Abstract
High-diversity genes represent an important class of loci in organismal genomes. Since elevated levels of nucleotide variation are a key component of the molecular signature for balancing selection or local adaptation, high-diversity genes may represent loci whose alleles are selectively maintained as balanced polymorphisms. Comparison of 4300 random shotgun sequence fragments of the Arabidopsis thaliana Ler ecotype genome with the whole genomic sequence of the Col-0 ecotype identified 60 genes with putatively high levels of intraspecific variability. Eleven of these genes were sequenced in multiple A. thaliana accessions, 3 of which were found to display elevated levels of nucleotide polymorphism. These genes encode the myb-like transcription factor MYB103, a putative soluble starch synthase I, and a homeodomain-leucine zipper transcription factor. Analysis of these genes and 4–7 flanking genes in 14–20 A. thaliana ecotypes revealed that two of these loci show other characteristics of balanced polymorphisms, including broad peaks of nucleotide diversity spanning multiple linked genes and an excess of intermediate-frequency polymorphisms. Scanning genomes for high-diversity genomic regions may be useful in approaches to adaptive trait locus mapping for uncovering candidate balanced polymorphisms.
UNCOVERING the genetic basis of adaptation has been a central goal of evolutionary genetics for nearly a century (Orr and Coyne 1992), and recent advances in genetic analysis have permitted the identification and isolation of loci responsible for speciation (Greenberg et al. 2003; Barbash et al. 2004), species differences (Doebley et al. 1997; Gompel and Carroll 2003), and adaptive intraspecific variation (Johanson et al. 2000; Kroymann et al. 2003). Several approaches based on patterns of molecular evolution have been proposed to scan genomes for genes associated with adaptation (Nielsen 2001; Swanson et al. 2001a,b; Schlotterer 2002; Bamshad and Wooding 2003; Barrier et al. 2003). These methods provide opportunities to analyze evolutionary diversification at both molecular genetic and phenotypic levels.
Approaches for mapping adaptive trait loci are based on detecting regions of the genome in which intraspecific sequence variation and/or interspecific divergence deviate either from predictions of a neutral-equilibrium model (Nielsen 2001) or from the norm of a genome-wide distribution (Otto 2000; Luikart et al. 2003). Evolutionary expressed sequence tag (EST) (Swanson et al. 2001a,b; Barrier et al. 2003) and comparative genomic approaches (Clark et al. 2003), for example, use interspecific patterns of nonsynonymous/synonynmous substitution ratios (Ka/Ks) to identify candidate adaptive genes on the basis of accelerated rates of protein evolution (Ka/Ks > 1). Genes and genomic regions associated with directional selection have also been identified by scanning dense sets of genome-wide molecular markers for reduced levels of variation (Harr et al. 2002; Payseur et al. 2002; Schlotterer 2002; Vigouroux et al. 2002; Wootton et al. 2002; Storz et al. 2004). The latter approach, referred to as hitchhiking mapping, is based on the premise that a beneficial mutation that rapidly spreads in a population will also reduce nucleotide variation at linked neutral loci. Hitchhiking mapping has successfully identified several genomic regions containing putative adaptive trait loci that were thought to contribute to the worldwide colonization of Drosophila melanogaster out of Africa ∼10,000 years ago (Harr et al. 2002). Although genome scanning for putative adaptive trait loci on the basis of levels of molecular diversity has focused largely on identifying genes associated with directional selection, this approach could also be employed in identifying genes and/or genomic regions that harbor balanced polymorphisms. This could complement other genome-scanning approaches, such as screens for elevated FST estimates in marker loci between two populations (Akey et al. 2002), in identifying genes under this selective regime.
