Abstract
The yeast Saccharomyces cerevisiae and other members of the genus Saccharomyces are descendants of an ancient whole-genome duplication event. Although most of the duplicate genes have since been deleted, many remain, and so there are many pairs of related genes. We have found that poorly expressed genes diverge rapidly from their paralog, while highly expressed genes diverge little, if at all. This lack of divergence of highly expressed paralogous gene pairs seems to involve gene correction: one member of the pair “corrects” the sequence of its twin, and so the gene pair evolves as a unit. This correction presumably involves gene conversion and could occur via a reverse-transcribed cDNA intermediate. Such correction events may also occur in other organisms. These results support the idea that copies of poorly expressed genes are preserved when they diverge to take on new functions, while copies of highly expressed genes are preserved when they are needed to provide additional gene product for the original function.
IT is generally believed that selection for preferred codons (codon bias) increases the sequence conservation of highly expressed genes relative to poorly expressed genes (the “selection hypothesis”) (Powell and Moriyama 1997). Both highly expressed and poorly expressed genes are selected for function, which means that many nonsynonymous codon changes are selected against; but in addition, for highly expressed genes, many synonymous changes are also selected against to maintain codons preferred for translational efficiency and accuracy. A corollary of this argument is that when an organism has two similar copies of a highly expressed gene, these copies should be preserved in evolution as a gene pair sharing high homology, because selection for both function and codon bias prevents the members of the pair from drifting apart.
In this study, we propose a parallel hypothesis for the conservation of duplicated highly expressed genes and show that the new hypothesis not only plays a significant role, at least in yeast, but also may be more important than selection under certain conditions.
We call our hypothesis the correction hypothesis. It consists of three proposals: first, that one copy of a gene can correct the sequence of a second copy; second, that correction depends on high sequence identity; and third, that the probability of correction depends on the level of gene expression. We propose two possible mechanisms of correction. In the first, correction happens through the occasional copying back of mature mRNA into cDNA using reverse transcriptase and a subsequent recombinational interaction between the cDNA and the second copy of the chromosomal gene. In the second mechanism, correction is due to a direct recombinational interaction between the two genes of a duplicate pair.
In the following sections, we first show that correction indeed plays a significant role in the conservation of gene pairs in yeast and that correction is correlated with the level of gene expression. Evidence for correction is, first, that the conservation between the members of a highly expressed gene pair is too high to be explained by selection alone, and second, that the pattern of nucleotide substitution within and between species of Saccharomyces is much more compatible with the correction hypothesis than with the selection hypothesis. We examine some properties of correction and consider whether correction might play a role in other organisms.
MATERIALS AND METHODS
Saccharomyces sequences were obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org/), from Washington University (http://www.genome.wustl.edu/projects/yeast/ and http://www.genome.wustl.edu/blast/yeast_client.cgi), and from the Massachusetts Institute of Technology (http://www-genome.wi.mit.edu/seq/Saccharomyces/). ClustalW-based end-to-end fungal alignments of the Saccharomyces cerevisiae genes and their analogous sequences in the other Saccharomyces species were obtained from the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org) whenever available.
Genes and gene pairs from the ancient duplication were selected using the “blocks menu” page of the website of Wolfe and colleagues at http://acer.gen.tcd.ie/cgi-bin/khwolfe/blocks.pl?block=ALL. All gene pairs from the above site whose coding sequences appear at SGD under the same systematic/standard name were downloaded in an automated manner. Any gene, and thus its pair, with a name that led to confusion in its recognition, was discarded. Note especially that we have not analyzed all duplicated genes in S. cerevisiae. We have restricted our study to those genes thought to have been duplicated as part of a single, ancient, genome-wide duplication event. For example, many duplications near telomeres appear to be recent duplications. We have not included any of these recently duplicated, telomere-associated genes in our analysis.
We used a modified version of the Jukes-Cantor model for measuring divergence between a pair of gene sequences. The conventional Jukes-Cantor model for computing evolutionary divergence between two sequences is extended to account for “internal” indels in the pairwise alignment of sequences. (An internal indel is not a part of the continuous batch of indels that might be present at either extremity of a pairwise alignment, perhaps owing to the difference in the lengths of the two sequences.) The modified measure treats an indel as a substitution of weight c between 0 and 1, inclusive, whereas a substitution of any kind is, as in the conventional model, of weight 1. Upon making the usual approximations, the divergence is given by −d = − ((3 + c)/(4 + c)) × ln(1 − (4 + c) × p/(3 + c)), where p is the proportion of substitutions, which, in our model, is a 1:c weighted proportion of both substitutions and internal indels. Clearly, a weight of c = 0 reduces the new measure to the old one. Inheriting the property of the old model, the new measure is also a partial function; i.e., it is not defined for certain values of the valid input p. The divergence-expression figures are plotted with the above parameter c set to 1. Varying the value of c between 0 and 1 makes little difference to the plots.
For computing the counts and ratio of synonymous and nonsynonymous substitutions, we use the Synonymous Nonsynonymous Analysis Program (SNAP) available at the HIV Sequence Database (hiv-web.lanl.gov) (Nei and Gojobori 1986). Complete data are available at: http://www.cs.sunysb.edu/~compbio/Correction/.
RESULTS
High expression, conservation, and correction:
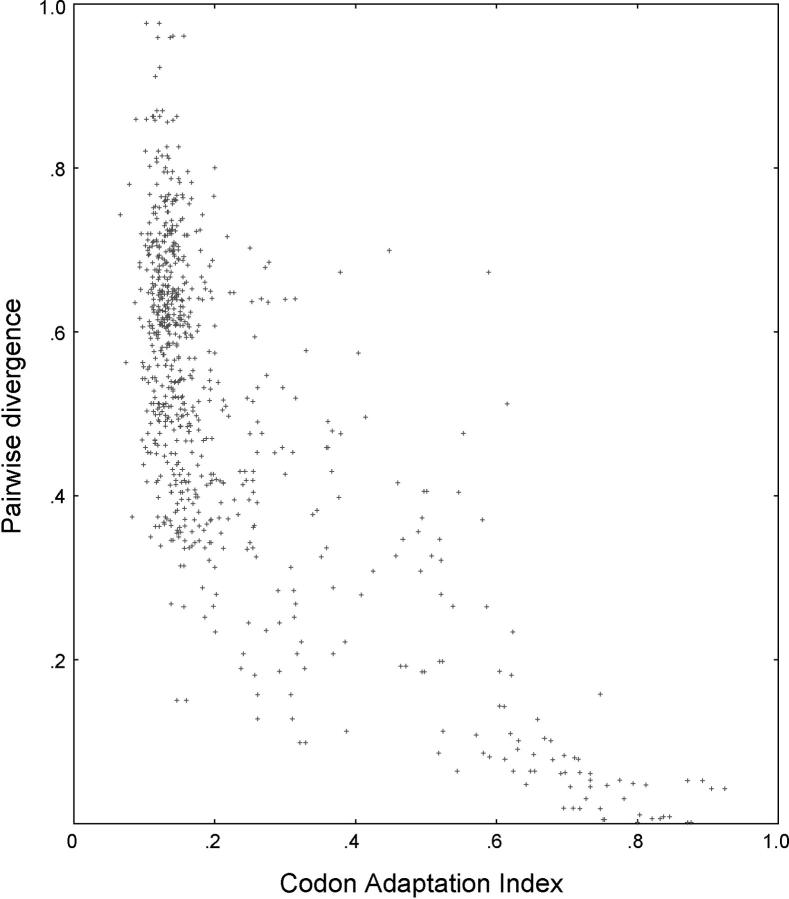
The genus Saccharomyces arose from an ancient whole-genome duplication event, shortly after Saccharomyces diverged from Kluyveromyces (Wolfe and Shields 1997; Kellis et al. 2004). Subsequent to the duplication, many individual deletion events deleted one of the copies of most of the duplicate genes. Nevertheless, the S. cerevisiae of today has up to 450 gene pairs (16% of the proteome) remaining from the ancient duplication (Seoighe and Wolfe 1999). While studying a 382-pair subset of these duplicates, we found a remarkably strong negative correlation between sequence divergence and the codon adaptation index (CAI; Figure 1). CAI (Sharp and Li 1987) is used as a surrogate for gene expression (Futcher et al. 1999). That is, the highly expressed genes have diverged less from their duplicates than the poorly expressed genes. The Pearson correlation is r = −0.72 (P < 10−16 for the null hypothesis that r = 0). The majority of gene pairs have low codon bias (i.e., low expression) and high DNA sequence divergence, while a substantial subgroup has high codon bias (i.e., high expression) and low DNA sequence divergence.
