Abstract
The expansion of Africanized honeybees from South America to the southwestern United States in <50 years is considered one of the most spectacular biological invasions yet documented. In the American tropics, it has been shown that during their expansion Africanized honeybees have low levels of introgressed alleles from resident European populations. In the United States, it has been speculated, but not shown, that Africanized honeybees would hybridize extensively with European honeybees. Here we report a continuous 11-year study investigating temporal changes in the genetic structure of a feral population from the southern United States undergoing Africanization. Our microsatellite data showed that (1) the process of Africanization involved both maternal and paternal bidirectional gene flow between European and Africanized honeybees and (2) the panmitic European population was replaced by panmitic mixtures of A. m. scutellata and European genes within 5 years after Africanization. The post-Africanization gene pool (1998–2001) was composed of a diverse array of recombinant classes with a substantial European genetic contribution (mean 25–37%). Therefore, the resulting feral honeybee population of south Texas was best viewed as a hybrid swarm.
THE evolutionary significance of natural hybridization has been debated for decades (Mayr 1942; Anderson 1949; Harrison 1993; Arnold 1997). At one extreme, it has been argued that natural hybridization is an evolutionary dead end due to formation of inviable and/or infertile hybrids (Mayr 1942; Barton and Hewitt 1985, 1989). At the other extreme, it has been suggested that natural hybridization may lead to new evolutionary lineages due to formation of relatively fit hybrids that expand into novel habitats (Anderson 1948; Arnold 1997; Ellstrand and Schierenbeck 2000; Bleeker 2003). A third potential evolutionary outcome is expansion of an intermixed form within the resident progenitor's habitat, in which case the degree of mixing between hybridizing forms may range from formation of a hybrid swarm to genetic assimilation of one form by the other (Childs et al. 1996; Rhymer and Simberloff 1996; Perry et al. 2001). Natural hybridization and introgression have been reported in a growing number of biological invasions (Rhymer and Simberloff 1996). Such invasive events are of great interest to the discipline of evolutionary biology because they provide unique opportunities to study evolutionary processes at initial stages of secondary contact of divergent genomes.
Our study of hybridization, which deals with one of the most spectacular biological invasions yet documented, that of the Africanized honeybees (Apis mellifera L.), builds on the previous studies of Africanization. Africanized honeybees derive from a founder population of the tropical-evolved African subspecies Apis mellifera scutellata brought from South Africa into Brazil in 1956 to interbreed with previously imported temperate-evolved European honeybees. The purpose of the introduction was to create a honeybee better adapted to tropical conditions, because the European honeybee colonies were poor honey producers and would survive only when intensively managed (Nogueira-Neto 1964; Kerr 1967). Soon after the introduction, queens of A. m. scutellata were accidentally released into the natural environment (Spivak et al. 1991) and their descendants have since expanded throughout South and Central America and have established large feral populations where European colonies could not thrive (Taylor 1988; Roubik and Boreham 1990; Winston 1992). The leading edge of the expanding front reached the southern United States in 1990, only 33 years after initial release (Sugden and Williams 1990). Prior to arrival of Africanized honeybees, the United States sustained large feral and managed populations of honeybees predominantly derived from eastern (A. m. ligustica, A. m. carnica, and A. m. caucasia) and western (A. m. mellifera) European subspecies (Rubink et al. 1990; Schiff et al. 1994; Schiff and Sheppard 1995, 1996; Loper et al. 1999). With the arrival and subsequent expansion of Africanized honeybees into Texas, New Mexico, Arizona, Nevada, and California, the European honeybee gene pool in those areas is being converted to one with a substantial African component (Rubink et al. 1996; Loper et al. 1999; Pinto et al. 2004).
The underlying mechanism for dispersal and the genetic composition of Africanized honeybee populations has been intensively studied in the American tropics (Del Lama et al. 1988; Hall and Muralidharan 1989; Lobo et al. 1989; Smith et al. 1989; Hall 1990; Rinderer et al. 1991; Lobo and Krieger 1992; Quezada-Euán 2000; Clarke et al. 2002) and to a lesser degree in more temperate regions of South (Sheppard et al. 1991a; Diniz et al. 2003; Quezada-Euán et al. 2003) and North America (Loper et al. 1999; Pinto et al. 2004). Studies of neotropical feral populations suggest that Africanized honeybees expanded by maternal migration (Taylor 1988; Hall and Muralidharan 1989; Smith et al. 1989; Hall and Smith 1991) and have introgressed unequal nuclear and mitochondrial proportions of European genes (Hall 1990; Sheppard et al. 1991b; Hall and McMichael 2001; Clarke et al. 2002). Surveys of allozymes (Del Lama et al. 1988; Lobo et al. 1989; Sheppard et al. 1991b; Diniz et al. 2003), restriction fragment length polymorphisms (Hall 1990; McMichael and Hall 1996; Hall and McMichael 2001), randomly amplified polymorphic DNA (Suazo et al. 1998), and microsatellites (Clarke et al. 2002) showed low-to-intermediate levels of European alleles in the Africanized gene pool, whereas surveys of mitochondrial DNA (mtDNA) showed fixed-to-low levels of African-type mitochondria (Hall and Muralidharan 1989; Smith et al. 1989; Hall and Smith 1991; Sheppard et al. 1991b; Clarke et al. 2001; Diniz et al. 2003). The varying proportions of European markers found in different studies seem to be dependent on (1) the size of the preexisting European honeybee population; (2) the time since secondary contact; and possibly (3) the nature of the population, i.e., managed vs. feral (Hall and Muralidharan 1989; Smith et al. 1989; Rinderer et al. 1991; Clarke et al. 2002). The overall pattern of Africanization that emerged from these studies is that Africanized honeybees may hybridize extensively when (managed) European honeybees are common. However, over time European alleles largely disappear mainly because of continued migration of African genotypes into the area and possibly because hybrid workers exhibit reduced fitness (Harrison and Hall 1993; Schneider and Hall 1997; Schneider et al. 2003).
As the Africanized honeybees are expanding in southwestern United States, a hybrid zone, which potentially mirrors a hybrid zone of South America, is forming. Studies of the stable hybrid zone in South America reported increasing and persisting proportions of European genes in north-south clines. In the subtropical-temperate transitional areas, a diverse array of associations of European and African markers was found, suggesting that the breakdown of the African genome was occurring (Lobo et al. 1989; Sheppard et al. 1991a; Diniz et al. 2003). While the hybrid zone in North America has not been studied, it is possible that its dynamics have been changed by the parasitic mite Varroa destructor. Since introduction in 1987 (De Guzman et al. 1997), Varroa has caused severe losses in managed and feral European populations (Kraus and Page 1995; Loper et al. 1999; Pinto et al. 2004). Further, several studies have shown that European honeybees are more susceptible to Varroa than Africanized honeybees (Moretto et al. 1991, 1993; Message and Gonçalves 1995; Guzman-Novoa et al. 1996). Therefore, the combined effect of a depauperate population and differential susceptibility to Varroa could have changed the rate and extent of hybridization between European and Africanized honeybees.
We completed an 11-year study of mtDNA of a feral population of European honeybees from the southern United States (Texas) undergoing successive invasions of both the Africanized honeybee and Varroa. This population displayed a rapid decline in the number of colonies of honeybees possessing non-A. m. scutellata mitotypes, but not in colonies of honeybees with the A. m. scutellata mitotype (Pinto et al. 2004). In spite of the initial trend, the authors found a substantial contribution of non-A. m. scutellata mitotypes several years after both invasions, suggesting a persistence of European genes as predicted in a more temperate setting. While the finding suggested that the feral population lies within a hybrid zone, it did not rule out complete genetic assimilation of the European genome by Africanized honeybees. In the present study, nuclear DNA (microsatellites) markers were screened from the 11-year honeybee collection and used concurrently with the mtDNA to better understand the underlying mechanisms of Africanization and the genetic composition of the feral population over time. This study reports, on a fine temporal scale, the pattern of hybridization and introgression involved in this event by examining estimates of single-locus disequilibria, linkage disequilibria, cytonuclear associations, and admixture proportions. This is the first comprehensive study concurrently using nuclear and mtDNA markers of a feral population in a more temperate setting encompassing the whole invasion process.
MATERIALS AND METHODS
Sample collection:
A total of 335 honeybee workers was sampled from feral colonies between 1991 and 2001 at the Welder Wildlife Refuge (WWR), which is located at 28° N latitude, San Patricio County, Texas. Each honeybee worker represented a different and a new colony; i.e., no colonies were resampled. The 11-year collection comprised honeybees from colonies obtained from tree cavities (244 samples) and swarm traps (91 samples) (see Pinto et al. 2004 for details on sampling procedure).
Honeybee workers of two additional populations (one European derived and one A. m. scutellata derived) were sampled for use as reference populations. The European-derived population (referred to as “Eur.” throughout this article) was sampled from 50 feral colonies caught in swarm traps between 1988 and early 1990 (prior to Africanization). The swarm traps were deployed across an east-west 120-km-long transect, established in the lower Rio Grande Valley (between the cities of Brownsville and La Joya) of South Texas (Rubink et al. 1996). The A. m. scutellata-derived population from Brazil (referred to as “Braz.” throughout this article) was sampled in 2002 from 43 colonies maintained in the apiary of the University of São Paulo (Ribeirão Preto, São Paulo, Brazil). The sampled colonies, for the most part, originated as feral swarms collected in the Luiz Antonio area (∼100 km north of the initial release site of A. m. scutellata in Rio Claro). The swarms were maintained without subsequent queen management practices and therefore were considered representative of the local feral population (D. De Jong, personal communication). We assumed that the sample from South Texas was representative of the pre-Africanized WWR population. The sample from Brazil was our best representative of the A. m. scutellata-derived founder population and was regarded as the source of the migrants. The honeybees were either frozen on dry ice (samples from South Texas and WWR) or stored in 95% ethanol (samples from Brazil) and transported back to the laboratory where they were stored at −80° and 4°, respectively, until DNA extraction.