Balanced polymorphisms are characterized by two or more alleles that are selectively maintained at intermediate frequencies within populations or species. Frequency-dependent selection (Charlesworth and Awadalla 1998; Bergelson et al. 2001), heterozygote advantage (Bamshad and Wooding 2003), and local adaptation (Kohn et al. 2000; Schulte et al. 2000; Gilad et al. 2002; Harr et al. 2002; Kohn et al. 2003; Storz et al. 2004) are some of the major selective mechanisms that can maintain balanced polymorphisms. These polymorphisms are also associated with specific levels and patterns of nucleotide variation at the selective target and in linked genomic regions (Strobeck 1983; Hudson and Kaplan 1988) and thus provide a molecular signature of adaptation that can aid in their identification. This signature can include increased levels of within-species diversity as well as intermediate-frequency polymorphisms (Bamshad and Wooding 2003), which, when maintained for long periods of time, are thought to result in trans-specific polymorphism (Schierup et al. 1998). Models of balancing or spatially heterogeneous selection also predict peaks of increased nucleotide diversity centered on a balanced polymorphism, which decrease symmetrically with distance (Hudson and Kaplan 1988; Nordborg 1997). Additional, less definitive features of a balanced polymorphism include high levels of linkage disequilibrium and a deficiency in the number of observed haplotypes (Charlesworth 2003), which result from selective hitchhiking. These characteristics of the signature of a balanced polymorphism have been observed in studies of several genes and/or gene regions in diverse species, including the human class I and II MHC (Garrigan and Hedrick 2003), Drosophila Adh (Kreitman and Aguade 1986; Kreitman and Hudson 1991), Fundulus Ldh (Schulte et al. 2000), Arabidopsis thaliana MAM (Kroymann et al. 2003), and disease resistance loci in plants (Stahl et al. 1999; Bergelson et al. 2001; Tian et al. 2002; Charlesworth et al. 2003).
Although balanced polymorphisms have been observed in various organisms, highly self-fertilizing species, like the model organism A. thaliana, are thought to be especially well suited for the identification and analysis of genes subject to balancing selection (Nordborg 1997; Tian et al. 2002; Shepard and Purugganan 2003). A. thaliana outcrosses at a rate of ∼1%, resulting in a low effective rate of recombination (Abbot and Gomes 1989), and linkage disequilibrium that can extend over genomic regions spanning ∼50–250 kb (Nordborg et al. 2002). This reduced effective recombination rate in A. thaliana should maintain correlations between nucleotide polymorphisms over larger distances and longer persistence times and facilitate the discovery of balanced polymorphisms.
The pattern of nucleotide variation associated with a balanced polymorphism in A. thaliana is illustrated by the RPS5 disease resistance locus (Tian et al. 2002). A genomic area centered on RPS5 ∼5.8 kb in length shows increased sequence variability, with silent-site levels of nucleotide diversity (π) at 0.025. In this instance, the balanced polymorphism appears to result in persistent haplotype dimorphism across this genomic region, although the dimorphic allele classes of closely linked genes are no longer associated with particular disease resistance alleles. Several of the features of the balanced polymorphism at RPS5 are also shared with the genomic region of CLAVATA2, a gene encoding a leucine-rich repeat involved in A. thaliana shoot meristem development (Shepard and Purugganan 2003), which also may be subject to balancing selection.
The predominantly selfing nature of A. thaliana, the availability of large-scale genome sequences from two Arabidopsis ecotypes, Columbia (Col-0) (Arabidopsis Genome Initiative 2000) and Landsberg erecta (Ler) (Jander et al. 2002), and current efforts to develop a species-wide haplotype map (http://walnut.usc.edu/2010/) provide a unique opportunity to scan the whole genome of this model plant for high-diversity genes that may arise from balanced polymorphisms. It is unclear, however, whether such an approach can identify these adaptive polymorphisms, since other evolutionary forces such as mutation, gene duplication, and population structuring could also result in high-diversity genes. Disentangling these alternative possibilities and determining the utility of a genome-scanning approach for identifying balanced polymorphisms requires a better understanding of the levels and patterns of nucleotide variation for high-diversity genes and their associated genomic regions.