Figure 1.—
Divergence as a function of expression in S. cerevisiae. For each of 764 genes (the members of qualified gene pairs from the ancient duplication; see materials and methods ), the closest homolog is found. The modified Jukes-Cantor divergence score for the pair, if defined (materials and methods), is plotted on the y-axis, on the basis of the ClustalW pairwise alignment of the genes. The CAI of the chosen gene is plotted on the x-axis. CAI is a surrogate for the level of mRNA produced. Note that the divergence of each gene pair (g1, g2) is plotted twice, once against the CAI of g1 and once against the CAI of g2. There is a strong negative correlation between divergence and CAI (i.e., level of expression), with correlation coefficient r = −0.72 (P < 10−16 for the null hypothesis that r = 0).
The correction model:
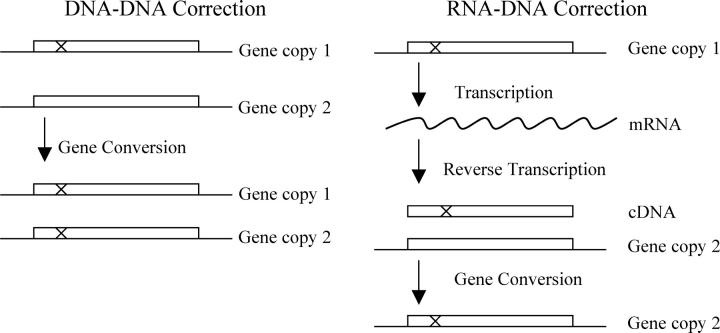
The correlation seen in Figure 1 is not necessarily inconsistent with the “selection” model. However, the very strong correlation and the very large differences in sequence identity were so striking that we wondered whether there might be some other explanation. In particular, some highly expressed gene pairs were >95% identical in DNA sequence despite apparently diverging many millions of years ago (Wolfe and Shields 1997). This striking conservation might be explained if DNA sequence correction occurred between members of the pair. S. cerevisiae has a very active homologous recombination system, and recombination or gene conversion could account for pairs with very high identity.
In the most obvious model of sequence correction (Figure 2, “DNA-DNA Correction”), the two gene copies interact, and gene conversion events occur directly between the two chromosomal copies. There is some evidence that very highly transcribed genes are particularly active in recombinational events, while repressed genes are relatively inactive (Saxe et al. 2000).
Figure 2.—
The correction model. In DNA-DNA Correction (left), one gene interacts with a second gene and corrects it by gene conversion. In RNA-DNA Correction (right), one gene is transcribed and then copied into cDNA (or a cDNA/RNA hybrid), and this cDNA molecule interacts with a second gene and corrects it by gene conversion.
A second model of sequence correction (Figure 2, “RNA-DNA Correction”) invokes an RNA intermediate. This model depends upon the fact that S. cerevisiae contains a retrotransposon, Ty, which encodes a reverse transcriptase. Occasionally, this reverse transcriptase makes cDNA copies of normal, cellular genes (Xu and Boeke 1990), and these cDNA copies can recombine with their chromosomal homologs (Derr et al. 1991; Derr and Strathern 1993). It has been proposed that reverse transcription followed by homologous recombination explains why so few genes in S. cerevisiae have introns (Baltimore 1985; Fink 1987). In these proposals, a gene with an intron produces a transcript; the transcript is spliced; the spliced transcript is converted to cDNA by Ty reverse transcriptase; and then the cDNA interacts with the chromosomal gene and removes the intron by gene conversion. Similarly, in our model of correction via an RNA intermediate (Figure 2), an mRNA produced by gene copy 1 is converted to cDNA by Ty reverse transcriptase. This cDNA then interacts with gene copy 2, and by gene conversion corrects gene copy 2 into an exact duplicate of gene copy 1. Over succeeding generations, gene copies 1 and 2 may again drift apart, but then will undergo another round of correction, again making the two genes identical. An attractive feature of this model is that the probability of correction is obviously directly proportional to the level of gene expression: the more mRNA that is made, the higher the probability that some of it will be converted to cDNA. This could explain the strong correlation between pairwise homology and expression level. However, a weakness of this model is that RNA-mediated gene conversion is much rarer than DNA-DNA events (Derr and Strathern 1993).
A feature of both of these correction models is that gene conversion will occur only between sequences that have a very high percentage of identity; even a small number of mismatches drastically reduces the frequency of gene conversion (Modrich and Lahue 1996; Datta et al. 1997; Chen and Jinks-Robertson 1998). Once members of a gene pair have drifted sufficiently far apart, they would no longer be able to correct each other.
In either model, genes expressed at a high level correct each other frequently because highly transcribed genes are recombinationally active (Saxe et al. 2000), as in the DNA-DNA model, or because highly transcribed genes make more RNA, as in the RNA-DNA model, and so do not drift apart. Because they do not drift apart, they remain eligible for future correction events. Genes expressed at a low level correct each other infrequently, and so sometimes drift far apart between correction events, greatly reducing the probability of future correction. Thus, there would be two groups of gene pairs—a highly expressed, highly conserved group and a poorly expressed, poorly conserved group.
Correction vs. selection:
We wished to distinguish the selection model from the correction models and took advantage of the fact that five other species of Saccharomyces (castelli, kluyveri, mikatae, paradoxus, and bayanus) have recently been sequenced (Cliften et al. 2003; Kellis et al. 2003). These species diverged from S. cerevisiae at various times (paradoxus, 10 MYA; byanus, 20 MYA; mikatae, 20 MYA; castelli, 50 MYA; kluyveri, very roughly 75 MYA), whereas the duplication of the Saccharomyces genome preceded most of these speciation events (the probable exception being the speciation of S. kluyveri). We reasoned as follows: if the sequence of a gene is maintained solely by selection, and not by correction, then the rate at which the similarity of gene 1 copy 1 and gene 1 copy 2 drift apart within S. cerevisiae will be roughly the same as that of gene 1 copy 1 of S. cerevisiae drifting from their orthologs in each of the other species. In other words, if there is no correction, then each gene in each species will diverge independently and at roughly the same rate, regardless of whether there is a duplicate gene in the same cell. If anything, divergence will be faster when there is a duplicate copy in the same cell, since the duplicate can provide important functions lost by its mutating partner. Alternatively, if correction is a significant force, then gene 1 copy 1 and gene 1 copy 2 in S. cerevisiae will drift apart more slowly (if at all) than gene 1 copy 1 of S. cerevisiae and its nearest ortholog in each of the other species, since of course there will not be any correction between species. (Note that for purposes of this argument, it does not matter whether the genes are unique or duplicate pairs in the other species.) Thus, for each S. cerevisiae gene in a list of 382 S. cerevisiae gene pairs, we found the closest ortholog in each of the other five species and compared the divergence of these five orthologs and of the S. cerevisiae paralog. Divergence was examined as a correlate of the codon adaptation index (Sharp and Li 1987). Results are shown in Figure 3.