Microsatellite analysis:
Total DNA was extracted from the honeybee mesosoma (thorax plus the first fused abdominal segment) using a QIAamp DNA mini kit (QIAGEN, Valencia, CA) and stored at −20° until analyzed. A total of 428 honeybee samples were scored at 12 microsatellite loci (A14, A7, A88, A107, A113, A35, A28, A79, A43, A8, IM, and ED1; see Estoup et al. 1994, 1995; Oldroyd et al. 1995; and Rowe et al. 1997 for primer sequences). Single polymerase chain reaction (PCR) amplifications were performed in 8 μl total volume containing 0.5× Taq DNA polymerase buffer, 1.5 mm MgCl2, 0.2 mm of each dNTPs, 2 pm of each primer, 1 μl of template DNA, and 0.4 units of Taq DNA polymerase (Promega, Madison, WI) for all primer pairs. The forward primer for each marker was labeled with one of three fluorescent dyes (HEX, 6-FAM, or NED; Applied Biosystems, Foster City, CA). The PCR conditions consisted of an initial 3-min denaturation step at 94° followed by 30 cycles of 94° for 15 sec, annealing temperature at 55° (except for A79 at 60°) for 15 sec, and 72° for 5 sec, plus a final extension at 72° for 10 min. The PCR products were loaded on Long Ranger polyacrylamide gels (BioWhittaker Molecular Applications, Rockland, ME) and run on an ABI Prism 377 automated DNA sequencer using HD Rox 400 (Applied Biosystems) as the internal size standard. The software Genotyper 2.5 (Applied Biosystems) was used for allele identification and comparison.
Statistical analysis:
Genetic diversity was assessed using unbiased estimates of gene diversity (Nei 1987) and allelic richness (Petit et al. 1998) for each microsatellite locus in each sample. Allele frequencies, number of alleles (A), proportion of heterozygotes (Hp), unbiased gene diversity (Hd), and allelic richness (Rs, a measure of the number of alleles independent of sample size) per locus and population sample were computed using the FSTAT version 2.9.3. package (Goudet 2001). Differences in average unbiased gene diversity and allelic richness between pairs of samples were assessed by Wilcoxon's signed rank test (Snedecor and Cochran 1978) using SPSS version 11.0 (Norušis 1993).
Departure from Hardy-Weinberg equilibrium (HWE) was tested for each locus and population sample using the Hardy-Weinberg exact test. A global probability value over all loci was obtained for each population sample using the procedure of Raymond and Rousset (1995a). Heterozygote excess and deficiency were tested at each locus and across loci for each sample using the score test (U-test) according to Rousset and Raymond (1995). Fisher's exact tests were performed to assess genotypic linkage disequilibrium for all locus pairs within each population sample (Raymond and Rousset 1995a). Population sample pairwise comparisons were performed to test for genic (Raymond and Rousset 1995b) and genotypic (Goudet et al. 1996) differentiation across loci. These analyses were performed by the Markov chain method (Guo and Thompson 1992) with 10,000 dememorization steps, 1000 batches, and 10,000 iterations per batch using GENEPOP version 3.4 (Raymond and Rousset 1995a).
Reference and WWR population sample pairwise values for FST were computed according to Weir and Cockerham (1984) using GENEPOP and then departures from zero were tested by 1000 permutations using GENETIX version 4.04 (Belkhir et al. 2002). Distance measures, termed proportion of shared alleles (DPS, Bowcock et al. 1994), between pairs of populations were generated using the program MSAT. These distances were used to build a neighbor-joining tree (Saitou and Nei 1987) using PAUP version 4.0b 10 (Swofford 2002). Where applicable throughout the analysis, statistical significance levels were adjusted for multiple comparisons using the sequential Bonferroni procedure to correct for type I error (Rice 1989).
The European and Brazilian reference population samples were used as parentals to compute admixture proportions in the WWR population using the estimators mR and mY. The least-squares estimator mR is based on a comparison of allele frequencies between parental and admixed populations and assumes identity by descent (Roberts and Hiorns 1965). The estimator mY is based on a coalescent approach that takes into account molecular information as well as allele frequencies. This estimator, which assumes the single stepwise mutation model, was computed using the squared difference in allele size as molecular distance. Both estimators were computed using the program ADMIX version 1.0. The bootstrap average and bootstrap standard deviation of the admixture coefficients were estimated over 1000 replications (Bertorelle and Excoffier 1998). Differences in mR and mY over time were assessed by a paired t-test using SPSS.
The European and Brazilian reference samples were also used as parentals to simulate 100 sets of F1, European backcross, and Brazilian backcross population samples of 50 individuals each. The multilocus genotype of each simulated individual was generated by randomly taking one allele per locus from each of the parental populations, according to their frequencies, to form the respective “gametes” using a program written by J. B. Patton (personal communication). The simulated F1 and reciprocal backcrosses were used as additional reference samples to build the above-mentioned neighbor-joining tree and to classify individual honeybees from WWR over time using GENECLASS (Cornuet et al. 1999), a software package that assigns individuals of unknown origin to a set of reference samples using either likelihood (frequency or Bayesian) or distance-based methods. To compare the performance of frequency, Bayesian, Cavalli-Sforza and Edwards (1967) chord distance, and (δμ)2 Goldstein et al. (1995) distance methods, multilocus genotypes of 1000 European, 1000 Brazilian, 1000 F1, 1000 European backcross, and 1000 Brazilian backcross individuals were randomly generated by crossing European × European, Brazilian × Brazilian, European × Brazilian, F1 × European, and F1 × Brazilian, respectively. Using one random set (among 100) of reference samples, each simulated individual was assigned and the percentage of misclassification was calculated for each method. The most accurate method was examined over all 100 sets of simulated reference samples and then used to classify individual honeybees from the WWR. Assignment scores for each WWR honeybee were computed 100 times, using the 100 sets of simulated reference samples. Classification of individual honeybees was based on the predominant assignment given for the 100 replications.
RESULTS
Genetic diversity:
Allele frequencies, number of alleles, allelic richness, proportion of heterozygotes, and unbiased gene diversity are given for each locus and population sample in supplementary data at http://www.genetics.org/supplemental/. All loci showed high variability with the total number of alleles ranging from 11 (A8) to 38 (A7), the mean number of alleles ranging from 5.8 (WWR 1991) to 15.1 (WWR 1995), the mean allelic richness ranging from 5.833 (WWR 1991) to 9.168 (WWR 2000), the mean proportion of heterozygotes ranging from 0.700 (WWR 1994) to 0.876 (WWR 1999), and finally the mean gene diversity ranging from 0.681 (WWR 1991) to 0.874 (Brazilian) across all population samples.
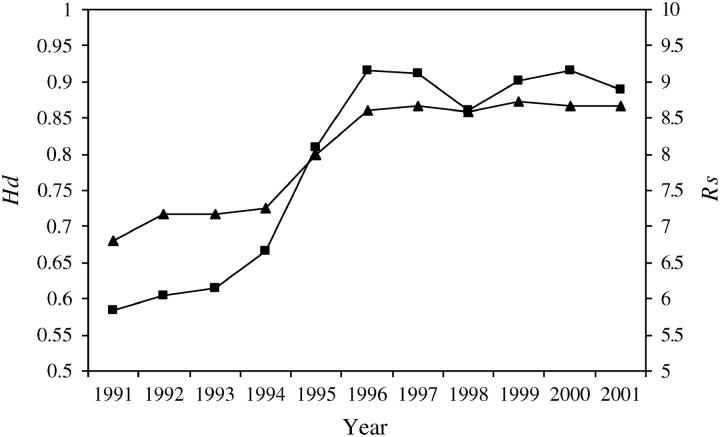
Genetic diversity was significantly higher in the Brazilian reference sample (Rs = 8.890; Hd = 0.874) than in the European (Rs = 5.934; Hd = 0.693) reference sample (P = 0.002 for both allelic richness and gene diversity). Pairwise comparisons between the reference and WWR samples showed, first, that mean allelic richness and mean gene diversity were significantly lower in the European (Rs = 5.934; Hd = 0.693) than in the WWR after 1994 (Rs = 6.653–9.168) and 1995 (Hd = 0.800–0.874), respectively (P ≤ 0.005); and, second, that mean allelic richness and mean gene diversity were significantly higher in the Brazilian (Rs = 8.890; Hd = 0.874) than in the WWR (P ≤ 0.006) prior to 1994 (Rs = 5.833–6.653) and 1995 (Hd = 0.681–0.800), respectively. No significant differences in genetic diversity were found for the remaining reference-WWR sample pairwise comparisons (P ≥ 0.060). During the 11-year study, the WWR population experienced a striking increase in gene diversity and allelic richness, with the greatest increment observed between 1994 and 1996 (Figure 1).
Figure 1.—
Temporal pattern of unbiased gene diversity Hd (▴) and allelic richness RS (▪) in the feral honeybee population of the WWR.
Hardy-Weinberg equilibrium and genotypic linkage equilibrium:
FIS estimates and respective significance, P-values for HWE, P-values for heterozygote deficiency, and P-values for heterozygote excess and their standard errors per locus are shown in supplementary data at http://www.genetics.org/supplemental/ and per population sample in Table 1. In the reference European sample, no significant departures from HWE were detected for any locus and across loci (0.085 ≤ P ≤ 0.963). Individual and multilocus U-tests for heterozygote deficiency and heterozygote excess yielded a single significant value at A107 (P = 0.036 for heterozygote excess), which became nonsignificant following sequential Bonferroni correction. In contrast, the Brazilian reference sample showed a deviation from HWE (P = 0.013), caused by a deficiency of heterozygotes (P = 0.001) across all loci. Single-locus P-values obtained with Fisher's exact test, U-tests, and/or FIS revealed a deficiency of heterozygotes at A88, A14, A35, and A79 and an excess of heterozygotes at ED1 in this reference population sample (supplementary data at http://www.genetics.org/supplemental/). However, when applying sequential Bonferroni correction, only A35 and A79 exhibited a significant deficiency of heterozygotes (P = 0.000). Fisher's exact tests for genotypic linkage disequilibrium between loci pairs produced eight (four for the European and four for the Brazilian reference sample) significant P-values from 132 comparisons. None of the loci pairs were shared by the two populations and none of the P-values was significant following sequential Bonferroni's correction.
