the Academy of Finland and Tampere University Hospital Medical Research Fund.
References
- Battersby, B. J., J. C. Loredo-Osti and E. A. Shoubridge, 2003. Nuclear genetic control of mitochondrial DNA segregation. Nat. Genet. 33: 183–186. [DOI] [PubMed] [Google Scholar]
- Blok, R. B., D. A. Gook, D. R. Thorburn and H. H. Dahl, 1997. Skewed segregation of the mtDNA nt 8993 (T→G) mutation in human oocytes. Am. J. Hum. Genet. 60: 1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron, T., D. Chretien, A. Rotig, A. Munnich and P. Rustin, 1993. Fate and expression of the deleted mitochondrial DNA differ between human heteroplasmic skin fibroblast and Epstein-Barr virus-transformed lymphocyte cultures. J. Biol. Chem. 268: 19369–19376. [PubMed] [Google Scholar]
- Chadalavada, R. S., J. Houldsworth, A. B. Olshen, G. J. Bosl, L. Studer et al., 2005. Transcriptional program of bone morphogenetic protein-2-induced epithelial and smooth muscle differentiation of pluripotent human embryonal carcinoma cells. Funct. Integr. Genomics 5: 59–69. [DOI] [PubMed] [Google Scholar]
- Chomyn, A., G. Meola, N. Bresolin, S. T. Lai, G. Scarlato et al., 1991. In vitro genetic transfer of protein synthesis and respiration defects to mitochondrial DNA-less cells with myopathy-patient mitochondria. Mol. Cell. Biol. 11: 2236–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn, A., A. Martinuzzi, M. Yoneda, A. Daga, O. Hurko et al., 1992. MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc. Natl. Acad. Sci. USA 89: 4221–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, T. W., and D. A. Clayton, 1988. A tridecamer DNA sequence supports human mitochondrial RNA 3′-end formation in vitro. Mol. Cell. Biol. 8: 4502–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar, D. R., P. A. Moonie, H. T. Jacobs and I. J. Holt, 1995. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc. Natl. Acad. Sci. USA 92: 6562–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Meziane, A., S. K. Lehtinen, N. Hance, L. G. Nijtmans, D. Dunbar et al., 1998. A tRNA suppressor mutation in human mitochondria. Nat. Genet. 18: 350–353. [DOI] [PubMed] [Google Scholar]
- Garrido, N., L. Griparic, E. Jokitalo, J. Wartiovaara, A. M. van der Bliek et al., 2003. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell 14: 1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, Y., I. Nonaka and S. Horai, 1990. A mutation in the tRNA (Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348: 651–653. [DOI] [PubMed] [Google Scholar]
- Hayashi, J., S. Ohta, A. Kikuchi, M. Takemitsu, Y. Goto et al., 1992. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. J. Inherit. Metab. Dis. 15: 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, I. J., A. E. Harding and J. A. Morgan-Hughes, 1988. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331: 717–719. [DOI] [PubMed] [Google Scholar]
- Holt, I. J., A. E. Harding, R. K. Petty and J. A. Morgan-Hughes, 1990. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet. 46: 428–433. [PMC free article] [PubMed] [Google Scholar]
- Holt, I. J., D. R. Dunbar and H. T. Jacobs, 1997. Behaviour of a population of partially duplicated mitochondrial DNA molecules in cell culture: segregation, maintenance and recombination dependent upon nuclear background. Hum. Mol. Genet. 6: 1251–1260. [DOI] [PubMed] [Google Scholar]
- Iborra, F. J., H. Kimura and P. R. Cook, 2004. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, H. T., S. K. Lehtinen and J. N. Spelbrink, 2000. No sex please, we're mitochondria: a hypothesis on the somatic unit of inheritance of mammalian mtDNA. BioEssays 22: 564–572. [DOI] [PubMed] [Google Scholar]
- King, M. P., and G. Attardi, 1989. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246: 500–503. [DOI] [PubMed] [Google Scholar]
- King, M. P., Y. Koga, M. Davidson and E. A. Schon, 1992. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol. Cell. Biol. 12: 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird, P. W., A. Zijderveld, K. Linders, M. A. Rudnicki, R. Jaenisch et al., 1991. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 19: 4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros, F., F. Malka, P. Frachon, A. Lombes and M. Rojo, 2004. Organization and dynamics of human mitochondrial DNA. J. Cell Sci. 117: 2653–2662. [DOI] [PubMed] [Google Scholar]