Figure 3.—
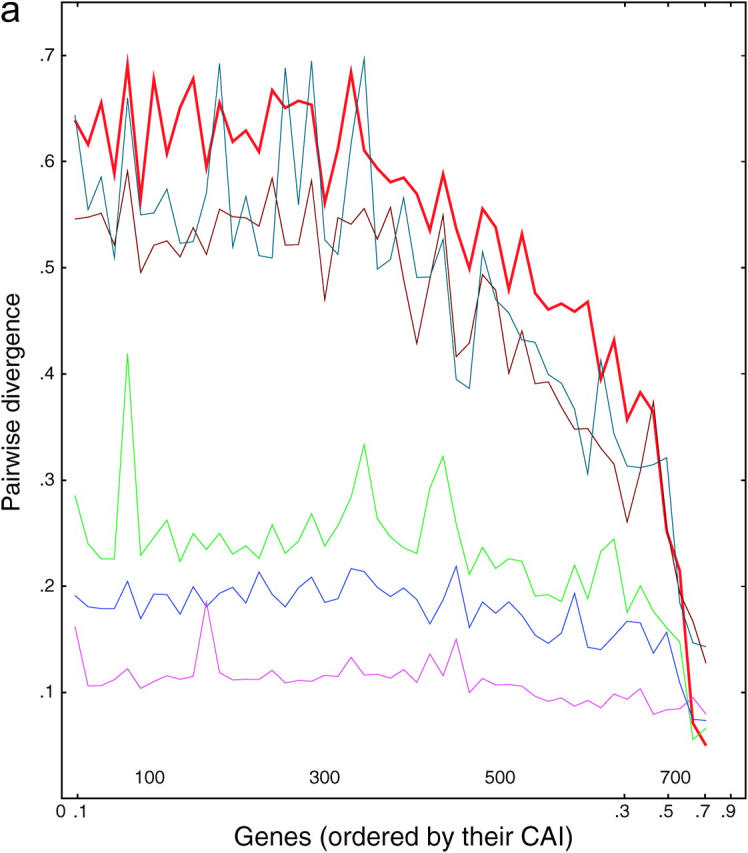
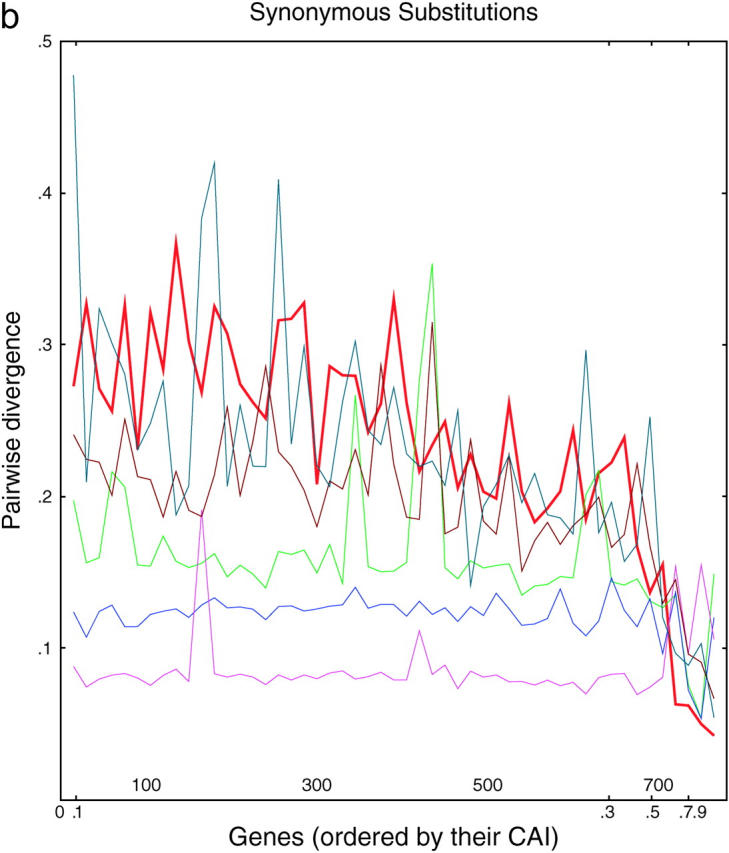
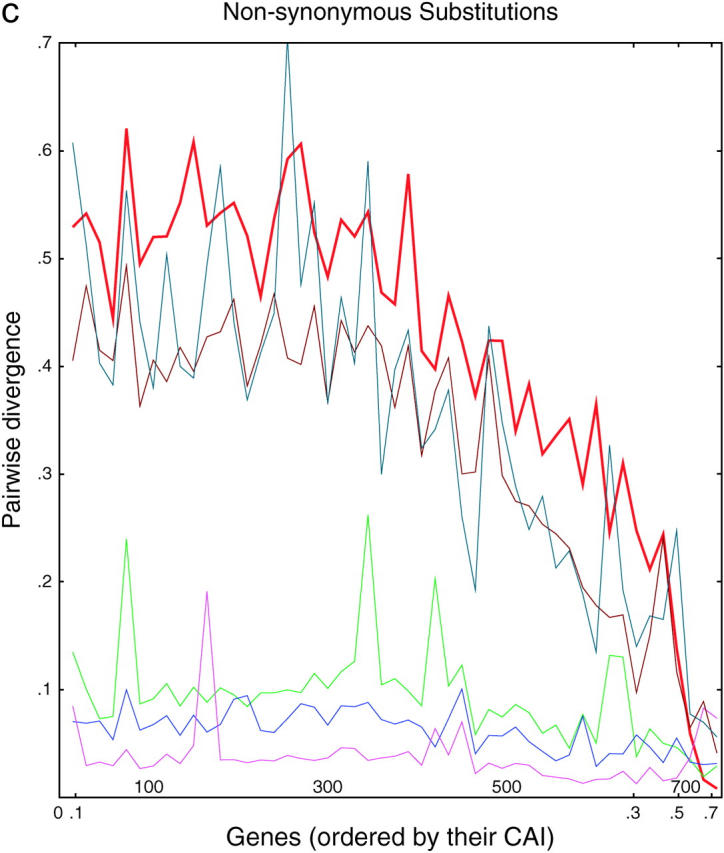
Divergence as a function of expression in six yeasts. (a) Each of 764 ancient duplicated genes in S. cerevisiae is compared to its closest homolog in S. cerevisiae (intraspecies comparison) or to its closest homolog in another species of Saccharomyces (interspecies comparisons). From top to bottom, on the left side of the graph, the lines are S. kluyveri (thin line, blue-green), S. cerevisiae (thick line, red), S. castellii (brown), S. bayanus (green), S. mikatae (blue), and S. paradoxus (purple). Divergence scores were calculated (materials and methods) for each pairwise comparison and plotted on the y-axis. Along the x-axis, the genes are sorted from left to right in the increasing order of the CAI of the chosen cerevisiae gene. Divergence scores were averaged over consecutive disjoint sets of 15 genes each to smooth the curves. The number of genes plotted in each curve varies from 752 to 758, depending on the existence/availability of interspecies homologs, whether the Jukes-Cantor score is defined, etc. b and c are the same as a, but are confined to synonymous and nonsynonymous substitutions, respectively.
There are several noteworthy points. First, for genes expressed at low and medium levels, the divergence is roughly proportional to the time since divergence. This is true both between species and within S. cerevisiae. Second, for genes expressed at high levels, the divergence is decreased; i.e., highly expressed genes tend to be more conserved. This conservation is consistent with the idea that selection is important in preserving highly expressed genes; presumably some of the effect is due to selection for preferred codons. Nevertheless, the divergence between the highly expressed S. cerevisiae genes and their castelli or kluyveri homologs is still considerable, showing that there is still sequence space into which these genes can diverge while still maintaining function. Third and most striking, for highly expressed genes, there is very little divergence between the two S. cerevisiae copies. The red line in Figure 3 for the intraspecies cerevisiae-cerevisiae comparisons initially (i.e., at lowest CAI) shows very high divergence scores, but then falls at higher CAIs, crossing through all the interspecies comparisons, until finally at the highest CAI the cerevisiae-cerevisiae comparisons have the lowest divergence. That is, two highly expressed cerevisiae copies may have only a few mismatches and >95% DNA sequence identity, despite the fact that the two genes diverged long ago, and despite the fact that many more mismatches are present between the same S. cerevisiae genes and their closest orthologs in all the other species, which diverged more recently. In summary, in intraspecies comparisons, we see a striking lack of divergence between pairs of very highly expressed genes, even though it is clear from interspecies comparisons that such genes can diverge. Since correction can occur within a species but not between species, we take this as evidence for correction.
The pattern of nucleotide substitution:
Figure 4 shows the evolution of the RPL4a/RPL4b genes (encoding ribosomal proteins) and the NTH1/NTH2 genes (encoding neutral trehalase). In S. cerevisiae, RPL4a and NTH2 are linked on chromosome 2, while their duplicates, RPL4b and NTH1, are linked on chromosome 4. These gene pairs are part of the “block 3” syntenic duplicated region defined by Wolfe and colleagues (http://acer.gen.tcd.ie/~khwolfe/yeast/nova/), and they are derived from the genome-wide duplication of 100 MYA. These gene duplicates have also survived in S. bayanus. Thus, we can now align four gene copies for each type of gene (S.c. RPL4a, S.c. RPL4b, S.b. RPL4a, and S.b. RPL4b, or S.c. NTH1, S.c. NTH2, S.b. NTH1, and S.b. NTH2) and ask about the patterns of nucleotide substitution.