TABLE 1.
Exact test probability of HWE,U-test probability of heterozygote deficiency and heterozygote excess, and respective standard errors performed across loci
| Braz. | Eur. | 1991 | 1992 | 1993 | 1994 | 1995 | 1996 | 1997 | 1998 | 1999 | 2000 | 2001 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P (HWE) | 0.013 | 0.562 | 0.994 | 0.190 | 0.059 | 0.212 | 0.003 | 0.002 | 0.109 | 0.748 | 0.142 | 0.005 | 0.371 |
| P (ht. def.) | 0.001 | 0.907 | 0.971 | 0.368 | 0.457 | 0.000 | 0.003 | 0.007 | 0.000 | 0.156 | 0.246 | 0.000 | 0.041 |
| ±SE | ±0.000 | ±0.002 | ±0.000 | ±0.002 | ±0.003 | ±0.000 | ±0.000 | ±0.000 | ±0.000 | ±0.002 | ±0.004 | ±0.000 | ±0.001 |
| P (ht. exc.) | 0.999 | 0.091 | 0.029 | 0.625 | 0.545 | 1.000 | 0.997 | 0.992 | 1.000 | 0.842 | 0.765 | 1.000 | 0.960 |
| ±SE | ±0.000 | ±0.002 | ±0.000 | ±0.002 | ±0.003 | ±0.000 | ±0.000 | ±0.000 | ±0.000 | ±0.002 | ±0.004 | ±0.000 | ±0.001 |
P (HWE), exact test probability of HWE; P (ht. def.), U-test probability of heterozygote deficiency; P (ht. exc.), U-test probability of heterozygote excess; SE, standard error. Braz. and Eur. refer to Brazilian and European reference population samples, respectively. The years 1991–2001 refer to the temporal WWR population samples. P-values in italics are significant following sequential Bonferroni correction (α = 0.05, k = 2 for the reference population samples, k = 11 for the reference WWR population samples).
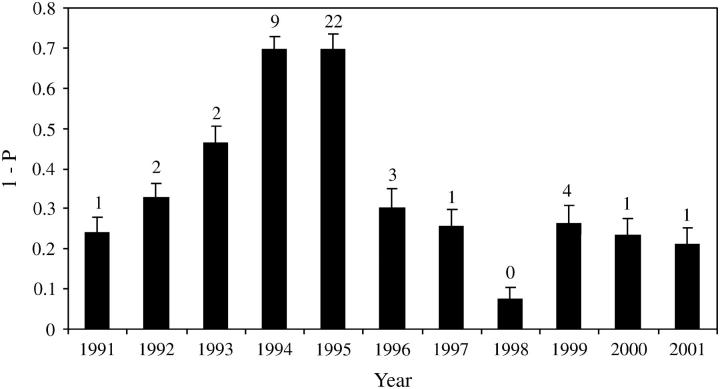
In the WWR population, significant departures from HWE across all loci were detected in 1995, 1996, and 2000 (0.002 ≤ P ≤ 0.005; Table 1). These deviations were caused by a deficiency of heterozygotes (0.0001 ≤ P ≤ 0.007). In addition to those years, multilocus U-tests revealed a deficiency of heterozygotes in 1994, 1997, and 2001 (0.000 ≤ P ≤ 0.041) and an excess of heterozygotes in 1991 (P = 0.029). However, when applying the sequential Bonferroni procedure, neither 1991 nor 2001 WWR population samples deviated from HWE. At the single-locus level, departures from HWE occurred at all loci except ED1 for at least one of the sampling years, as indicated by Fisher's exact test, U-tests, and/or FIS P-values (supplementarydataathttp://www.genetics.org/supplemental). The greatest number of significant P-values was observed in 1995 (8 values of P < 0.05 given by Fisher's exact test, U-tests, and FIS) and 1996 (8 values of P < 0.05 given by Fisher's exact test, U-tests, and FIS). The majority of the 16 P-values were marginally significant and subsequently became nonsignificant when corrected for type I error. Across all years, locus A79 exhibited the most consistent deficiency of heterozygotes, which was detected by Fisher's exact test, U-test, and/or FIS in 1995, 1996, 1997, 1999, 2000, and 2001. The hypothesis of null alleles at this locus was dismissed because no heterozygote deficiency was detected in the reference European and early WWR population samples. However, it was detected in the reference Brazilian. Because deficiency of heterozygotes seems to be correlated with Africanization, this locus deserves further investigation. Fisher's exact tests for linkage disequilibrium yielded 46 significant values from 704 comparisons (35 significant values are expected at the 5% level). The most significant values were in the 1995 (22) and 1994 (9) population samples (Figure 2). When applying sequential Bonferroni correction, only 5 (1 in 1994 and 4 in 1995) of the 46 above-mentioned P-values were significant. Two pairs of loci, A28/ED1 (mapping distance 23 cM) and A7/A14 (mapping distance 22 cM), are genetically linked, while the eight remaining loci are unlinked (M. Solignac, personal communication). However, all 12 loci behaved similarly and no single-locus pair produced consistently significant values for all years after population mixing. Across the 11-year study, the mean P-value was lowest in 1994 and 1995. P-values obtained from 1996 onward suggested a rapid dissipation of linkage disequilibrium (Figure 2).
Figure 2.—
Temporal pattern of genotypic linkage disequilibrium among 12 microsatellite loci in the feral honeybee population of the WWR. 1-P (where P is the average value of Fisher's exact tests for each locus pair) and their standard errors are given for each sampling year. The number of significant P-values (not corrected for type I error), provided by Fisher's exact tests for each locus pair, is shown above the error bars for each sampling year.
Genetic differentiation:
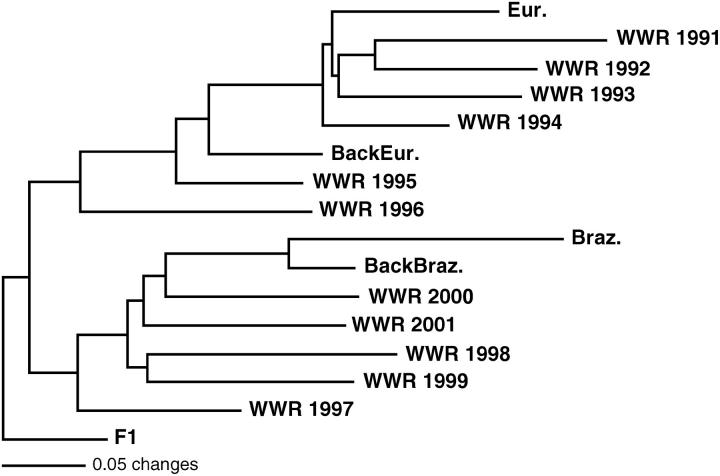
The multilocus distance measure (DPS), multilocus FST, and tests for homogeneity of genic and genotypic distributions (genic and genotypic differentiation) showed a high level of differentiation between Brazilian and European reference samples (P < 0.000 for genic and genotypic differentiation and FST; Tables 2 and 3). Pairwise comparisons between reference European and WWR samples revealed an increase in genic and genotypic differentiation over time, with FST values becoming significantly different from zero since 1995. In contrast, pairwise comparisons between reference Brazilian and WWR population samples showed a decrease in differentiation over time. In spite of the trend, all pairwise comparisons yielded significant values, suggesting that the WWR population was genetically distinct from the Brazilian. Values of DPS, FST, and genic and genotypic differentiation indicated that the WWR population samples could be divided into pre- and post-1996 populations. Between 1991 and 1994 pairwise comparisons of genic and genotypic differentiation were nonsignificant, and pairwise multilocus FST values were not significantly different from zero. The same pattern was shown by pairwise comparisons performed between 1997 and 2001 samples. In contrast, pre-1996 vs. post-1996 sample comparisons showed a high level of differentiation, suggesting that a profound genetic change occurred in a short time frame. A neighbor-joining tree confirmed this pattern (Figure 3). Until 1996, the WWR samples showed closer genetic affinities with the reference European sample. After 1996, the WWR population samples were clustered closer to the reference Brazilian population.
TABLE 2.
P-values for genotypic (above diagonal) and genic (below diagonal) differentiation
| Braz. | Eur. | 1991 | 1992 | 1993 | 1994 | 1995 | 1996 | 1997 | 1998 | 1999 | 2000 | 2001 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Braz. | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |
| Eur. | 0.0000 | 0.2601 | 0.0039 | 0.0005 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |
| 1991 | 0.0000 | 0.2204 | 0.3751 | 0.0182 | 0.8989 | 0.2523 | 0.0011 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |
| 1992 | 0.0000 | 0.0028 | 0.4668 | 0.1819 | 0.7963 | 0.0094 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |
| 1993 | 0.0000 | 0.0003 | 0.1010 | 0.1418 | 0.8130 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |
| 1994 | 0.0000 | 0.0001 | 0.9297 | 0.8414 | 0.8660 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |
| 1995 | 0.0000 | 0.0000 | 0.8650 | 0.1644 | 0.0045 | 0.0000 | 0.0282 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |
| 1996 | 0.0000 | 0.0000 | 0.0156 | 0.0000 | 0.0000 | 0.0000 | 0.0005 | 0.3519 | 0.0018 | 0.0001 | 0.0262 | 0.0063 | |
| 1997 | 0.0000 | 0.0000 | 0.0004 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.1379 | 0.1131 | 0.0911 | 0.4959 | 0.0219 | |
| 1998 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0008 | 0.1930 | 0.2181 | 0.3220 | 0.1723 | |
| 1999 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0501 | 0.4035 | 0.5132 | 0.0522 | |
| 2000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0046 | 0.1776 | 0.3057 | 0.4566 | 0.5245 | |
| 2001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0012 | 0.0079 | 0.1371 | 0.0314 | 0.3170 |
P-values were calculated between pairs of reference (Braz., Africanized from Brazil; Eur., European from South Texas) and WWR (from 1991 to 2001) honeybee population samples. P-values in italics are significant following sequential Bonferroni correction (α = 0.05, k = 78).