- Lehtinen, S. K., N. Hance, A. El Meziane, M. K. Juhola, K. M. Juhola et al., 2000. Genotypic stability, segregation and selection in heteroplasmic human cell lines containing np 3243 mutant mtDNA. Genetics 154: 363–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loktionov, A., M. A. Watson, M. Gunter, W. S. Stebbings, C. T. Speakman et al., 2001. Glutathione-S-transferase gene polymorphisms in colorectal cancer patients: interaction between GSTM1 and GSTM3 allele variants as a risk-modulating factor. Carcinogenesis 22: 1053–1060. [DOI] [PubMed] [Google Scholar]
- Lynn, S., G. M. Borthwick, R. M. Charnley, M. Walker and D. M. Turnbull, 2003. Heteroplasmic ratio of the A3243G mitochondrial DNA mutation in single pancreatic beta cells. Diabetologia 46: 296–299. [DOI] [PubMed] [Google Scholar]
- Mariotti, C., V. Tiranti, F. Carrara, B. Dallapiccola, S. DiDonato et al., 1994. Defective respiratory capacity and mitochondrial protein synthesis in transformant cybrids harboring the tRNA(Leu(UUR)) mutation associated with maternally inherited myopathy and cardiomyopathy. J. Clin. Invest. 93: 1102–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, P. M., R. M. Brown, K. Morten, D. Marchington, J. Poulton et al., 1995. Intracellular heteroplasmy for disease-associated point mutations in mtDNA: implications for disease expression and evidence for mitotic segregation of heteroplasmic units of mtDNA. Hum. Genet. 96: 261–268. [DOI] [PubMed] [Google Scholar]
- Poulton, J., S. O'Rahilly, K. J. Morten and A. Clark, 1995. Mitochondrial DNA, diabetes and pancreatic pathology in Kearns-Sayre syndrome. Diabetologia 38: 868–871. [DOI] [PubMed] [Google Scholar]
- Prezant, T. R., J. V. Agapian, M. C. Bohlman, X. Bu, S. Oztas et al., 1993. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 4: 289–294. [DOI] [PubMed] [Google Scholar]
- Rahman, S., J. Poulton, D. Marchington and A. Suomalainen, 2001. Decrease of 3243 A→G mtDNA mutation from blood in MELAS syndrome: a longitudinal study. Am. J. Hum. Genet. 68: 238–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1990 Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shoffner, J. M., M. T. Lott, A. M. Lezza, P. Seibel, S. W. Ballinger et al., 1990. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell 61: 931–937. [DOI] [PubMed] [Google Scholar]
- Shoubridge, E. A., 1995. Segregation of mitochondrial DNAs carrying a pathogenic point mutation (tRNA(leu3243)) in cybrid cells. Biochem. Biophys. Res. Commun. 213: 189–195. [DOI] [PubMed] [Google Scholar]
- Spelbrink, J. N., R. Zwart, M. J. Van Galen and C. Van den Bogert, 1997. Preferential amplification and phenotypic selection in a population of deleted and wild-type mitochondrial DNA in cultured cells. Curr. Genet. 32: 115–124. [DOI] [PubMed] [Google Scholar]
- van den Ouweland, J. M., H. H. Lemkes, W. Ruitenbeek, L. A. Sandkuijl, M. F. de Vijlder et al., 1992. Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat. Genet. 1: 368–371. [DOI] [PubMed] [Google Scholar]
- Vergani, L., R. Rossi, C. H. Brierley, M. Hanna and I. J. Holt, 1999. Introduction of heteroplasmic mitochondrial DNA (mtDNA) from a patient with NARP into two human rho degrees cell lines is associated either with selection and maintenance of NARP mutant mtDNA or failure to maintain mtDNA. Hum. Mol. Genet. 8: 1751–1755. [DOI] [PubMed] [Google Scholar]
- Weber, K., J. N. Wilson, L. Taylor, E. Brierley, M. A. Johnson et al., 1997. A new mtDNA mutation showing accumulation with time and restriction to skeletal muscle. Am. J. Hum. Genet. 60: 373–380. [PMC free article] [PubMed] [Google Scholar]
- Wong, A., L. Cavelier, H. E. Collins-Schramm, M. F. Seldin, M. McGrogan et al., 2002. Differentiation-specific effects of LHON mutations introduced into neuronal NT2 cells. Hum. Mol. Genet. 11: 431–438. [DOI] [PubMed] [Google Scholar]
- Yasukawa, T., T. Suzuki, N. Ishii, S. Ohta and K. Watanabe, 2001. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 20: 4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda, M., A. Chomyn, A. Martinuzzi, O. Hurko and G. Attardi, 1992. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyopathy. Proc. Natl. Acad. Sci. USA 89: 11164–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younkin, D. P., C. M. Tang, M. Hardy, U. R. Reddy, Q. Y. Shi et al., 1993. Inducible expression of neuronal glutamate receptor channels in the NT2 human cell line. Proc. Natl. Acad. Sci. USA 90: 2174–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]