Figure 4.—
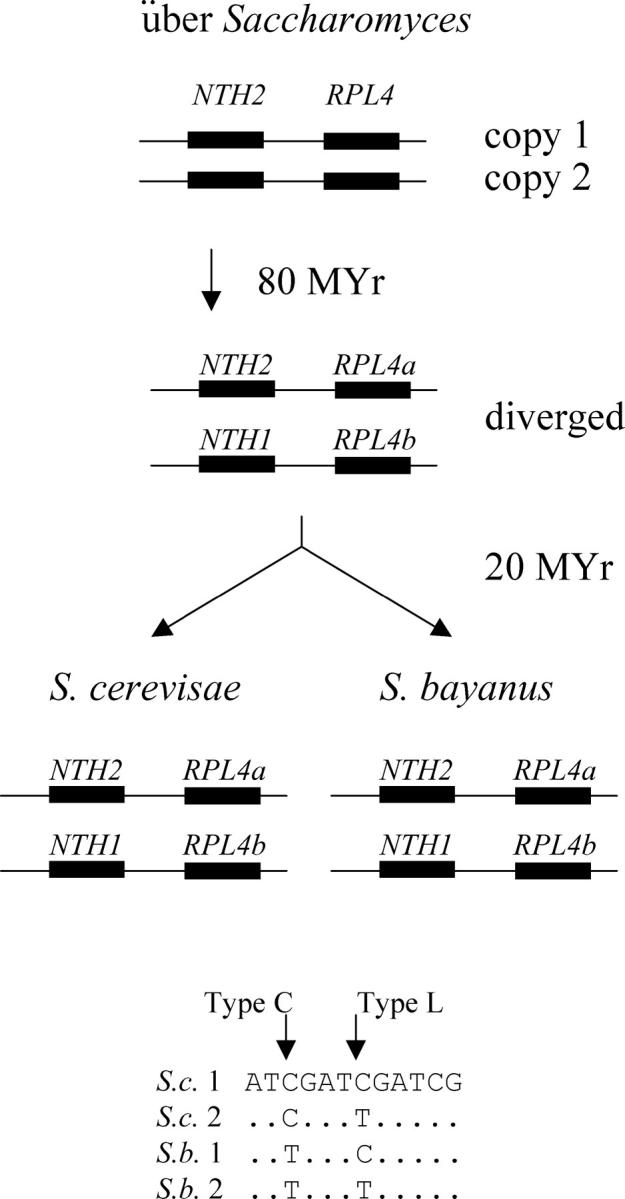
Gene lineages and patterns of nucleotide substitution. The evolutionary lineages of the NTH1/2 and RPL4a/b genes are shown. At a type C (correction) position, the intraspecies duplicates share a distinguishing nucleotide, while at a type L (lineage) position, the interspecies homologs, which are most closely related by descent, share a distinguishing nucleotide.
Any pair of duplicated genes from the ancient duplication event has had ∼80 MY in which to diverge before the separation of S. cerevisiae from S. bayanus ∼20 MYA. Thus, in the absence of correction, one would expect cerevisiae copy 1 and its ortholog bayanus copy 1 to share certain nucleotide changes, while cerevisiae copy 2 and its ortholog bayanus copy 2 to share other changes, because copy 1 and copy 2 have had ∼80 MY to diverge, while the two orthologs of copy 1 in the two species have had only 20 MY. We call this pattern of nucleotide substitution (where the orthologs in different species share a distinguishing nucleotide) the “L” pattern, for “lineage.” In contrast, if correction occurs, then cerevisiae copy 1 and cerevisiae copy 2 (i.e., the paralogs) will share certain nucleotide changes (because the change has been copied from 1 to 2 or vice versa), while bayanus copy 1 and bayanus copy 2 will share other changes. We call this pattern of substitution (where paralogs within the species share a distinguishing nucleotide) the “C” pattern, for “correction.” The bottom of Figure 4 shows these two different patterns of nucleotide substitution. Finally, mutational noise will sometimes generate a situation in which cerevisae copy 1 will share a distinguishing nucleotide with copy 2 (i.e., the nonorthologous gene) in bayanus. We call this a type N pattern, for “noise”; its frequency is important for estimating the number of C and L patterns that might be due to noise.
Figure 5 shows a sample four-way alignment of part of the RPL4a,b and NTH1,2 genes, and Table 1 shows results for the full-length four-way alignments. It is clear and striking that the highly expressed RPL4 genes show exclusively the type C pattern of nucleotide substitution, arguing that they have undergone correction, while the tightly linked NTH1,2 genes show mainly type L substitution (305 positions). Although 31 type C substitutions are seen in the NTH1 vs. NTH2 comparison, there are also 35 type N substitutions, arguing that the type C substitutions in these genes are simply mutational noise, and not correction. Thus, as predicted, this pair of highly expressed genes shows primarily (in this case, exclusively) the correction pattern of substitution, while the poorly expressed but linked genes show primarily or (after allowing for noise) exclusively the lineage pattern of substitution. We consider this very strong evidence for the correction model.
Figure 5.—
Alignment of the RPL4a,b and NTH1,2 genes. A representative 60 nucleotides of the RPL4a,b and NTH1,2 genes from S. cerevisiae (S.c.) and S. bayanus (S.b.) are aligned. Type C (correction) substitutions are underlined and in boldface type; type L (lineage) substitutions are in boldface type, and type N (noise) substitutions are underlined.
TABLE 1.
Correction and lineage substitutions in theRPL4 andNTH1/2 genes
| Length | Mismatch positions |
Type C |
Type L |
Type N |
|
|---|---|---|---|---|---|
| RPL4a,b | 1089 | 69 | 40 | 0 | 0 |
| NTH1,2 | 2355 | 970 | 31 | 305 | 35 |
“Length” is the length of the nucleotide alignment, and in these cases is the length of the open reading frame of the gene. “Mismatch positions” is the number of positions where all four nucleotides in the alignment are not identical, e.g., an alignment of T:T:T:C or T:T:C:C, etc. “Type C” is a mismatch position of the C type, characteristic of correction, where the two cerevisae genes share the same nucleotide and the two bayanus genes share a different nucleotide (e.g., T:T:C:C). “Type L” is a mismatch position of the L type, characteristic of the gene lineage, where cerevisiae gene 1 and its bayanus homolog share the same nucleotide, while cerevisiae gene 2 and its bayanus homolog share a different nucleotide (e.g., T:C:T:C). “Type N” is a mismatch position characteristic of mutational noise, where cerevisiae gene 1 and its bayanus nonhomolog share the same nucleotide, while cerevisiae gene 2 and its bayanus nonhomolog share a different nucleotide (e.g., T:C:C:T). Positions of the type T:T:T:C (i.e., one nucleotide at odds with the other three) are mismatch positions, but are not C or L or N.
We extended this analysis to most of the syntenic duplicated blocks defined by Wolfe and co-workers (http://acer.gen.tcd.ie/~khwolfe/yeast/nova/). We examined all blocks (a) that contained at least one gene pair with a high codon bias and (b) where we could find two different bayanus orthologs of the high-codon-bias gene(s). There were ∼28 eligible blocks. For all of these genes (both high and low bias) where two bayanus orthologs existed (168 genes total), we did the four-way alignments, as shown in Figure 5, and noted the number of C (correction), L (lineage), and N (noise) nucleotide substitutions. These results are shown in Table 2. The main results are as follows.
TABLE 2.