TABLE 3.
Proportion of shared-allele distance (above diagonal) and multilocus estimates ofFST (below diagonal)
| Braz. | Eur. | 1991 | 1992 | 1993 | 1994 | 1995 | 1996 | 1997 | 1998 | 1999 | 2000 | 2001 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Braz. | 0.6670 | 0.6900 | 0.6820 | 0.6520 | 0.6230 | 0.4960 | 0.4850 | 0.3940 | 0.4250 | 0.3660 | 0.3280 | 0.3540 | |
| Eur. | 0.1405 | 0.2570 | 0.2340 | 0.2220 | 0.1780 | 0.2780 | 0.4060 | 0.4040 | 0.4910 | 0.4960 | 0.5170 | 0.4850 | |
| 1991 | 0.1319 | 0.0060 | 0.2410 | 0.2890 | 0.2600 | 0.3390 | 0.4460 | 0.4770 | 0.5670 | 0.5550 | 0.5510 | 0.5450 | |
| 1992 | 0.1176 | 0.0079 | −0.0014 | 0.2210 | 0.2020 | 0.2830 | 0.4100 | 0.4370 | 0.5080 | 0.5100 | 0.5080 | 0.4930 | |
| 1993 | 0.1213 | 0.0088 | 0.0148 | 0.0025 | 0.2010 | 0.2830 | 0.3950 | 0.4470 | 0.4940 | 0.5070 | 0.5010 | 0.4760 | |
| 1994 | 0.1225 | 0.0053 | 0.0040 | 0.0009 | 0.0017 | 0.2160 | 0.3460 | 0.3740 | 0.4710 | 0.4660 | 0.4700 | 0.4470 | |
| 1995 | 0.0719 | 0.0181 | 0.0148 | 0.0068 | 0.0118 | 0.0079 | 0.2920 | 0.2900 | 0.4020 | 0.3610 | 0.3580 | 0.3550 | |
| 1996 | 0.0454 | 0.0420 | 0.0397 | 0.0310 | 0.0326 | 0.0273 | 0.0071 | 0.3090 | 0.3850 | 0.3630 | 0.3440 | 0.3360 | |
| 1997 | 0.0277 | 0.0561 | 0.0549 | 0.0464 | 0.0548 | 0.0472 | 0.0191 | 0.0025 | 0.2940 | 0.2610 | 0.2570 | 0.2810 | |
| 1998 | 0.0284 | 0.0768 | 0.0796 | 0.0657 | 0.0692 | 0.0686 | 0.0322 | 0.0104 | 0.0001 | 0.2770 | 0.3090 | 0.3220 | |
| 1999 | 0.0199 | 0.0793 | 0.0773 | 0.0654 | 0.0690 | 0.0686 | 0.0304 | 0.0127 | 0.0018 | −0.0028 | 0.2460 | 0.2730 | |
| 2000 | 0.0142 | 0.0866 | 0.0811 | 0.0649 | 0.0726 | 0.0722 | 0.0309 | 0.0116 | 0.0047 | 0.0006 | −0.0006 | 0.2570 | |
| 2001 | 0.0182 | 0.0755 | 0.0776 | 0.0620 | 0.0630 | 0.0617 | 0.0264 | 0.0089 | 0.0038 | 0.0030 | 0.0025 | −0.0011 |
Values of proportion of shared-allele distance and FST were calculated for pairs of reference (Braz., Africanized from Brazil; Eur., European from South Texas) and WWR (from 1991 to 2001) honeybee population samples. FST values in italics are significantly different from zero following sequential Bonferroni correction (α = 0.05, k = 78).
Figure 3.—
Rooted neighbor-joining tree constructed from proportion of shared-alleles distance for reference and temporal WWR population samples. The reference population samples are European (Eur.), Brazilian (Braz.), simulated backcross with European (BackEur.), simulated F1, and simulated backcross with Brazilian (BackBraz.). Each simulated reference sample comprises 100 randomly generated individuals.
Admixture proportions:
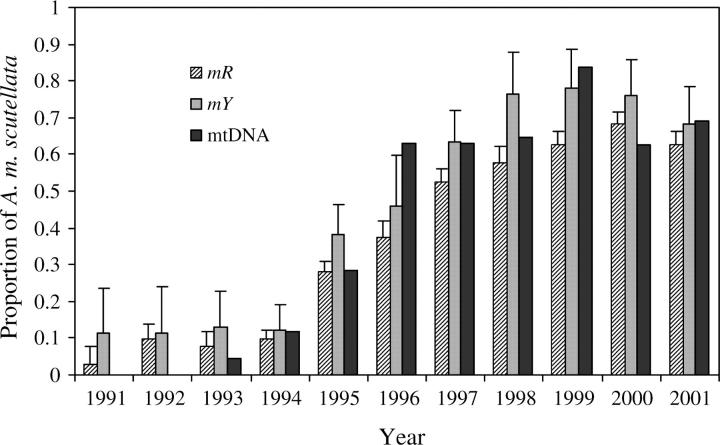
Both mR and mY estimators revealed a striking change in the admixture proportions of the WWR population over time (Figure 4). For all years, mY estimated a greater contribution of the parental Brazilian to the WWR population than did mR (P = 0.000). The proportion of A. m. scutellata introgressed alleles in the WWR population increased from 2 to 11% in 1991 to 63–68% in 2001 with mR and mY estimators, respectively. The steepest increase in the proportion of introgressed A. m. scutellata alleles occurred between 1994 (mR = 9.6%; mY = 12.1%) and 1997 (mR = 52.5%; mY = 63.4%). From 1998 on, the nuclear proportion of A. m. scutellata genes in the WWR population appeared to have stabilized at 62.8% ± 0.042 (SD) for mR and 74.7 ± 0.043 (SD) for mY. Interestingly, as shown in Figure 4, the temporal pattern of introgression of nuclear genes was paralleled by A. m. scutellata mitotype (Pinto et al. 2004).
Figure 4.—
Proportions of the A. m. scutellata mitotype and nuclear alleles in the WWR population over time. Nuclear admixture bootstrap average and standard deviation were computed for 1000 iterations using the gene-frequency-based estimator mR and the gene-frequency and molecular-based estimator mY.
Associations of nuclear and mtDNA markers:
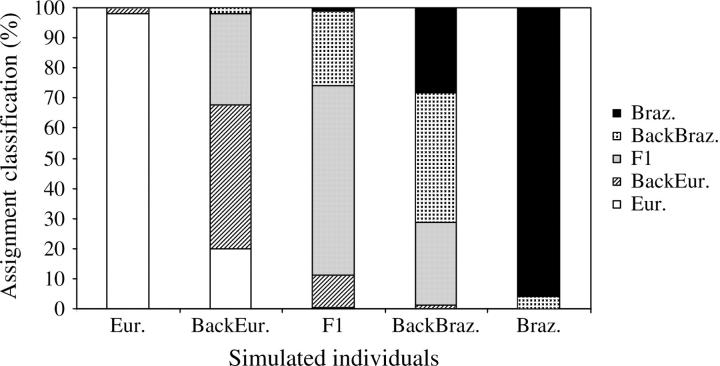
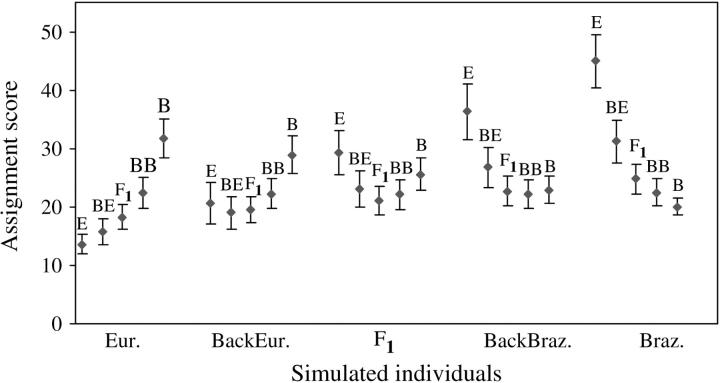
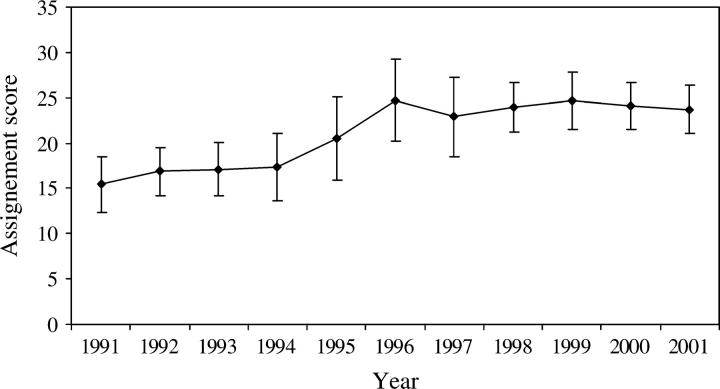
Among the assignment methods that were tested in GENECLASS, Bayesian showed the best performance with the highest rate (68.3%) of individuals correctly assigned (Table 4). Cornuet et al. (1999) also found this method to be the most accurate even when the assumptions of HWE and genotypic linkage equilibrium were violated. The percentage of misclassification given by the Bayesian method (and the other methods tested; data not shown) varied across types of simulated individuals. More than 96% of the simulated European and Brazilian individuals were correctly assigned whereas <50% of backcross individuals were correctly assigned (Figure 5). The average assignment scores (values produced by the Bayesian methodology of Rannala and Mountain 1997 as modified in GENECLASS), which were obtained for simulated individuals, are shown in Figure 6. The highest assignment scores were produced by reciprocal backcross (19.04 ± 2.77 for European backcross; 22.15 ± 2.39 for Brazilian backcross) and F1 (21.11 ± 2.45) individuals whereas the lowest were produced by European (13.64 ± 1.75) and Brazilian (20.03 ± 1.49) individuals. As crosses became more complex, the overlap among scores increased and the power of the assignment test decreased (Figure 6). The average assignment score obtained for the WWR population increased steeply between 1994 and 1996 (Figure 7), which suggests an increase in hybrid complexity of multilocus genotypes. After 1997, the average assignment score stabilized at high values. Comparisons of ranges of the assignment scores for simulated (Figure 6) and WWR individuals (Figure 7) demonstrate that crosses were characteristically more complex than simple backcrosses.