In this article we report on a molecular population genetic analysis of high-diversity genes and genomic regions in the model genetic organism A. thaliana. From a comparison of genome sequence data between the Col-0 and Ler A. thaliana ecotypes, we have identified three gene fragments that show divergence between these two ecotypes of >5%. The levels and patterns of nucleotide polymorphism in the genomic regions spanning these high-diversity gene fragments were also ascertained, thereby providing the foundation for determining the utility of large-scale intraspecific genome scans in identifying candidate genes that may harbor balanced polymorphisms.
MATERIALS AND METHODS
Identification of genes for analysis:
Approximately 4300 genome-wide shotgun sequence fragments from the A. thaliana Ler ecotype (Jander et al. 2002) were compared to the Col-0 ecotype full-genome sequence (Arabidopsis Genome Initiative 2000) through a large-scale BLAST analysis (Altschul et al. 1990). Sequences that were 2–10% divergent between these two ecotypes were identified. Transposable elements, genes producing proteins <150 amino acids in length, pseudogenes, and duplicate gene copies were excluded from this sequence list. Eleven of these high-diversity gene fragments were sequenced in five to six additional A. thaliana ecotypes to confirm the high levels of within-species polymorphism at these loci. Genes were included in further analysis if elevated levels of nucleotide diversity were validated and if the gene fragments displayed significantly positive Tajima's D or Fu and Li's D* value.
Isolation and sequencing of alleles:
Genomic DNA was isolated from young leaves of 21 A. thaliana accessions (supplementary Table S1 at http://www.genetics.org/supplemental/) and from one to three A. lyrata plants using the plant DNeasy mini kit (QIAGEN, Valencia, CA). The A. thaliana accessions primarily span the geographic range of this species in Europe, although some Asian and North African accessions are included (see supplementary Table S1 at http://www.genetics.org/supplemental/). A. lyrata seed from a Karhumaki, Russia, population was provided by O. Savolainen (University of Oulu) and Helmi Kuittinen (University of Barcelona). PCR primers were designed from Col-0 genomic BAC sequences using Primer3 (Rozen and Skaletsky 2000). Primers were designed to amplify ∼1-kb regions of the three confirmed high-diversity genes identified by our BLAST analysis (AT1G63910, AT5G24300, AT1G19700) (Table 1 and supplementary Tables S2 and S3 at http://www.genetics.org/supplemental/). Primers were also designed to amplify 0.5- to 1-kb regions of genes flanking each of our identified genes to assess the extent to which elevated levels of polymorphism reach into the flanking chromosomal region. Flanking genes were sampled from each side of our identified gene until levels of nucleotide diversity dropped near the A. thaliana mean.
TABLE 1.
Genes contained in each of the three identified high-diversity regions listed in 5′–3′ order from top to bottom
| Locus | Gene product description |
|---|---|
| High-diversity region 1 | |
| AT1G63940a | Monodehydroascorbate reductase (putative) |
| AT1G63930 | Expressed protein |
| AT1G63920 | Pseudogene, similar to putative AP endonuclease/reverse transcriptase |
| AT1G63910a | MYB family transcription factor (MYB103) |
| AT1G63900a | Zinc-finger (C3HC4-type RING finger) family protein |
| AT1G63880 | Disease resistance protein (TIR-NBS-LRR class) (putative) |
| AT1G63870 | Disease resistance protein (TIR-NBS-LRR class) (putative) |
| AT1G63860 | Pseudogene, disease resistance protein |
| AT1G63857 | Expressed protein |
| AT1G63855 | Expressed protein |
| AT1G63850a | PRLI-interacting factor related |
| AT1G63840a | Zinc-finger (C3HC4-type RING finger) family protein |
| High diversity region 2 | |
| AT5G24280a | Expressed protein |
| AT5G24290a | Integral membrane family protein |
| AT5G24300a | Starch synthase (putative) |
| AT5G24310a | Expressed protein |
| AT5G 24313 | Expressed protein |
| AT5G24314a | Expressed protein |
| AT5G24316 | Proline-rich family protein |
| MOP9.15a,b | Expressed protein |
| AT5G24320 | WD-40 repeat family protein |
| AT5G24330 | PHD finger family protein/SET domain-containing protein |
| AT5G24340a | 3′–5′ exonuclease domain-containing protein |
| AT5G24350a | Expressed protein |
| High diversity region 3 | |
| AT1G19715a | Jacalin lectin family protein |
| AT1G19710a | Glycosyl transferase family 1 protein |
| AT1G19700a | Homeobox-leucine zipper family protein |
| AT1G19690a | Expressed protein |
| AT1G19680a | Expressed protein |
Genes included in analysis.