Ratios of correction to lineage substitutions for duplicated genes
| Block no. | Gene pair | CAI | C | L | N | C/L |
|---|---|---|---|---|---|---|
| 7 | NHP6B-NHP6A | Low | 3 | 26 | 5 | |
| 7 | YMC2-YMC1 | Low | 9 | 132 | 10 | |
| 7 | TKL2-TKL1 | Low | 26 | 320 | 18 | |
| 7 | TEF2-TEF1 | High | 44 | 0 | 0 | High |
| 8 | SSE2-SSE1 | Medium | 26 | 252 | 25 | |
| 8 | SMY2-YPL105C | Low | 28 | 504 | 32 | |
| 8 | YBR177C-YPL095C | Low | 19 | 243 | 27 | |
| 8 | RPS6B-RPS6A | High | 12 | 0 | 0 | High |
| 8 | SMP1-RLM1 | Low | 6 | 74 | 4 | |
| 8 | YBR183W-YPL087W | Low | 18 | 203 | 9 | |
| 8 | RPS9B-RPS9A | High | 17 | 2 | 2 | High |
| 8 | RPL21A-RPL21B | High | 4 | 14 | 1 | Low |
| 8 | YBR197C-YPL077C | Low | 7 | 163 | 10 | |
| 8 | KTR4-KTR6 | Low | 15 | 426 | 9 | |
| 8 | KTR3-KTR6 | Low | 10 | 343 | 11 | |
| 10 | RPS14A-RPS14B | High | 8 | 0 | 0 | High |
| 12 | ARF2-ARF1 | Low | 9 | 19 | 2 | |
| 12 | RPL35B-RPL35A | High | 13 | 1 | 0 | High |
| 12 | PPH21-PPH22 | Low | 87 | 20 | 6 | High |
| 12 | LYS21-LYS20 | Low | 116 | 37 | 3 | High |
| 15 | BDF2-BDF1 | Low | 20 | 482 | 19 | |
| 15 | RPS29B-RPS29A | High | 2 | 8 | 1 | Low |
| 19 | PPZ2-PPZ1 | Low | 29 | 335 | 28 | |
| 19 | YDR438W-YML018C | Low | 15 | 251 | 8 | |
| 19 | YDR450W-RPS18B | High | 1 | 4 | 2 | Low |
| 19 | YDR451C-YOX1 | Low | 19 | 240 | 9 | |
| 26 | RPS26B-RPS26A | High | 4 | 12 | 0 | Low |
| 26 | PMD1-MDS3 | Low | 62 | 935 | 64 | |
| 29 | YGR221C-YHR149C | Low | 18 | 431 | 20 | |
| 29 | YGR230W-SPO12 | Low | 6 | 77 | 9 | |
| 29 | KEL2-KEL1 | Low | 36 | 499 | 34 | |
| 29 | YAP1802-YAP1801 | Low | 27 | 387 | 20 | |
| 29 | YGR243W-YHR162W | Low | 4 | 64 | 5 | |
| 29 | SOL4-SOL3 | Low | 16 | 147 | 13 | |
| 29 | ENO1-ENO2 | High | 48 | 14 | 7 | High |
| 29 | GND2-GND1 | Medium | 20 | 100 | 14 | |
| 30 | YGR004W-YLR324W | Low | 14 | 365 | 14 | |
| 30 | STF2-YLR327C | Low | 2 | 37 | 4 | |
| 30 | YGR010W-YLR328W | Low | 17 | 175 | 11 | |
| 30 | RPS25A-RPS25B | High | 2 | 3 | 0 | Low |
| 30 | ORM1-YLR350W | Low | 8 | 109 | 5 | |
| 30 | BUD9-BUD8 | Low | 15 | 210 | 24 | |
| 30 | YGR043C-TAL1 | Medium | 13 | 164 | 8 | |
| 30 | SCM4-YLR356W | Low | 6 | 158 | 4 | |
| 30 | RSC1-RSC2 | Low | 26 | 553 | 33 | |
| 30 | ROM1-ROM2 | Low | 54 | 673 | 46 | |
| 30 | YGR071C-YLR373C | Low | 35 | 671 | 31 | |
| 32 | YGL139W-YPL221W | Low | 33 | 371 | 26 | |
| 32 | RPL1B-RPL1A | High | 29 | 1 | 0 | High |
| 32 | PCL10-PCL8 | Low | 21 | 324 | 14 | |
| 32 | YGL133W-YPL216W | Low | 50 | 724 | 50 | |
| 33 | YGL084C-YPL189W | Low | 35 | 273 | 33 | |
| 33 | YGL082W-YPL191C | Low | 18 | 217 | 13 | |
| 33 | RPL7A-RPL7B | High | 37 | 5 | 1 | High |
| 33 | AFT1-YPL202C | Low | 9 | 388 | 10 | |
| 33 | PUS2-PUS1 | Low | 19 | 266 | 14 | |
| 34 | RPL11B-RPL11A | High | 30 | 7 | 0 | High |
| 34 | DBF2-DBF20 | Low | 23 | 215 | 18 | |
| 34 | ASK10-YPR115W | Low | 46 | 628 | 44 | |
| 34 | CLB1-CLB2 | Low | 13 | 100 | 17 | |
| 34 | CLB6-CLB5 | Low | 14 | 220 | 22 | |
| 34 | RPS23A-RPS23B | High | 5 | 0 | 3 | High |
| 34 | MEP1-MEP3 | Low | 16 | 188 | 21 | |
| 34 | ASN2-ASN1 | Low | 45 | 112 | 20 | |
| 34 | YGR131W-NCE102 | Low | 7 | 95 | 8 | |
| 34 | YGR136W-YPR154W | Low | 12 | 88 | 8 | |
| 34 | YGR141W-YPR157W | Low | 26 | 230 | 21 | |
| 34 | SKN1-KRE6 | Low | 43 | 290 | 31 | |
| 35 | YHL017W-PTM1 | Low | 6 | 176 | 7 | |
| 35 | YHL012W-UGP1 | Low | 17 | 365 | 8 | |
| 35 | LAG1-YKL008C | Low | 12 | 126 | 15 | |
| 35 | RPL14B-RPL14A | High | 13 | 5 | 0 | High |
| 35 | YHR001W-YKR003W | Low | 18 | 180 | 24 | |
| 37 | YHR115C-YNL116W | Low | 25 | 228 | 18 | |
| 37 | TOM72-TOM70 | Low | 25 | 385 | 24 | |
| 37 | EPT1-CPT1 | Low | 15 | 215 | 13 | |
| 37 | YHR131C-YNL144C | Low | 32 | 477 | 36 | |
| 37 | YHR133C-YNL156C | Low | 6 | 220 | 9 | |
| 37 | YCK1-YCK2 | Low | 25 | 243 | 22 | |
| 37 | SPS100-YGP1 | Medium | 12 | 212 | 10 | |
| 37 | RPL42B-RPL42A | High | 3 | 2 | 0 | High |
| 38 | UBP7-UBP11 | Low | 29 | 426 | 28 | |
| 38 | YIL151C-YKR096W | Low | 57 | 637 | 52 | |
| 38 | RPL40A-RPL40B | High | 0 | 8 | 3 | Low |
| 39 | TPM2-TPM1 | Low | 5 | 86 | 4 | |
| 39 | RPL16A-RPL16B | High | 5 | 14 | 3 | Low |
| 39 | FKH1-FKH2 | Low | 23 | 291 | 17 | |
| 39 | SIM1-SUN4 | Low | 5 | 11 | 4 | |
| 39 | YIL121W-YNL065W | Low | 21 | 390 | 17 | |
| 39 | YIL120W-YNL065W | Low | 21 | 406 | 19 | |
| 39 | POR2-POR1 | Low | 17 | 153 | 9 | |
| 39 | YIL113W-MSG5 | Low | 11 | 100 | 10 | |
| 39 | COX5B-COX5A | Low | 9 | 51 | 3 | |
| 39 | SEC24-YNL049C | Low | 41 | 449 | 29 | |
| 39 | YIL105C-YNL047C | Low | 29 | 373 | 28 | |
| 39 | PRK1-ARK1 | Low | 24 | 336 | 15 | |
| 40 | RPL17B-RPL17A | High | 5 | 14 | 1 | Low |
| 40 | HAL5-KKQ8 | Low | 31 | 459 | 32 | |
| 40 | TPK1-TPK3 | Low | 22 | 162 | 13 | |
| 40 | CIS3-PIR3 | High | 5 | 99 | 9 | |
| 41 | YUR1-KTR2 | Low | 25 | 212 | 13 | |
| 41 | TIF2-TIF1 | High | 58 | 1 | 0 | High |
| 41 | GLG2-GLG1 | Low | 22 | 261 | 20 | |
| 41 | RPS21B-RPS21A | High | 5 | 2 | 5 | High |
| 41 | LCB3-LBP2 | Low | 14 | 133 | 9 | |
| 41 | MRS3-MRS4 | Low | 7 | 140 | 9 | |
| 41 | TRK1-TRK2 | Low | 42 | 456 | 35 | |
| 41 | NCA3-UTH1 | Low | 22 | 147 | 9 | |
| 41 | YJL112W-CAF4 | Low | 27 | 423 | 18 | |
| 41 | GZF3-DAL80 | Low | 7 | 169 | 7 | |
| 41 | YJL105W-YKR029C | Low | 15 | 459 | 13 | |
| 41 | CHS6-YKR027W | Low | 22 | 408 | 23 | |
| 41 | SAP185-SAP190 | Low | 46 | 594 | 36 | |
| 41 | YJL084C-YKR021W | Low | 39 | 623 | 30 | |
| 41 | YJL083W-IRS4 | Low | 0 | 2 | 0 | |
| 41 | YJL082W-YKR018C | Low | 32 | 354 | 35 | |
| 44 | CNA1-CNA2 | Low | 25 | 258 | 22 | |
| 44 | RPS1A-RPS1B | High | 5 | 30 | 3 | Low |
| 44 | SIR3-ORC1 | Low | 30 | 743 | 30 | |
| 44 | RPL6B-RPL6A | High | 6 | 14 | 3 | Low |
| 44 | FPR4-FPR3 | Low | 10 | 188 | 15 | |
| 44 | HMG2-HMG1 | Low | 47 | 575 | 41 | |
| 45 | YLR266C-YRR1 | Low | 38 | 505 | 38 | |
| 45 | YLR270W-YOR173W | Low | 12 | 179 | 10 | |
| 45 | BRR5-YOR179C | Low | 9 | 90 | 13 | |
| 45 | RPS30A-RPS30B | High | 7 | 0 | 0 | High |
| 45 | GSP1-GSP2 | Medium | 4 | 47 | 4 | |
| 45 | EXG1-SPR1 | Low | 17 | 197 | 10 | |
| 47 | YMR222C-YOR280C | Low | 8 | 151 | 13 | |
| 47 | RPS10B-RPS10A | High | 3 | 10 | 0 | Low |
| 47 | YMR233W-YOR295W | Low | 11 | 168 | 14 | |
| 47 | YMR237W-BUD7 | Low | 32 | 342 | 22 | |
| 47 | RPL20A-RPL20B | High | 12 | 10 | 0 | High |
| 47 | ZRC1-COT1 | Low | 14 | 223 | 18 | |
| 47 | FAA4-FAA1 | Low | 51 | 307 | 35 | |
| 48 | MMT1-MMT2 | Low | 16 | 245 | 18 | |
| 48 | YMR180C-CET1 | Low | 14 | 229 | 15 | |
| 48 | YMR181C-YPL229W | Low | 6 | 120 | 5 | |
| 48 | RGM1-YPL230W | Low | 14 | 117 | 9 | |
| 48 | SSO2-SSO1 | Low | 10 | 119 | 12 | |
| 48 | YMR192W-YPL249C | Low | 26 | 500 | 24 | |
| 48 | RPL36A-RPL36B | High | 3 | 11 | 1 | Low |
| 48 | YMR195W-YPL250C | Low | 6 | 85 | 2 | |
| 48 | CIK1-VIK1 | Low | 21 | 484 | 26 | |
| 48 | CLN1-CLN2 | Low | 12 | 284 | 14 | |