TABLE 4.
Percentage of individuals misclassified using different assignment methods
| Assignment method | % misclassification |
|---|---|
| Likelihood | |
| Bayesiana | 31.7 |
| Frequencyb | 34.4 |
| Distance | |
| Cavalli-Sforzac | 48.8 |
| Goldsteind | 66.2 |
Modified by Cornuet et al. (1999) from Rannala and Mountain (1997).
Modified by Cornuet et al. (1999) from Paetkau et al. (1995).
Modified by Cornuet et al. (1999) from Cavalli-Sforza and Edwards (1967) chord distance.
(δμ)2 distance of Goldstein et al. (1995).
Figure 5.—
Proportion of simulated individuals assigned to the reference European (Eur.), Brazilian (Braz.), simulated European backcross (BackEur.), simulated F1, and simulated Brazilian backcross (BackBraz.) using the Bayesian method. Multilocus genotypes of European, Brazilian, F1, European backcross, and Brazilian backcross individuals (1000 of each type) were simulated by crossing European × European, Brazilian × Brazilian, European × Brazilian, F1 × European, and F1 × Brazilian, respectively. Each set of 1000 simulated individuals was then assigned for 100 sets of simulated reference samples.
Figure 6.—
Average assignment scores and their standard deviations for simulated European (Eur.), European backcross (BackEur.), F1, Brazilian backcross (BackBraz.), and Brazilian (Braz.) individuals. Assignment scores, given for each reference sample (E, European; BE, backcross with European; F1; BB, backcross with Brazilian; B, Brazilian), were produced by the Bayesian method. Each individual score was computed for 100 sets of simulated reference samples.
Figure 7.—
Average assignment scores and their standard deviations for the temporal WWR population sample. Using the Bayesian method, individual honeybees were assigned to one of the five reference samples (European, Brazilian, simulated European backcross, simulated F1, and simulated Brazilian backcross). For each individual honeybee, the average value was obtained from 100 data sets of the simulated reference samples of 50 individuals each.
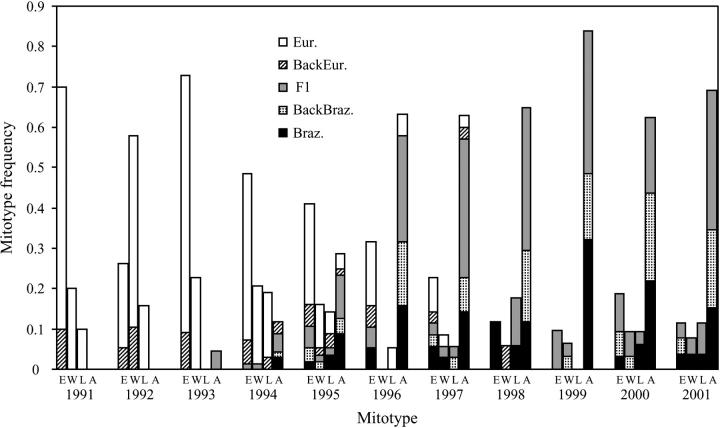
Figure 8 shows the temporal frequency of non-A. m. scutellata (eastern European, western European, and A. m. lamarckii; Pinto et al. 2004) and A. m. scutellata mitotypes and corresponding nuclear assignment classes (European, Brazilian, F1, European backcross, or Brazilian backcross). The proportion of individuals of eastern European, western European, A. m. lamarckii, and A. m. scutellata mitotypes that were genotypically closer to European, Brazilian, F1, European backcross, or Brazilian backcross reference population changed radically in WWR over time. Prior to 1994 most individuals exhibited a non-A. m. scutellata mitotype and were classified as either European or (less frequently) European backcrosses. Between 1994 and 1996, individuals of non-A. m. scutellata maternal descent were genotypically more homogeneous than those with A. m. scutellata mitotype and an association between nuclear and mtDNA genes of non-A. m. scutellata origin was apparent (Figure 8). Significant heterogeneity in genic and genotypic distributions between both groups of mitotypes was found in that 3-year period (0.000 ≤ P ≤ 0.005; the three non-A. m. scutellata mitotypes were pooled to increase sample size for added statistical power). However, a breakdown of non-A. m. scutellata cytonuclear associations appeared to have occurred as no significant differences in genic and genotypic distributions were found from 1997 onward (0.117 ≤ P ≤ 0.932; the three non-A. m. scutellata mitotypes were pooled). After 1997, most individuals exhibited a mixed ancestry with no observed association between mitotype and nuclear composition.
Figure 8.—
Associations of nuclear and mitochondrial markers. Proportion of WWR honeybees assigned to the reference European (Eur.), Brazilian (Braz.), simulated backcross with European (BackEur.), simulated F1, and simulated backcross with Brazilian (BackBraz.) per mitotype (E, eastern European; W, western European; L, A. m. lamarckii; A, A. m. scutellata) across time. Bayesian assignment classification was computed for 100 replications of simulated population samples. The assignment of each individual was based on the predominant assignment given for the 100 replications. The simulated reference samples represent a subset of a vast array of possible recombinant classes. It should be noted that individuals possessing recombinant genotypes rather than F1 and reciprocal backcrosses would be assigned to one of the five reference populations as the Bayesian method always designates a population of origin.
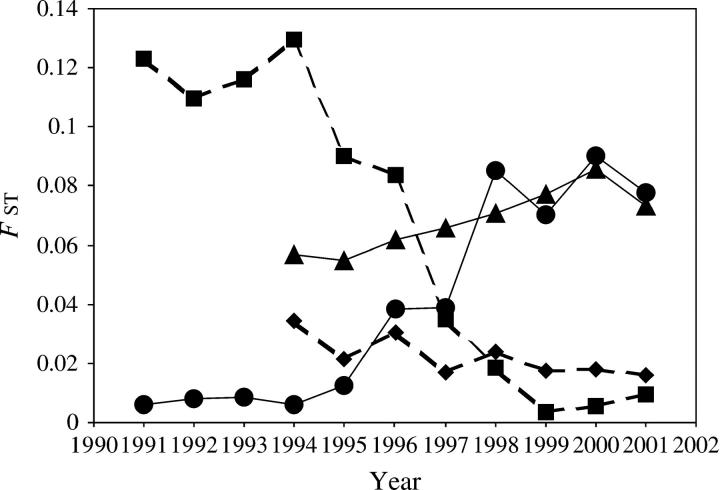
While Figure 8 allows a detailed look at mitotype by nuclear genome interactions, it does not allow a concise summary statistic of those interactions. Therefore, we generated Figure 9 using the multilocus FST values of the genomic composition of the WWR honeybees characterized by possession of either the A. m. scutellata or the non-A. m. scutellata mitotype by year as compared to the two reference populations. The nuclear genomes of individuals possessing either the A. m. scutellata or the non-A. m. scutellata mitotype began to merge at least as early as 1995. By 1997 the nuclear genomic composition associated with the respective mitotypes had converged to a point where the multilocus test for both genic and genotypic distributions was nonsignificant as previously discussed. Multilocus FST values for comparisons of the years from 1998 to 2001 virtually overlay one another, indicating the nonexistence of cytonuclear associations (Figure 9).
Figure 9.—
Temporal pattern of multilocus FST values. The FST values were computed between the reference Brazilian (Braz) and European (Eur.) population samples and the WWR subsamples separated by mitotype (A. m. scutellata vs. non-A. m. scutellata). It should be noted that no FST value was calculated for 1993 because the A. m. scutellata mitotype subsample was composed of a single individual. (▪) Braz. vs. WWR with non-A. m. scutellata mitotype, (♦) Braz. vs. WWR with A. m. scutellata mitotype, (•) Eur. vs. WWR with non-A. m. scutellata mitotype, (▴) Eur. vs. WWR with A. m. scutellata mitotype.
DISCUSSION
Mechanism of Africanization:
Initial Africanization:
mtDNA data revealed the year 1993 as the time of Africanization onset in the WWR population (Pinto et al. 2004), whereas estimates of nuclear racial admixture indicated a contribution of A. m. scutellata alleles beginning in 1991 (Figure 4). This result suggests that Africanization could have started prior to 1993 through matings of migrant drones with resident European queens. Alternatively, the early evidence of admixture could be an artifact of the analysis because for 1991 the error bars for the estimated admixture proportion, as calculated by both mR and mY, included zero. Hunter et al. (1993) documented the presence of morphologically defined Africanized honeybees in a county adjacent to San Patricio county, where WWR is located, in 1991. Therefore, it is possible that Africanized drones may have hybridized with European queens, which established colonies at WWR as early as 1991. In 1992 the error bars around the mR estimator do not overlap with zero, whereas those of the mY estimator do. While the results are not entirely statistically significant, the observation of 14 swarms of Africanized honeybees in San Patricio county in 1992 (Hunter et al. 1993) supports the contention that paternal gene flow did occur and was the likely cause for the African component of the admixture proportions. Additional support is given by Figure 8, which shows an excess of individuals assigned as European backcrosses for both 1991 and 1992 relative to the simulations to estimate the frequency of misassignment illustrated in Figure 5 for the European reference population. To conclude, the body of evidence suggests that Africanization in WWR started by matings of Africanized drones rather than by immigration of Africanized queens.