BAC locus annotation for a predicted gene with EST support.
PCR of A. thaliana and A. lyrata samples was performed using either Taq DNA polymerase (Roche, Indianapolis) or ExTaq DNA polymerase (Takara, Madison, WI). Amplified DNA fragments were purified using QIAquick PCR purification or gel extraction kits (QIAGEN). A. thaliana PCR products were cycle sequenced directly with Big Dye terminators and run on Prism 3700 96 capillary automated sequencers (Applied Biosystems, Foster City, CA) at the North Carolina State University Genome Research Laboratory. Amplified A. lyrata products were cloned using the TOPO TA PCR cloning kit (Invitrogen, San Diego), and plasmid DNA was isolated using the QIAaprep spin miniprep (QIAGEN). The presence of inserts in plasmid clones was confirmed by restriction digests using EcoRI and five to six independent clones were identified for sequencing. The PHRED and PHRAP functions (Ewing and Green 1998; Ewing et al. 1998) of Biolign (Tom Hall, North Carolina State University) were used in base calling and creating sequence contigs. All polymorphisms were visually confirmed, and questionable polymorphisms were rechecked through PCR reamplification and sequencing. Nucleotide sequence alignments and tables of polymorphic sites are available upon request.
Molecular population genetic analysis:
Sequences were visually aligned using the A. thaliana Col-0 sequence as a reference. DnaSP 3.99 (Rozas et al. 2003) was used for intraspecific analysis of polymorphism data. Nucleotide diversity was estimated for silent sites as both π (Tajima 1983) and θw (Watterson 1975). MEGA2.0 (Kumar et al. 2001) was used to calculate interspecific silent-site nucleotide divergence (K) between Col-0 and one A. lyrata individual for each gene, using the Kimura two-parameter model. Tajima's D (Tajima 1993) and both Fu and Li's D and D* (with and without outgroup, respectively) (Fu and Li 1993), haplotype number, and the intragenic linkage disequilibrium statistic ZnS (Kelly 1997) were also estimated for each gene. Statistical significance of these estimates was determined by coalescent simulations with 10,000 runs, conditioning on the number of segregating sites and under the conservative assumption of no recombination. Levels of linkage disequilibrium both within and between genes in a region were estimated using the r2 statistic based on informative sites (Hill and Robertson 1968), and significant associations were determined using Fisher's exact test.
The HKA (Hudson et al. 1987) test of selection was applied using the multilocus HKA program available from Jody Hey (http://lifesci.rutgers.edu/~heylab/HeylabSoftware.html). Only exon sequences were used in HKA tests for the expressed protein-encoding gene AT5G24310 since, for this gene, intron sequences between species were difficult to align with confidence. Complete sequences were analyzed in all other cases. Tests were based on silent sites and comparisons were made between each of our sequenced genes and a set of three neutral reference genes. These three reference loci, PI, AP1, and FAH, all have θ vs. K values that fall within the 95% confidence limit of the regression of genome-wide nucleotide diversity on interspecific divergence (Schmid et al. 2005; R. C. Moore and P. Awadalla, personal communication). Bonferroni corrections for multiple testing were applied for all tests of selection that were conducted across multiple linked genes in a given region. Allelic relationships were inferred using the neighbor-joining algorithm in MEGA 2.0 under the Kimura two-parameter substitution model and handling gaps and missing data as pairwise deletions.