| 49 | MCK1-YOL128C | Low | 15 | 305 | 13 | |
| 49 | RPS19B-RPS19A | High | 8 | 4 | 0 | High |
| 49 | TRF5-TRF4 | Low | 21 | 331 | 22 | |
| 49 | CLA4-SKM1 | Low | 30 | 369 | 25 | |
| 49 | MSB3-MSB4 | Low | 21 | 294 | 25 | |
| 49 | RFC3-RFC4 | Low | 12 | 172 | 7 | |
| 51 | DED1-DBP1 | Low | 20 | 291 | 18 | |
| 51 | YOR222W-YPL134C | Low | 12 | 163 | 4 | |
| 51 | YOR226C-YPL135W | Low | 5 | 65 | 5 | |
| 51 | YOR227W-YPL137C | Low | 60 | 707 | 42 | |
| 51 | YOR229W-UME1 | Low | 12 | 322 | 12 | |
| 51 | WTM1-UME1 | Low | 15 | 308 | 11 | |
| 51 | MKK1-MKK2 | Low | 28 | 260 | 24 | |
| 51 | KIN4-YPL141C | Low | 27 | 482 | 35 | |
| 51 | RPL33B-RPL33A | High | 2 | 7 | 2 | Low |
| 51 | HES1-KES1 | Low | 18 | 197 | 12 | |
| IV:VIII | STP1-STP2 | Low | 24 | 319 | 17 | |
| IV:VIII | RPL27B-RPL27A | High | 4 | 19 | 0 | Low |
| VII:VII | TIF4631-TIF4632 | Low | 35 | 492 | 29 | |
| VII:VII | RPL24B-RPL24A | High | 15 | 2 | 4 | High |
| VII:X | RNR4-RNR2 | Medium | 25 | 215 | 3 | |
| VII:X | BUB1-MAD3 | Low | 20 | 351 | 24 | |
| VII:X | TDH3-TDH2 | High | 10 | 0 | 7 | High |
| VIII:X | RPS4B-RPS4A | High | 26 | 1 | 0 | High |
“Block no.” is from Wolfe and colleagues (http://acer.gen.tcd.ie/~khwolfe/yeast/nova/). Blocks were analyzed only if they contained at least one gene pair of high CAI (see below), and gene pairs in such blocks were analyzed only if two different homologs could be found in S. bayanus (i.e., analysis was carried out only when it was possible to make the four-way alignment). CAI was considered “high” if both of the cerevisiae genes had a CAI >0.80; CAI was considered “medium” if at least one of the cerevisiae genes had a CAI >0.45 but at least one was <0.80; otherwise, CAI was considered “low.” C is the number of correction substitutions in the four-way alignment; L, the number of lineage substitutions in the four-way alignment; N, the number of noise substitutions in the four-way alignment. For all gene pairs with high CAI, the C/L ratio is noted as “high” (>1) or “low” (<1). With two exceptions, all gene pairs with a low CAI had low C/L ratios, and so are not noted. The two exceptions are noted by italics. Correction, lineage, and noise-type substitutions are defined in Figures 4 and 5 and Table 1.
First, we compared the low-bias genes to the high-bias genes. The high-bias genes tend to have a high C/L ratio (weighted mean C/L ratio = 2.1), while the low-bias genes tend to have a low C/L ratio (weighted mean C/L ratio = 0.078), and these ratios differ significantly between the two groups (P < 10−15 by a chi-square test).
Second, of 128 low-bias gene pairs, 126 have a low C/L ratio, as predicted. The two exceptions are LYS20-LYS21 and PPH21-PPH22, both members of block 12. These two genes in block 12 are close together on chromosome 4 and in the same orientation. That is, LYS20 is close to LYS21, and PPH21 is close to PPH22, all on chromosome 4. This situation is perhaps favorable for DNA-DNA conversion (e.g., during meiotic mispairing of these tightly linked tandem syntenic regions), and this may explain the high C/L ratio for these two gene pairs. That is, DNA-DNA-based correction may have occurred in the recent past, and DNA-DNA correction may not be as dependent on expression level as the RNA-DNA correction that we suggest for the majority of genes.
Third, of 33 high-bias genes, 20 have a C/L ratio >1, as predicted, but 13 have a C/L ratio <1; for instance, in the most extreme case, the RPS1A, RPS1B gene pair has 5 correction substitutions, but 30 lineage substitutions. Thus a substantial minority of the high-bias genes do not seem to be undergoing correction. For the 20 high-bias genes with C/L >1, the total number of events is 400 C, 57 L, and 29 N, for a C/L ratio of 7.0; and for the 13 high-bias genes with C/L <1, the total number of events is 40 C, 154 L, and 17 N, for a C/L ratio of 0.26. The distribution of the normalized number of C substitutions for the 33 high-bias genes appears to be bimodal, and statistical tests reject the hypothesis of a single normally distributed population (P < 10−7 using a Shapiro-Wilk normality test). Thus, there seem to be two kinds of high-bias genes: one kind that undergoes concerted evolution via correction and a second kind that does not. Possible reasons for these two populations are considered in the discussion.
Fourth, the 20 high-bias gene pairs with a C/L ratio >1 had a mean of 3.8% base-pair mismatches, while the 13 high-bias genes with a C/L ratio <1 had a mean of 8.2% base-pair mismatches, confirming that correction is associated with high sequence identity, while lack of correction is associated with divergence.
Fifth, we judged six gene pairs to be of medium codon bias. All six pairs showed the low C/L ratio typical of low-bias genes.
Counterarguments:
We have considered several alternative explanations for the unexpectedly high conservation between highly expressed S. cerevisiae gene pairs. One obvious alternative is that these gene pairs are not derived from the ancient genome-wide duplication event, but instead are the result of a much more recent chromosomal duplication. This is the case for several duplications found near telomeres, which we do not consider here. However, it appears not to be the case for the gene pairs that we consider here, because the unexpectedly high degree of homology among the gene pairs that we are considering ends abruptly at the boundary of the gene's open reading frame. The 5′ and 3′ noncoding regions of these genes do not show a strikingly high level of conservation. Furthermore, these genes are typically embedded in syntenic, duplicated regions, and there are typically poorly expressed, poorly conserved duplicated genes flanking the highly expressed, highly conserved genes. The exceptional genes in block 12, LYS20-LYS21 and PPH21-PPH22, could be a recent duplication.
A second alternative is that the preferred codons are different in different species. In this case, the lack of intra-cerevisiae divergence at high CAI would be explained by the need to maintain preferred codons, while the presence of interspecies divergence would be explained by highly expressed genes evolving to conform to a different, species-specific codon bias. However, three findings argue against this possibility. First, the preferred codons seem to be the same in each of the six species (Table 3). Second, a substantial proportion of the interspecies divergence (at least between S. cerevisiae and S. bayanus) is due to nonsynonymous base changes (Table 4), which of course is not explainable by differences in codon bias. Third, this argument does not explain gene families with a high proportion of correction nucleotide substitutions.