In the neotropics, genetic studies show that in feral populations maternal gene flow was the primary driving force of Africanization (Taylor 1988; Hall and Muralidharan 1989; Smith et al. 1989; Hall and Smith 1991), whereas in managed populations the primary force was paternal gene flow (Rinderer et al. 1991; Clarke et al. 2002). In contrast with the neotropics, where a virtually “pure African” expanding front was observed, migrants arriving at WWR were of mixed ancestry (Figure 8). This had the effect of diluting the input of the African nuclear loci relative to the African mtDNA, which could account for maintenance of Hardy-Weinberg equilibrium prior to 1994. Regardless of the mechanism of initial Africanization, the most dramatic genetic change of the WWR population occurred after the arrival of the Africanized queen in 1993.
Turnover in the population:
A 4-year period of Hardy-Weinberg disequilibrium due to deficiency of heterozygotes was observed in WWR between 1994 and 1997 (Table 1). Hardy-Weinberg disequilibrium was anticipated upon arrival of migrants as mixing of distinct gene pools prior to reproduction would cause a Wahlund effect. While the Wahlund effect is expected to disappear after one generation of random mating, the persistence of heterozygote deficiency for 4 years could have been due to: (1) positive assortative mating, (2) continuing influx of genetically distinct migrants, and/or (3) selection. In early reports of Africanization, positive assortative mating, due to asynchrony in mating flight times, was suggested as an explanation for apparent partial reproductive isolation between European and Africanized honeybees (reviewed in Michener 1975). This hypothesis has long been disregarded because findings of associations between European and African mitotypes with a range of morphological and allozymic phenotypes demonstrated interbreeding between the honeybee types (Rinderer et al. 1991; Sheppard et al. 1991a). The patterns depicted in Figures 8 and 9 clearly show bidirectional gene flow in the WWR population. As a result of gene exchange, associations between non-A. m. scutellata mitotypes and European nuclear classification gradually decayed and by 1997 there were no nuclear multilocus genotypic and genic differences between non-A. m. scutellata and A. m. scutellata mitotypes (P ≥ 0.117).
The A. m. scutellata mitotype increased in frequency between 1993 and 1995 from 4.5 to 28.6% (Figure 4). This could have been due to in situ reproduction of recently arrived migrants and immigration of additional Africanized swarms that further contributed to heterozygote deficiency. By 1997 the genetic composition of the population had become predominantly A. m. scutellata. The turnover resulted primarily from a dramatic reduction in the number of colonies of non-A. m. scutellata matrilines that was not observed for colonies of A. m. scutellata matrilines (Pinto et al. 2004). The authors attributed this differential loss to the parasitic mite V. destructor, which was first detected in WWR in early 1995. The observed mortality was consistent with the observation that European honeybees are more susceptible to Varroa than Africanized honeybees (Moretto et al. 1991, 1993; Message and Gonçalves 1995; Guzman-Novoa et al. 1996). In a crossbreeding experiment between Africanized and European honeybees Guzman-Novoa et al. (1996) found that (1) susceptibility to becoming infested by Varroa was least likely in the brood of Africanized honeybees, followed by European, F1 of European mother, and F1 of Africanized mother, respectively, and (2) adult European honeybees were more likely to become infested with Varroa than were adult Africanized and hybrid honeybees. In conclusion, because of the differential susceptibility of honeybees to Varroa and because the WWR population was hit at an early stage of Africanization, when European and likely F1 individuals were abundant, we suggest that differential selection pressures, possibly combined with the ongoing influx of migrants and some assortative mating, contributed to the observed disequilibrium throughout 1996 and 1997.
The temporal pattern of linkage disequilibrium experienced by the WWR population, as given by the average P-values of Fisher's exact test, is shown in Figure 2. Contrary to expectations, linkage disequilibrium decayed more rapidly than heterozygote deficiency. This circumstance is even more striking, given that two pairs of loci are genetically linked (A28/ED1 and A7/A14; M. Solignac, personal communication). Typically, more generations are required to reach linkage equilibrium than to reach Hardy-Weinberg equilibrium. In addition, both equilibria are approached at a slower pace when mating is assortative (Hartl and Clark 1997).
It is possible that the rapid dissipation of linkage disequilibrium (LD) results from Fisher's exact test for LD being less powerful than the U-test for heterozygote deficiency in detecting small deviations from expectations. Reduced statistical power for the Fisher's exact tests is not unexpected, given that European and A. m. scutellata subspecies share relatively common alleles at several loci (supplementary data at http://www.genetics.org/supplemental/; Estoup et al. 1995; Clarke et al. 2002) and given the moderate sample size (especially from 1996 onward), together with high levels of polymorphism. Nonetheless, given that Hardy-Weinberg proportions were reached in 1998 and linkage disequilibrium decayed even more quickly (Figure 2), heterozygotes (intermating between migrants and resident honeybees) must have been formed in significant numbers.
Mechanism of population turnover:
In the American tropics, feral colonies of Africanized honeybees that exhibited little or no introgression of European genes are reported to have built up to high densities within 2 to 3 years after initial colonization (Taylor 1985). It has been suggested that rapid expansion of a nearly “pure” neotropical African population has resulted from (1) high fitness of Africanized honeybees exhibiting virtually no introgression of European alleles, (2) reduced competition from a small and poorly adapted European population, and (3) cytonuclear incompatibility in hybrids of European mothers (Taylor 1985, 1988; Rinderer et al. 1991; Sheppard et al. 1991a; Harrison and Hall 1993; Schneider et al. 2003).
The WWR, which lies near the subtropical-temperate interface, supported a high density of European colonies at the arrival of the first wave of Africanized swarms (11.7 colonies/km2 in 1995; Baum 2003). Therefore, our prediction was that rate and extent of Africanization in the WWR population would be slower and lower than in neotropical populations. We expected (1) increased fitness of European honeybees under these more temperate climatical and ecological conditions, (2) higher frequency of matings with European honeybees due to the larger resident population size, and (3) strong competition of feral European colonies with the Africanized colonies for the density-limited food and nest resources. Five years after the initial observation of honeybees with African mitotypes at WWR, “pure” European honeybees were replaced by Africanized honeybees exhibiting varying degrees of introgressive hybridization (Figure 8). This level of hybridization approached the nuclear composition of the honeybees of Yucatan 13 years after the onset of Africanization (Rinderer et al. 1991; Quezada-Euán et al. 1996; Clarke et al. 2002). Complete and rapid replacement of resident European honeybees by Africanized honeybees in WWR would be expected if the latter exhibited higher fitness than the former. Whether European honeybees were equally or better fit than Africanized honeybees in the ecological and climatical conditions of southern Texas may never be known. Regardless of the relative fitnesses in a Varroa-free environment, Africanized and hybrid honeybees were likely more successful in the WWR environment that contained Varroa (Guzman-Novoa et al. 1996). This appears the most logical explanation for the unexpectedly steep increase in A. m. scutellata genetic proportions observed between 1996 and 1999 (Figure 4). Freed of competition for limited food and nest resources, expansion of the Africanized honeybees was greatly facilitated. During this period the available cavities were rapidly filled, approaching carrying capacity by 1999 (Baum 2003). Therefore, we suggest that the nearly coincidental arrival of Varroa with expansion of the Africanized front hastened the demise of “pure” European honeybees and thereby had a major role in restructuring the post-Africanization WWR population. Had the resident European population not collapsed, the rate of Africanization would have been slower. At the same time, hybridization may have prevented the loss of European alleles if F1 and later-generation hybrids were more tolerant of Varroa than “pure” European honeybees.
In contrast with neotropical feral populations, the nuclear and mitochondrial makeup of the WWR population changed symmetrically over time (Figure 4). The result does not support the hypothesis of cytonuclear incompatibilities in hybrids of European mothers, as was suggested to explain the paucity of European mitochondria in neotropical populations (Harrison and Hall 1993; Schneider et al. 2003). However, under the temperate conditions the selective pressure generating cytonuclear incompatibility may not be operative. Definitive conclusions regarding cytonuclear asymmetry in WWR cannot be drawn, given the very recent secondary contact. The long-term evolutionary outcome of nuclear and mitochondrial introgression is uncertain because European alleles may be disfavored in an A. m. scutellata genetic background. If negative selection is occurring, then European alleles may gradually be eliminated from the WWR population. Alternatively, if selection favors individuals of mixed ancestry, then the European contribution will persist.
We suggest that the latter scenario is more likely for two reasons. First, Africanized honeybee populations living in subtropical-temperate transitional zones of South America have retained a substantial component of European origin several decades after initial colonization (Lobo et al. 1989; Sheppard et al. 1991a; Lobo and Krieger 1992; Diniz et al. 2003; Quezada-Euán et al. 2003). Second, the WWR data of 2000 and 2001 indicated an influx of European alleles. In 2000, a strong heterozygote deficiency (P = 0.000) was accompanied by a reduction in the A. m. scutellata proportions, as shown by two of the three estimators depicted in Figure 4. In 2001, both estimators of nuclear admixture and the genetic distance measure (Figures 3 and 4) indicated continued reduction in the level of Africanization. In both years linkage disequilibrium was detected for the linked loci A28/ED1 (P < 0.05; data not shown). This trend was also mirrored by a reduction in the number of individuals assigned as “pure Brazilian” (Figure 8). It appears that when honeybees of largely African genetic composition began to compete for more limited resources, it was the more European component of the population that become favored. The reappearance of disequilibrium in 2000 and 2001 would be most parsimoniously explained by migration of honeybees with higher European proportions back into the WWR hybrid zone. Estimates of the genetic makeup of the new colonies appearing at WWR lend support to this assumption. In fact it suggests that movement into the zone likely occurred by both more strongly Africanized honeybees and honeybees with a more European component. This can be readily seen in Figure 8 in comparison with Figure 5.