Estimation of local recombination rates:
Genetic markers were obtained from the Lister and Dean Col × Ler recombinant inbred map (Lister and Dean 1993), and marker physical distances were obtained from the The Arabidopsis Information Resource database (ftp://tairpub:tairpub@ftp.arabidosis.org/home/tair/Maps/mapviewer_data). Markers with known genetic and physical map positions were ordered according to their physical positions and noncollinear markers were removed. Recombination rates were calculated between each pair of adjacent markers. Local recombination rate estimates were taken as the estimated rate between the two closest markers flanking the region of interest.
RESULTS
Genome scanning for high-diversity genes in A. thaliana:
A large-scale comparison of ∼2.5 Mb of Ler genomic shotgun sequence fragments against the Col-0 whole-genome sequence provides a preliminary scan for high-diversity genes in the A. thaliana genome. In this species, silent-site nucleotide diversity has been estimated to be ∼0.7% (Yoshida et al. 2003). On the basis of this consideration, we chose gene fragments with an interecotype divergence range of 2–10% as representing putative high-diversity loci. The low end of this range is comparable to the nucleotide diversity estimate for the RPS5 disease resistance gene (π = 2.5%), which has been shown to be under balancing selection (Tian et al. 2002). The high end of the range is slightly less than the interspecific divergence estimate (K = 12%) between A. thaliana and its sister species, A. lyrata (Barrier et al. 2003). We also removed repetitive sequences (e.g., transposable elements, duplicate genes), pseudogenes, and short, hypothetical genes from the list of putative high-diversity loci.
We identified a list of 60 functionally annotated genes ranging from 2–10% divergence between Col and Ler. From this list we chose 11 genes that spanned the specified range of divergence; these were arbitrarily chosen on the basis of functional annotation (e.g., transcription factor genes). Since the divergence estimates are based on raw shotgun genome sequence data from Ler, it is possible that several of these putative high-diversity estimates arose from sequencing errors or the presence of a rare divergent allele. We conducted another round of screening to confirm which genes truly represent high-diversity loci that could be candidates for further study. Fragments of ∼0.5–1.0 kb in length were sequenced in each of the 11 putative high-diversity genes in five to six additional ecotypes to confirm the observed elevated levels of polymorphism for these loci. Silent-site nucleotide diversity and Tajima's D and Fu and Li's D* were estimated for the genes in this second screen. Genes were included for further analysis if they had (i) silent-site nucleotide diversity (π) >3% and (ii) positive Tajima's D or Fu and Li's D* that were significantly higher than neutral-equilibrium expectations.
Of the 11 genes examined in the secondary screen, 3 were confirmed to have high levels of silent-site nucleotide diversity and significantly positive Tajima's D and/or Fu and Li's D* test statistics. These three genes are: (i) AT1G63910, which encodes the myb-like transcription factor MYB103; (ii) AT5G24300, which encodes a putative soluble starch synthase I enzyme; and (iii) AT1G19700, which encodes a member of the homeobox-leucine zipper transcription factor gene family.
High-diversity genomic regions:
One known signature of a balanced polymorphism is a peak of elevated nucleotide diversity centered on the target of selection (Tian et al. 2002). The high-diversity gene fragments identified in this study may represent this peak of elevated nucleotide diversity or may represent the effect of genetic hitchhiking with the target of selection at a linked locus. Alternatively, the observed high diversity may not be due to a balanced polymorphism, but may arise from alternative genetic/genomic or demographic factors. Discriminating among these alternatives requires a detailed examination of the levels and patterns of nucleotide variation not only in the three high-diversity genes identified in the genome scan, but also in an extended genomic region surrounding these loci. If these high-diversity genes are associated with balanced polymorphisms, one might expect a broad region of elevated nucleotide polymorphism spanning several genes, given the reduced effective recombination rate in the predominantly selfing A. thaliana. We thus isolated and sequenced ∼0.5- to 1.2-kb fragments from these high-diversity genes and from five to eight flanking loci in 14–20 A. thaliana accessions across each of these genomic regions (Table 1). The orthologous gene fragments in the sister species A. lyrata were also isolated and sequenced to serve as an outgroup for comparison.