TABLE 3.
Preferred codons in six species of Saccharomyes
| Amino acid | S. bayanus | S. castelli | S. kluyveri | S. mikatae | S. paradoxus | S. cerevisiae |
|---|---|---|---|---|---|---|
| Ile | ATC 0.66 | ATT 0.51 | ATC 0.70 | ATC 0.54 | ATC 0.56 | ATC 0.58 |
| Asn | AAC 0.96 | AAC 1.00 | AAC 1.00 | AAC 0.92 | AAC 0.93 | AAC 0.94 |
| Asp | GAC 0.65 | GAT 0.50 | GAC 0.70 | GAC 0.63 | GAC 0.66 | GAC 0.65 |
| Gln | CAA 0.99 | CAA 0.98 | CAA 1.00 | CAA 0.98 | CAA 0.99 | CAA 1.00 |
| Ala | GCT 0.68 | GCT 0.69 | GCT 0.73 | GCT 0.74 | GCT 0.78 | GCT 0.80 |
| His | CAC 0.82 | CAC 0.71 | CAC 1.00 | CAC 0.84 | CAC 0.88 | CAC 0.90 |
| Thr | ACT 0.52 | ACC 0.50 | ACC 0.60 | ACT 0.51 | ACC 0.52 | ACT 0.50 |
| Tyr | TAC 0.91 | TAC 0.90 | TAC 1.00 | TAC 0.85 | TAC 0.91 | TAC 0.92 |
| Glu | GAA 0.97 | GAA 0.99 | GAA 0.94 | GAA 0.95 | GAA 0.98 | GAA 0.98 |
| Pro | CCA 0.91 | CCA 0.92 | CCA 0.96 | CCA 0.90 | CCA 0.92 | CCA 0.94 |
| Leu | TTG 0.83 | TTG 0.74 | TTG 0.95 | TTG 0.80 | TTG 0.83 | TTG 0.89 |
| Phe | TTC 0.84 | TTC 0.85 | TTC 0.86 | TTC 0.84 | TTC 0.83 | TTC 0.81 |
| Gly | GGT 0.94 | GGT 0.96 | GGT 0.95 | GGT 0.96 | GGT 0.96 | GGT 0.96 |
| Lys | AAG 0.88 | AAG 0.89 | AAG 1.00 | AAG 0.84 | AAG 0.84 | AAG 0.85 |
| Trm | TAA 0.90 | TAA 1.00 | TAA 1.00 | TAA 0.62 | TAA 0.80 | TAA 0.90 |
| Arg | AGA 0.84 | AGA 0.89 | AGA 0.93 | AGA 0.88 | AGA 0.84 | AGA 0.85 |
| Cys | TGT 0.86 | TGT 0.83 | TGT 1.00 | TGT 1.00 | TGT 1.00 | TGT 1.00 |
| Val | GTC 0.51 | GTT 0.50 | GTT 0.52 | GTT 0.60 | GTT 0.55 | GTT 0.55 |
| Ser | TCC 0.51 | TCT 0.58 | TCT 0.56 | TCT 0.53 | TCT 0.52 | TCT 0.50 |
Ten highly expressed genes of S. cerevisae (RPL11A, ENO1, TDH1, RPL4A, RPL8A, RPL9A, RPL15A, RPS2, RPS3, and RPS5) were selected, and their full-length closest homologs were identified in the other yeasts whenever possible. For each amino acid, the preferred codon and its frequency is listed for each yeast over the 10 selected proteins. Amino acids with only one codon (Met, Trp) are omitted. For most amino acids, all yeasts had the same preferred codon. For Ile, Asp, Thr, Val, and Ser, there were minor differences (indicated by italics). All of these minor differences occur when there are two commonly used codons, each with a frequency of close to 50%. For instance, in S. castelli, the preferred codon for Ile is ATT (frequency of 0.50), but the second-most preferred codon is ATC (frequency of 0.48). Similarly, in S. castelli, the preferred codon for Asp is GAT (frequency 0.50), but the second-most preferred codon is GAC (frequency 0.50). In the case of Thr, only two codons are substantially used, ACT and ACC, and these have nearly an equal frequency in each yeast. Similarly, for Ser, only TCC and TCT are substantially used, each at ∼0.5 in each yeast.
TABLE 4.
Synonymous and nonsynonymous nucleotide changes
| Gene | Length | C-C Syn | C-C Non | C-B Syn | C-B Non | C-M Syn | C-M Non |
|---|---|---|---|---|---|---|---|
| eno1 | 1314 | 41 | 27 | 68 | 30 | 40 | 2 |
| rpl11a | 525 | 14 | 2 | 34 | 4 | 19 | 0 |
| rpl1a | 654 | 4 | 0 | 31 | 5 | 31 | 0 |
| rps8a | 603 | 11 | 0 | 25 | 7 | 10 | 1 |
| tef1 | 1377 | 2 | 0 | 32 | 14 | 19 | 1 |
| tif51a | 474 | 28 | 15 | 30 | 10 | 16 | 6 |
Six highly expressed genes of S. cerevisiae were compared to their closest homolog in S. cerevisiae (“C-C” comparisons), in S. bayanus (“C-B” comparisons), or in S. mikatae (“C-M” comparisons). The number of synonymous (“Syn”) and nonsynonymous (“Non”) changes are tabulated.
Patterns and properties of correction:
Assuming that the unexpectedly high degree of conservation between pairs of highly expressed genes does reflect recombinational correction, we can draw some inferences about the properties of correction. First, correction ends abruptly at the boundaries of identity. This can be seen most easily at the beginning and end of each open reading frame. Within the open reading frame, the percentage identity can be 95% or more, but, immediately outside the open reading frame, identity decays to essentially random levels (data not shown). The same effect can be seen in genes with multiple exons. Although identity may be high within each exon, the introns show roughly random levels of identity.
If correction ends at the end of a tract of high homology, then the correction of different exons of the same gene may be independent events. In this case, the frequency of correction should be proportional to the length of the exon (since longer exons have an increased chance of interacting with each other). Indeed, in pairs of genes with multiple exons we have found that the degree of sequence identity between the first exons of a pair of genes can be different from the sequence identity between the second exons of the same pair of genes. Furthermore, longer exons typically have higher degrees of identity than shorter exons (although there are exceptions; data not shown).
Exon length and RNA-DNA correction:
If correction occurs via a cDNA intermediate, then this cDNA can correct the same gene that originally generated the cDNA, as well as any copy of the gene. Such self-correction would not have any effect on the nucleotide sequence of the open reading frame, since this would be identical between the cDNA and the gene. However, on (rare?) occasions when correction proceeded past a boundary of high sequence identity, it could remove an intron from the gene. Indeed, it has been proposed that this kind of self-correction is responsible for removing most of the (presumed) originally existing introns from the genome of S. cerevisiae (Fink 1987). The few introns that remain tend to be at the extreme 5′-end of the gene, suggesting that correction begins, or is more probable, at the 3′-end of the gene.
Interestingly, in many species, exons at the 3′-end of a gene tend to be longer than exons at the 5′-end of a gene (Table 5; see also Xia et al. 2003). This is consistent with the idea that, in many species, self-correction occurs via a cDNA intermediate, beginning at the 3′-end of the gene. Such self-correction would tend to remove 3′ introns, thus generating abnormally long 3′ exons.
TABLE 5.
Distribution of exon lengths
| 1-exon genes: |
2-exon genes
|
3-exon genes
|
||||
|---|---|---|---|---|---|---|
| Length of exon 1 |
Length of exon 1 |
Length of exon 2 |
Length of exon 1 |
Length of exon 2 |
Length of exon 3 |
|
| Drosophila | 934 | 403 | 643 | 271 | 454 | 458 |
| Arabidopsis | 970 | 441 | 489 | 346 | 284 | 402 |
| S. cerevisiae | 1419 | 313 | 1260 | 69 | 149 | 238 |
| S. pombe | 1462 | 301 | 946 | 206 | 279 | 721 |
| Caenorhabditis elegans | 611 | 229 | 327 | 178 | 267 | 244 |
Mean exon lengths for genes of 1, 2, and 3 exons are given.