Genetic composition of the post-Africanization population:
Within 5 years of detection of the first migrant colony of the A. m. scutellata mitotype, a panmitic European population was replaced by panmitic mixtures of African and European subspecies. As for feral neotropical populations, the WWR Africanized gene pool was predominantly of A. m. scutellata origin (mR = 62.8%; mY = 74.7%; mtDNA = 69%). However, both mitochondrial (Pinto et al. 2004) and nuclear European markers were more frequent in the WWR population than in most feral neotropical populations (Hall and Muralidharan 1989; Smith et al. 1989; Hall 1990; Sheppard et al. 1991b; Quezada-Euán and Hinsull 1995; McMichael and Hall 1996; Clarke et al. 2001, 2002; Hall and McMichael 2001).
The WWR post-Africanization gene pool was composed of a diverse array of recombinant classes with a substantial European genetic contribution. On the basis of data from the last two years of the study we expect that European genes will be maintained at moderate frequencies. As Hardy-Weinberg equilibrium has been largely maintained, the WWR population is best viewed as a “hybrid swarm” (Rinderer 1986; Rinderer et al. 1991). Given the current data, we suggest, as in Pinto et al. (2004), that this hybrid swarm represents what will likely become part of the more southern limit of the developing hybrid zone.
Acknowledgments
We are indebted to D. De Jong for providing the sample collection from Brazil; to A. Cavazos, R. Medrano, J. Teer, L. Drawe, T. Blankenship, and S. Glasscock for their help in the field work at the Welder Wildlife Refuge; to L. Ross for technical assistance in molecular biology; to J. B. Patton for writing the program used to simulate the hybrid populations; to E. Saillant and C. McKenna for assistance in the data analysis; to J. W. Bickham for graciously providing laboratory space for the genetic analysis. Microsatellite analyses were performed in the Center for Biosystematics and Biodiversity at Texas A&M University. The United States Department of Agriculture-Agriculture Research Service, Honey Bee Laboratory at Weslaco, Texas, and the Welder Wildlife Foundation were instrumental in the initialization and long-term support of many aspects of this research project. M. A. Pinto thanks Escola Superior Agrária de Bragança and the European Union program PRODEP II (Programa de Desenvolvimento Educativo para Portugal; Medida 5/Acção 5.3) for financial support. This is Welder Wildlife Foundation contribution no. 597. The Texas Legislative Initiative: Protection and Management of Honey Bees—Pollinators of Agricultural Crops, Orchards and Natural Landscapes funded this research.
References
- Anderson, E., 1948. Hybridization of the habitat. Evolution 2: 1–9. [Google Scholar]
- Anderson, E., 1949 Introgressive Hybridization. John Wiley & Sons, New York.
- Arnold, M. L., 1997 Natural Hybridization and Evolution. Oxford University Press, New York.
- Barton, N. H., and G. M. Hewitt, 1985. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 16: 113–148. [Google Scholar]
- Barton, N. H., and G. M. Hewitt, 1989. Adaptation, speciation and hybrid zones. Nature 341: 497–503. [DOI] [PubMed] [Google Scholar]
- Baum, K. A., 2003 Feral Africanized honey bee ecology in a coastal prairie landscape. Ph.D. Thesis, Texas A&M University, College Station, TX.
- Belkhir, K. P., P. Borsa, L. Chikhi, N. Raufaste and F. Bonhomme, 2002 Genetix 4.04, logiciel sous windows TM pour la génétic des populations. Laboratoire Génome, Populations, Interactions, Université de Montpellier, Montpellier, France.
- Bertorelle, G., and L. Excoffier, 1998. Inferring admixture proportions from molecular data. Mol. Biol. Evol. 15: 1298–1311. [DOI] [PubMed] [Google Scholar]
- Bleeker, W., 2003. Hybridization and Rorippa austriaca (Brassicaceae) invasion in Germany. Mol. Ecol. 12: 1831–1841. [DOI] [PubMed] [Google Scholar]
- Bowcock, A. M., A. Ruiz-Linares, J. Tomfohrde, E. Minch, J. R. Kidd et al., 1994. High resolution of human evolution with polymorphic microsatellites. Nature 388: 455–457. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza, L. L., and A. W. F. Edwards, 1967. Phylogenetic analysis: models and estimation procedures. Am. J. Hum. Genet. 19: 233–257. [PMC free article] [PubMed] [Google Scholar]
- Childs, M. R., A. A. Echelle and T. E. Dowling, 1996. Development of the hybrid swarm between Pecos pupfish (Cyprinodontidae: Cyprinodon pecosensis) and sheepshead minnow (Cyprinodon variegatus): a perspective from allozymes and mtDNA. Evolution 50: 2014–2022. [DOI] [PubMed] [Google Scholar]
- Clarke, K. E., B. P. Oldroyd, J. Javier, G. Quezada-Euán and T. E. Rinderer, 2001. Origin of honeybees (Apis mellifera L.) from the Yucatan peninsula inferred from mitochondrial DNA analysis. Mol. Ecol. 10: 1347–1355. [DOI] [PubMed] [Google Scholar]
- Clarke, K. E., T. E. Rinderer, P. Franck, J. G. Quezada-Euán and B. P. Oldroyd, 2002. The Africanization of honey bees (Apis mellifera L.) of the Yucatan: a study of a massive hybridization event across time. Evolution 56: 1462–1474. [DOI] [PubMed] [Google Scholar]
- Cornuet, J. M., S. Piry, G. Luikart, A. Estoup and M. Solignac, 1999. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153: 1989–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman, L. I., T. E. Rinderer and J. A. Stelzer, 1997. DNA evidence of the origin of Varroa jacobsoni Oudemans in the Americas. Biochem. Genet. 35: 327–335. [DOI] [PubMed] [Google Scholar]
- Del Lama, M. A., R. A. Figueiredo, A. E. E. Soares and S. N. Del Lama, 1988. Hexokinase polymorphism in Apis mellifera and its use for Africanized honeybee identification. Rev. Bras. Genet. 11: 287–297. [Google Scholar]
- Diniz, M. N., A. E. E. Soares, W. S. Sheppard and M. A. Del Lama, 2003. Genetic structure of honey bee populations from southern Brazil and Uruguay. Genet. Mol. Biol. 26: 47–52. [Google Scholar]
- Ellstrand, N. C., and K. A. Schierenbeck, 2000. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc. Natl. Acad. Sci. USA 97: 7043–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estoup, A., M. Solignac and J.-M. Cornuet, 1994. Precise assessment of the number of patrilines and of genetic relatedness in honeybee colonies. Proc. R. Soc. Lond. Ser. B 258: 1–7. [Google Scholar]
- Estoup, A., L. Garney, M. Solignac and J.-M. Cornuet, 1995. Microsatellite variation in honey bee (Apis mellifera L.) populations: hierarchical genetic structure and test of the infinite allele and stepwise mutation models. Genetics 140: 679–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, D. B., A. Ruiz-Linares, L. L. Cavalli-Sforza and M. W. Feldman, 1995. Genetic absolute dating based on microsatellites and origin of modern humans. Proc. Natl. Acad. Sci. USA 92: 6723–6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet, J., 2001 FSTAT, A Program to Estimate and Test Gene Diversities and Fixation Indices (version 2.9.3). http://www2.unil.ch/izea/softwares/fstat.html.
- Goudet, J., M. Raymond, T. De Meeüs and F. Rousset, 1996. Testing differentiation in diploid populations. Genetics 144: 1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S. W., and E. A. Thompson, 1992. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48: 361–372. [PubMed] [Google Scholar]
- Guzman-Novoa, E., A. Sanchez, R. E. Page and T. Garcia, 1996. Susceptibility of European and Africanized honeybees (Apis mellifera L) and their hybrids to Varroa jacobsoni Oud. Apidologie 27: 93–103. [Google Scholar]
- Hall, H. G., 1990. Parental analysis of introgressive hybridization between African and European honeybees using nuclear DNA RFLPs. Genetics 125: 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, H. G., and M. A. McMichael, 2001. Frequencies of restriction fragment-length polymorphisms indicate that neotropical honey bee (Hymenoptera: Apidae) populations have African and west European origins. Ann. Entomol. Soc. Am. 94: 670–676. [Google Scholar]
- Hall, H. G., and K. Muralidharan, 1989. Evidence from mitochondrial DNA that African honey bees spread as continuous maternal lineages. Nature 339: 211–213. [DOI] [PubMed] [Google Scholar]
- Hall, H. G., and D. R. Smith, 1991. Distinguishing African and European honeybee matrilines using amplified mitochondrial DNA. Proc. Natl. Acad. Sci. USA 88: 4548–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, J. F., and H. G. Hall, 1993. African-European honey bee hybrids have low non-intermediate metabolic capacities. Nature 363: 258–260. [Google Scholar]
- Harrison, R. G., 1993 Hybrid Zones and the Evolutionary Process. Oxford University Press, New York.
- Hartl, D. L., and A. G. Clark, 1997 Principles of Population Genetics. Sinauer Associates, Sunderland, MA.
- Hunter, L. A., J. A. Jackman and E. A. Sugden, 1993. Detection records of Africanized honey bees in Texas during 1990, 1991, and 1992. Southwest. Entomol. 18: 79–89. [Google Scholar]
- Kerr, W. E., 1967. The history of the introduction of African bees to Brazil. S. Afr. Bee J. 2: 3–5. [Google Scholar]
- Kraus, B., and R. E. Page, 1995. Effect of Varroa jacobsoni (Mesostigmata: Varroidae) on feral Apis mellifera (Hymenoptera: Apidae) in California. Environ. Entomol. 24: 1473–1480. [Google Scholar]
- Lobo, J. A., and H. Krieger, 1992. Maximum likelihood estimates of gene frequencies and racial admixture in Apis mellifera L. (Africanized honey bees). Heredity 58: 441–448. [Google Scholar]
- Lobo, J. A., M. A. Del Lama and M. A. Mestriner, 1989. Population differentiation and racial admixture in the Africanized honeybee (Apis mellifera L.). Evolution 43: 794–802. [DOI] [PubMed] [Google Scholar]
- Loper, G. M., J. Fewell, D. R. Smith, W. S. Sheppard and N. Schiff, 1999 Genetic changes of a population of feral honey bees in Sonoran desert of southern Arizona following the arrival of Acarapis woodi, Varroa jacobsoni, and Africanization, pp. 47–51 in Apiculture for the 21st Century, edited by R. Hoopingarner and L. J. Conner. Wicwas, Cheshire, CT.