Our genome scan initially identified the gene MYB103 (Li et al. 1999), located in the middle of the bottom arm of chromosome I, as a high-diversity locus. We have designated a 44.2-kb genomic region associated with MYB103 (AT1G63910) as high-diversity region 1. This region contains 12 annotated loci, and, aside from MYB103, we sequenced and analyzed genes encoding a putative monodehydroascorbate reductase (AT1G63940), two C3HC4-type zinc-finger proteins (AT1G68900 and AT1G63840), and a PRLI-interacting factor-related protein (AT1G63850) that together span this region. One striking feature of this genomic region is the presence of two putative TIR-NBS-LRR-type disease-resistance genes that encode proteins with Toll and interleukin-1 receptor (TIR), nucleotide-binding site (NBS), and leucine-rich repeat (LRR) domains and one disease-resistance pseudogene tandemly located together in a gene block in the Col-0 accession. PCR primers designed from the Col-0 sequence to amplify the second TIR-NBS-LRR putative disease resistance gene were successful in only 9 of the 19 ecotypes attempted. Previous work has suggested that balancing selection can act on the presence/absence of alleles of disease resistance genes; our results suggest that this TIR-NBS-LRR cluster may be a target of selection, and the elevated nucleotide variation in this genomic region may result from genetic hitchhiking.
Elevated nucleotide diversity at a putative soluble starch synthase I gene on chromosome V was used to identify high-diversity region 2. The region we analyzed encompasses 59.5 kb and includes 12 annotated loci. We examined an additional seven genes in this region, including five annotated genes that have no known function, but which all show evidence of being transcriptionally expressed. EST analyses indicate that two of these genes are associated with full-length cDNAs (AT5G24280 and AT5G24214); one gene is supported by a near full-length cDNA, which lacks only the first 20 bases of sequence (MOP9.15), and the remaining two genes are associated with an EST hit at least 500 bp in length (AT5G24210 and AT5G24250). Other genes in high-diversity region 2 that were sequenced include one encoding an integral membrane family protein (AT5G24290) and a protein containing a 3′–5′ exonuclease domain (AT5G24340).
High-diversity region 3, in the middle of the top arm of chromosome I, was identified in the genome scan by elevated nucleotide polymorphism in a gene encoding a homeobox-leucine zipper family protein. We analyzed this region, which spans 21.3 kb and includes five annotated loci. Other sequenced genes in this region include a jacalin lectin family protein (AT1G19715), a glycosyl transferase family 1 protein (AT1G19710), and two other expressed proteins of unknown function (AT1G19690 and AT1G19680).
Silent-site nucleotide variation across high-diversity genomic regions:
Increased nucleotide variation in high-diversity genomic regions 1 and 2 was not confined to the initially identified high-diversity gene. In both of these genomic regions, other genes also displayed elevated levels of nucleotide polymorphism. In high-diversity region 1, which spans 44.2 kb, we sequenced a total of 3441 nucleotide sites, including 1559 silent sites. A total of 195 single nucleotide polymorphisms (SNPs) were identified by the analysis, including 164 silent-site polymorphisms. Four of the five genes surveyed in high-diversity region 1 have silent-site nucleotide diversity levels (π) ranging from 0.022 to 0.089 (Table 2), which is 3- to 12-fold higher than the mean level of 0.007 observed from previously studied A. thaliana nuclear genes (Yoshida et al. 2003).
TABLE 2.