DISCUSSION
For highly expressed gene pairs, within-species divergence is significantly less than between-species divergence, even though the within-species pairs have had a longer time to diverge (Figure 3). Furthermore, for the majority of highly expressed genes, the correction pattern of nucleotide substitution is much more common than the lineage pattern of substitution, while the opposite is true for poorly expressed genes (Figures 4 and 5; Tables 1 and 2). These observations are very difficult to explain by selection alone. We believe that selection and correction are synergistic with each other for highly expressed genes; selection for both function and codon bias tends to minimize the rate of drift, and the resulting high level of sequence identity keeps the gene pairs eligible for correction, which fully restores sequence identity between duplicates. There is less selection in poorly expressed genes, since codon bias is of little or no importance. In addition, there is less selection in the 5′ or 3′ regions of genes or within introns, since many base changes in these regions have little or no impact on gene function. Thus, poorly expressed genes, 5′ and 3′ untranslated regions, and introns drift more rapidly and soon diverge to the point where sequence identity is too low to allow a recombinational interaction. After this point, they are no longer eligible for correction and continue to drift apart with time.
Surprisingly, we found a substantial minority of highly expressed genes that have a low C/L ratio, i.e., that appear not to have corrected in the 20 MY since the split between cerevisiae and bayanus (Table 2). Why should some gene pairs fail to correct? In the context of the RNA-DNA correction model, one possibility is that some genes are more readily reverse transcribed than others. Xu and Boeke (1990) found that some cellular mRNAs copurified with Ty virus-like particles (VLPs; TRP1, HIS3, RPS17a), while other mRNAs did not (ACT1, GAL1, PYK1). The mRNAs copurifying with Ty VLPs may actually have been packaged within the particles, and so these mRNAs would presumably be more likely to be reverse transcribed than mRNAs not so packaged. Interestingly, RPS17a, which copurifies with Ty VLPs, is highly similar to its paralog RPS17b, while PYK1, which has a CAI similar to RPS17a, but which does not copurify with Ty VLPs, is highly diverged from its paralog, PYK2. A second possibility is that the genes that fail to correct are those where the two paralogs diverged significantly by chance before the cerevisiae-bayanus split and, because of the divergence, were no longer eligible for gene conversion and correction. A third possibility (see below) is that the two copies have taken on somewhat different cellular roles, and so both genes are needed.
Correction could occur by a DNA-DNA interaction between the two genes of the pair or by a cDNA-DNA (the RNA-DNA model) interaction occurring after reverse transcription of an mRNA. Our evidence does not distinguish these two models. The RNA-DNA model has several appealing features. First, it gives a clear expectation that correction should be more prevalent for highly expressed genes. Second, it is widely believed that self-correction via a cDNA does occur in S. cerevisiae, and if self-correction can occur, then correction of a copy should also occur. Third, it explains why the introns of highly expressed genes are not conserved, whereas a DNA-DNA interaction between two chromosomal genes might tend to correct these introns as well as flanking exons. On the other hand, DNA-DNA events seem to be much more frequent than RNA-DNA events (Derr and Strathern 1993), and so one would expect correction to be dominated by the DNA-DNA mechanism, even if RNA-DNA events sometimes occurred.
It is unclear to what extent similar events may occur in other organisms. S. cerevisiae has a highly active system for homologous recombination and thus is especially suited to correction. However, most other organisms also have homologous recombination, and many or most other eukaryotes contain reverse transcriptases. We therefore imagine that correction could occur at some level in many or most other organisms. Table 5 shows that 3′ exons are typically longer than 5′ exons for many organisms, and this is consistent with correction. Zhang et al. (2002) have shown that there are >2000 reverse-transcribed ribosomal protein pseudogenes in the human genome, showing that the conversion of a transcript to a cDNA is a reasonably common event in humans. Zhang et al. (2002) have also shown the existence of a number of duplicate ribosomal protein genes. If any of these duplicates predate the divergence between, e.g., humans and mice, then analysis of these duplicates, such as we have done here with Saccharomyces, may show whether these pairs are maintained by correction in mammals.
We note that correction could occur between a highly expressed gene and an unexpressed pseudogene. We have preliminary data from S. cerevisiae suggesting that, in a few cases, one member of a pair of ribosomal protein genes is expressed poorly; possibly such poorly expressed genes are maintained by correction from their highly expressed twin. Furthermore, this idea could explain the maintenance of the large number of nonmutated ribosomal protein pseudogenes that are present in the human genome (Zhang et al. 2002).
About 90% of the genes originally duplicated in the ancient duplication event have since been deleted, while ∼10% remain as duplicates. Why do these 10% remain? Our results and the recent results of Kellis et al. (2004) allow us to point to two kinds of reasons. First, for poorly expressed genes, one member of the gene pair seems to evolve quickly, gaining many substitutions rapidly and acquiring a new biological role (Kellis et al. 2004). Thus, for poorly expressed genes, sequence divergence is favorable for maintaining the copy. Second, for highly expressed genes, we now argue that the duplication aids in making large amounts of protein in cases where large amounts of protein are needed. Thus, for highly expressed genes, sequence conservation is favorable for maintaining the copy. Figure 3, b and c, supports this view, because it shows that for poorly expressed genes, nonsynonymous substitutions are relatively favored (i.e., promoting divergence of protein function), while for highly expressed genes, synonymous substitutions are relatively favored (i.e., conserving protein function). Correction fits into this scheme well, since correction seems to work only on highly expressed genes, which are precisely the genes where sequence conservation, and not divergence, leads to preservation of the copy. It is interesting to speculate that the rare, highly expressed genes not showing copy correction are highly expressed genes that have nevertheless evolved to take on new cellular roles. It is interesting to note that deletion of either copy of RPS1a/b (the most extremely diverged high-bias gene pair) leads to severe growth defects (Saccharomyces Genome Database), suggesting that the two copies may have nonoverlapping roles.
Acknowledgments
This work was sponsored by National Institutes of Health grants GM39978 and GM648131 to B.F. and National Science Foundation grant EIA0325123 to S.S.
References
- Baltimore, D., 1985. Retroviruses and retrotransposons: the role of reverse transcription in shaping the eukaryotic genome. Cell 40: 481–482. [DOI] [PubMed] [Google Scholar]
- Chen, W., and S. Jinks-Robertson, 1998. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol. Cell. Biol. 18: 6525–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton et al., 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71–76. [DOI] [PubMed] [Google Scholar]
- Datta, A., M. Hendrix, M. Lipsitch and S. Jinks-Robertson, 1997. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc. Natl. Acad. Sci. USA 94: 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr, L. K., and J. N. Strathern, 1993. A role for reverse transcripts in gene conversion. Nature 361: 170–173. [DOI] [PubMed] [Google Scholar]
- Derr, L. K., J. N. Strathern and D. J. Garfinkel, 1991. RNA-mediated recombination in S. cerevisiae. Cell 67: 355–364. [DOI] [PubMed] [Google Scholar]
- Fink, G. R., 1987. Pseudogenes in yeast? Cell 49: 5–6. [DOI] [PubMed] [Google Scholar]
- Futcher, B., G. I. Latter, P. Monardo, C. S. McLaughlin and J. I. Garrels, 1999. A sampling of the yeast proteome. Mol. Cell. Biol. 19: 7357–7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., N. Patterson, M. Endrizzi, B. Birren and E. S. Lander, 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254. [DOI] [PubMed] [Google Scholar]
- Kellis, M., B. W. Birren and E. S. Lander, 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624. [DOI] [PubMed] [Google Scholar]
- Modrich, P., and R. Lahue, 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65: 101–133. [DOI] [PubMed] [Google Scholar]
- Nei, M., and T. Gojobori, 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Powell, J. R., and E. N. Moriyama, 1997. Evolution of codon usage bias in Drosophila. Proc. Natl. Acad. Sci. USA 94: 7784–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe, D., A. Datta and S. Jinks-Robertson, 2000. Stimulation of mitotic recombination events by high levels of RNA polymerase II transcription in yeast. Mol. Cell. Biol. 20: 5404–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe, C., and K. H. Wolfe, 1999. Updated map of duplicated regions in the yeast genome. Gene 238: 253–261. [DOI] [PubMed] [Google Scholar]
- Sharp, P. M., and W. H. Li, 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K. H., and D. C. Shields, 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713. [DOI] [PubMed] [Google Scholar]
- Xia, X., Z. Xie and W. H. Li, 2003. Effects of GC content and mutational pressure on the lengths of exons and coding sequences. J. Mol. Evol. 56: 362–370. [DOI] [PubMed] [Google Scholar]
- Xu, H., and J. D. Boeke, 1990. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: analysis of mini-Ty1 elements. Mol. Cell. Biol. 10: 2695–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., P. Harrison and M. Gerstein, 2002. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res. 12: 1466–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]