- Mayr, E., 1942 Systematics and the Origin of Species. Columbia University Press, New York.
- McMichael, M., and H. G. Hall, 1996. DNA RFLPs at a highly polymorphic locus distinguish European and African subspecies of the honey bee Apis mellifera L. and suggest geographical origins of New World honey bees. Mol. Ecol. 5: 403–416. [Google Scholar]
- Message, G., and L. S. Gonçalves, 1995. Effect of the size of worker brood cells of Africanized honey bee on infestation and reproduction of the ectoparasitic mite Varroa jacobsoni. Apidologie 26: 381–386. [Google Scholar]
- Michener, C. D., 1975. The Brazilian bee problem. Ann. Rev. Entomol. 20: 399–416. [DOI] [PubMed] [Google Scholar]
- Moretto, G., L. S. Gonçalves, D. De Jong and M. Z. Bichuette, 1991. The effects of climate and bee race on Varroa jacobsoni Oud infestations in Brazil. Apidologie 22: 197–203. [Google Scholar]
- Moretto, G., L. S. Gonçalves and D. De Jong, 1993. Heritability of Africanized and European honey bee defensive behavior against the mite Varroa jacobsoni. Rev. Bras. Genet. 16: 71–77. [Google Scholar]
- Nei, M., 1987 Molecular Evolutionary Genetics. Columbia University Press, New York.
- Nogueira-Neto, P., 1964. The spread of a fierce African bee in Brazil. Bee World 45: 119–121. [Google Scholar]
- Norušis, M. J., 1993 SPSS for Windows Base System User's Guide Release 6.0. SPSS, Chicago.
- Oldroyd, B. P., A. J. Smolenski, J.-M. Cornuet, S. Wongsiri, A. Estoup et al., 1995. Levels of polyandry and intra-colonial genetic relationships in Apis florea. Behav. Ecol. Sociobiol. 37: 329–335. [Google Scholar]
- Paetkau, D., W. Calvert, I. Stirling and C. Strobeck, 1995. Microsatellite analysis of population-structure in Canadian polar bears. Mol. Ecol. 4: 347–354. [DOI] [PubMed] [Google Scholar]
- Perry, W. L., J. L. Feder, G. Dwyer and D. M. Lodge, 2001. Hybrid zone dynamics and species replacement between Orconectes crayfishes in a northern Wisconsin lake. Evolution 55: 1153–1166. [DOI] [PubMed] [Google Scholar]
- Petit, R. J., A. El Mousadik and O. Pons, 1998. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 12: 844–855. [Google Scholar]
- Pinto, M. A., W. L. Rubink, R. N. Coulson, J. C. Patton and J. S. Johnston, 2004. Temporal pattern of Africanization in a feral honey bee population from Texas inferred from mitochondrial DNA. Evolution 58: 1047–1055. [DOI] [PubMed] [Google Scholar]
- Quezada-Euán, J. J. G., 2000. Hybridization between European and Africanized honeybees in tropical Yucatan, Mexico. II. Morphometric, allozymic and mitochondrial DNA variability in feral colonies. Apidologie 31: 443–453. [Google Scholar]
- Quezada-Euán, J. J. G., and S. M. Hinsull, 1995. Evidence of continued European morphometrics and mtDNA in feral colonies of honey bees (Apis mellifera L.) from the Yucatan peninsula, Mexico. J. Apic. Res. 34: 161–166. [Google Scholar]
- Quezada-Euán, J. J. G., C. M. Echazarreta and R. J. Paxton, 1996. The distribution and range expansion of Africanized honey bees (Apis mellifera) in the state of Yucatan, Mexico. J. Apic. Res. 35: 85–95. [Google Scholar]
- Quezada-Euán, J. J. G., E. E. Perez-Castro and W. D. May-Itza, 2003. Hybridization between European and African-derived honeybee populations (Apis mellifera) at different altitudes in Peru. Apidologie 34: 217–225. [Google Scholar]
- Rannala, B., and J. L. Mountain, 1997. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA 94: 9197–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, M., and F. Rousset, 1995. a Population genetics software for exact test and ecumenism. J. Hered. 86: 248–250. [Google Scholar]
- Raymond, M., and F. Rousset, 1995. b An exact test for population differentiation. Evolution 49: 1280–1283. [DOI] [PubMed] [Google Scholar]
- Rhymer, J. M., and D. Simberloff, 1996. Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 27: 83–109. [Google Scholar]
- Rice, W. R., 1989. Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Rinderer, T. E., 1986. Africanized bees: the Africanization process and potential range in the United States. Bull. Entomol. Soc. Am. 32: 222–227. [Google Scholar]
- Rinderer, T. E., J. A. Stelzer, B. P. Oldroyd, S. M. Buco and W. L. Rubink, 1991. Hybridization between European and Africanized honey bees in the neotropical Yucatan Peninsula. Science 253: 309–311. [DOI] [PubMed] [Google Scholar]
- Roberts, D., and R. Hiorns, 1965. Methods of analysis of the genetic composition of a hybrid population. Hum. Biol. 37: 38–43. [PubMed] [Google Scholar]
- Roubik, D. W., and M. W. Boreham, 1990. Learning to live with Africanized honeybees. Interciencia 15: 146–153. [Google Scholar]
- Rousset, F., and M. Raymond, 1995. Testing heterozygote excess and deficiency. Genetics 140: 1413–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe, D. J., T. E. Rinderer, J. A. Stelzer, B. P. Oldroyd and R. H. Crozier, 1997. Seven polymorphic microsatellite loci in honeybees (Apis mellifera). Insectes Soc. 44: 85–93. [Google Scholar]
- Rubink, W. L., W. T. Wilson, J. J. Resendez and D. L. Maki, 1990. Pre-Africanized Apis mellifera (Hemenoptera: Apidae) swarming dynamics in Northeastern Mexico and Southern Texas. J. Kans. Entomol. Soc. 63: 288–297. [Google Scholar]
- Rubink, W. L., P. Luevano-Martinez, E. A. Sugden, W. T. Wilson and A. M. Collins, 1996. Subtropical Apis mellifera (Hymenoptera: Apidae) swarming dynamics and Africanization rates in northeastern Mexico and southern Texas. Ann. Entomol. Soc. Am. 89: 243–251. [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–426. [DOI] [PubMed] [Google Scholar]
- Schiff, N. M., and W. S. Sheppard, 1995. Genetic analysis of commercial honey bees (Hymenoptera: Apidae) from the southeastern United States. J. Econ. Entomol. 88: 1216–1220. [Google Scholar]
- Schiff, N. M., and W. S. Sheppard, 1996. Genetic differentiation in the queen breeding population of the western United States. Apidologie 27: 77–86. [Google Scholar]
- Schiff, N. M., W. S. Sheppard, G. M. Loper and H. Shimanuki, 1994. Genetic diversity of feral honey bee (Hymenoptera: Apidae) populations in the southern United States. Ann. Entomol. Soc. Am. 87: 842–848. [Google Scholar]
- Schneider, S. S., and H. G. Hall, 1997. Diet selection and foraging distances of African and European-African hybrid honey bee colonies in Costa Rica. Insectes Soc. 44: 171–187. [Google Scholar]
- Schneider, S. S., L. J. Leamy, L. A. Lewis and G. Degrandi-Hoffman, 2003. The influence of hybridization between African and European honeybees, Apis mellifera, on asymmetries in wing size and shape. Evolution 57: 2350–2364. [DOI] [PubMed] [Google Scholar]
- Sheppard, W. S., T. E. Rinderer, J. A. Mazzolli, J. A. Stelzer and H. Shimanuki, 1991. a Gene flow between African- and European-derived honey bee populations in Argentina. Nature 349: 782–784. [Google Scholar]
- Sheppard, W. S., A. E. E. Soares, D. DeJong and H. Shimanuki, 1991. b Hybrid status of honey bee populations near the historic origin of Africanization in Brazil. Apidologie 22: 643–652. [Google Scholar]
- Smith, D. R., O. R. Taylor and W. M. Brown, 1989. Neotropical Africanized honey bees have African mitochondrial DNA. Nature 339: 213–215. [DOI] [PubMed] [Google Scholar]
- Snedecor, G. W., and W. G. Cochran, 1978 Statistical Methods. Iowa State University Press, Ames, IA.
- Spivak, M. D., J. C. Fletcher and M. D. Breed, 1991 Introduction, pp. 1–9 in The “African” Honey Bee, edited by M. Spivak, D. J. C. Fletcher and M. D. Breed. Westview Press, Boulder, CO.
- Suazo, A., R. McTiernan and H. G. Hall, 1998. Differences between African and European honey bees (Apis mellifera L.) in random amplified polymorphic DNA (RAPD). J. Hered. 89: 32–36. [Google Scholar]
- Sugden, E. A., and K. R. Williams, 1990. October 15: the day the bee arrived. Glean. Bee Cult. 119: 18–21. [Google Scholar]
- Swofford, D. L., 2002 PAUP*. Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4.0b 10. Sinauer Associates, Sunderland, MA.
- Taylor, O. R., 1985. African bees: potential impact in the United States. Bull. Entomol. Soc. Am. 31: 14–24. [Google Scholar]
- Taylor, O. R., 1988 Ecology and economic impact of African and Africanized honey bees, pp. 29–41 in Africanized Honey Bees and Bee Mites, edited by G. R. Needham, R. E. Page, M. Delfinado-Baker and C. E. Bowman. Ellis Horwood, Chichester, UK.
- Weir, B. S., and C. C. Cockerham, 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Winston, M. L., 1992. The biology and management of Africanized honey bees. Annu. Rev. Entomol. 37: 173–193. [Google Scholar]