Measures of diversity for the sampled genes in each high-diversity region
| Gene | na | Length (bp)b | No. of silent sites | Sc | S (silent)d | πe | θwe | Kf |
|---|---|---|---|---|---|---|---|---|
| High-diversity region 1 | ||||||||
| AT1G63940 | 16 | 919 | 642.17 | 34 | 34 | 0.022 | 0.016 | 0.094 ± 0.012 |
| AT1G63910 | 18 | 411 | 82.18 | 54 | 24 | 0.089 | 0.085 | 0.236 ± 0.049 |
| AT1G63900 | 14 | 846 | 483.69 | 80 | 80 | 0.068 | 0.052 | 0.130 ± 0.017 |
| AT1G63850 | 18 | 863 | 258.46 | 24 | 23 | 0.024 | 0.026 | 0.122 ± 0.021 |
| AT1G63840 | 19 | 402 | 92.50 | 3 | 3 | 0.005 | 0.009 | 0.149 ± 0.036 |
| High-diversity region 2 | ||||||||
| AT5G24280 | 17 | 711 | 340.2 | 6 | 2 | 0.002 | 0.001 | 0.173 ± 0.024 |
| AT5G24290 | 18 | 704 | 341.7 | 5 | 4 | 0.002 | 0.003 | 0.122 ± 0.019 |
| AT5G24300 | 18 | 1240 | 793.6 | 63 | 62 | 0.035 | 0.023 | 0.071 ± 0.008 |
| AT5G24310 | 18 | 681 | 395.61 | 23 | 22 | 0.024 | 0.016 | 0.179 ± 0.027 |
| AT5G24314 | 17 | 827 | 728.6 | 107 | 104 | 0.056 | 0.042 | 0.151 ± 0.017 |
| MOP9.15g | 15 | 761 | 250.89 | 45 | 29 | 0.044 | 0.036 | 0.143 ± 0.024 |
| AT5G24340 | 15 | 797 | 416.38 | 3 | 1 | 0.001 | 0.001 | 0.086 ± 0.015 |
| AT5G24350 | 16 | 807 | 594.24 | 18 | 16 | 0.006 | 0.008 | 0.124 ± 0.015 |
| High-diversity region 3 | ||||||||
| AT1G19715 | 18 | 646 | 237.4 | 7 | 5 | 0.002 | 0.006 | 0.132 ± 0.023 |
| AT1G19710 | 18 | 896 | 339.9 | 5 | 4 | 0.003 | 0.003 | 0.134 ± 0.019 |
| AT1G19700 | 20 | 990 | 411.3 | 50 | 43 | 0.051 | 0.032 | 0.124 ± 0.017 |
| AT1G19690 | 17 | 576 | 425.6 | 13 | 12 | 0.006 | 0.008 | 0.112 ± 0.017 |
| AT1G19680 | 17 | 542 | 127.3 | 5 | 2 | 0.002 | 0.005 | 0.114 ± 0.027 |
Number of samples.
Length of sequenced region.
Number of segregating sites in the sample.
Number of segregating silent sites in the sample.
Estimates are based on silent sites.
Divergence between the A. thaliana Colombia-0 accession and A. lyrata based on silent sites including standard errors.
BAC locus annotation for a predicted gene with EST support.
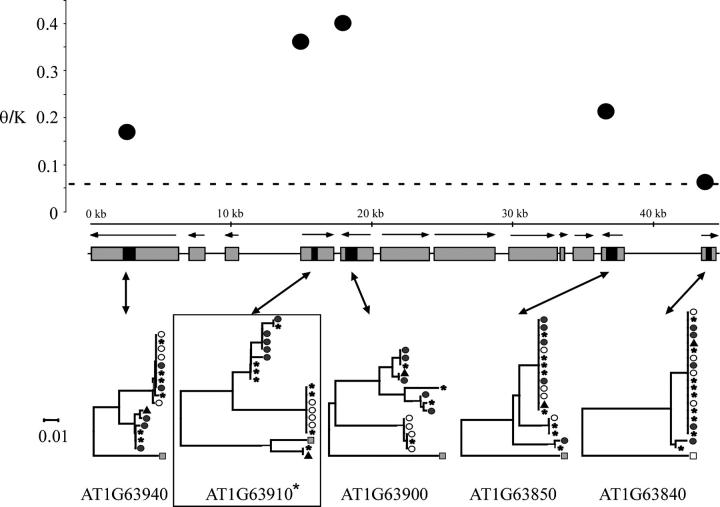
This increased intraspecific nucleotide diversity could reflect increased neutral mutation rates for these loci. Variation in neutral mutation rates should be mirrored by differences in interspecific nucleotide substitution rates, and we therefore examined the silent-site nucleotide divergence (K) between these A. thaliana genes and their A. lyrata orthologs. Silent-site interspecific nucleotide divergence (K) estimates for these genes range from 0.09 to 0.24; the mean K between A. thaliana and A. lyrata is 0.12 (Barrier et al. 2003). Figure 1 depicts the ratio of θ/K across this region; the mean value for previously studied A. thaliana genes is ∼0.06 (Barrier et al. 2003). The ratio θ/K is three- to sevenfold higher than the mean for A. thaliana nuclear genes, and there appears to be a peak of diversity surrounding the putative disease resistance genes.
