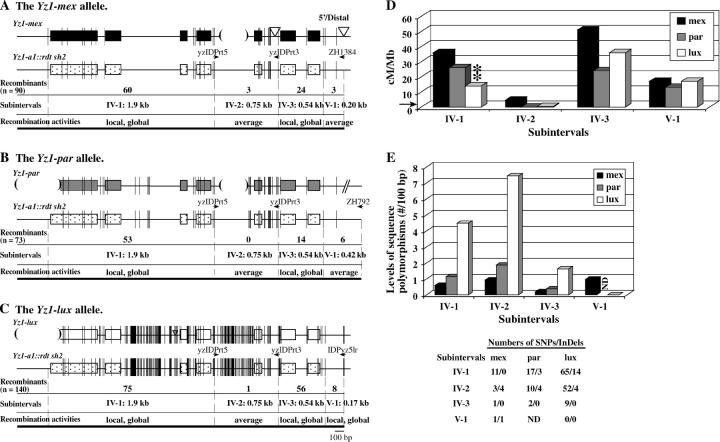
Figure 4.—
High-resolution mapping of the recombination breakpoints that resolved in the yz1 locus in the mex, par, and lux haplotypes. (A–C) Exons of the yz1 gene are shown as boxes. Short vertical lines represent sequence polymorphisms between each teosinte Yz1 allele and the Yz1 allele from the a1::rdt sh2 stock. The widths of these short vertical lines are proportional to the numbers of polymorphic nucleotides. Subintervals are defined by sequence polymorphisms. Haplotype-specific primers are indicated by horizontal arrows. The numbers of recombination breakpoints that mapped to each subinterval are shown for each haplotype. Each interval is classified as being an average recombination spot (average), a local recombination hot spot (local), or a local and global recombination hot spot (local, global; see legend of Table 3 for definitions). Large InDeLs are indicated by triangles (insertions) and parentheses (deletions). (D) Comparison of recombination rates per megabase across the yz1 locus among the mex, par, and lux haplotypes. The horizontal arrow indicates the average recombination rate per megabase of the maize genome (2.1 cM/Mb). (**) indicates that the recombination rate per megabase in the labeled haplotype at the corresponding subinterval is significantly different from the others at the 0.01 level. (E) Comparison of the levels of sequence polymorphisms (no./100 bp) at the yz1 locus among the mex, par, and lux haplotypes. Numbers of sequence polymorphisms were calculated by comparing each of the teosinte Yz1 alleles and the common Yz1 allele from the a1::rtdt sh2 stock. Numbers of SNPs/InDeLs in each of the subintervals are listed.