Figure 1.—
Nucleotide diversity and neighbor-joining trees of high-diversity region 1. The dashed line indicates the average level of θ/K for A. thaliana. Sequenced sites from each gene are solid. The MYB103 gene originally identified by our BLAST analysis is indicated by an asterisk. Symbols in neighbor-joining trees were applied to ecotypes on the basis of membership in a specific haplogroup in the most structured gene in the region, indicated by a boxed tree. Solid circles represent accessions possessing the Col-0-type allele, open circles represent Ler-type alleles, triangles are used if a third A. thaliana allele class is present, shaded squares represent the A. lyrata outgroup sequence, and stars indicate accessions that are not common across all genes in each region.
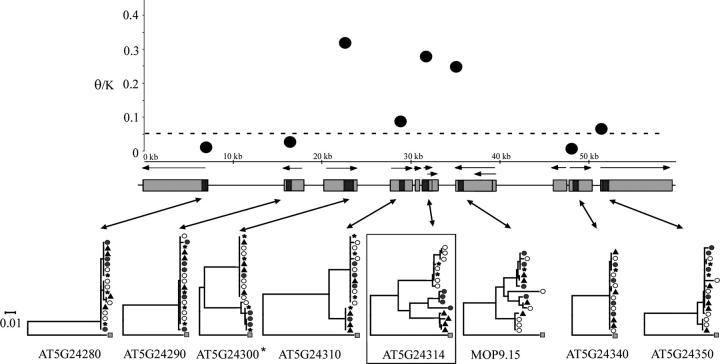
High-diversity region 2 contains the putative soluble starch synthase I gene (AT5G24300), and nucleotide variation in this region is also elevated across multiple linked loci. We sequenced 6528 nucleotide sites from eight genes, including 3861 silent sites across the 59.5-kb region. A total of 265 SNPs were identified by the analysis, including 240 silent-site polymorphisms. Values of silent site π for the putative soluble starch synthase I gene and the three loci immediately downstream (which all encode expressed proteins of unknown function) are all high. Estimates of π for these genes range from 0.024 to 0.056, values three- to eightfold higher than the mean for A. thaliana (Table 2). Interspecific nucleotide divergence estimates for the genes in this region range from 0.07 and 0.18, and the ratio of θ/K is shown in Figure 2. Like high-diversity region 1, a peak of elevated nucleotide polymorphism is also observed in this genomic region. An expressed protein gene (AT5G24310) within this putative diversity peak also has elevated levels of intraspecific nucleotide variation (silent site π = 0.024, θ = 0.016), but an elevated interspecific divergence estimate for this locus (silent site K = 0.179) results in a low level of θ/K. The four genes immediately upstream and downstream of this diversity peak, however, have θ/K ratios similar to or lower than the mean for A. thaliana. Both high-diversity genomic regions 1 and 2 share the pattern of multiple linked genes of elevated nucleotide polymorphism.
Figure 2.—
Nucleotide diversity and neighbor-joining trees of high-diversity region 2. The dashed line indicates the average level of θ/K for A. thaliana. Sequenced sites from each gene are solid. The putative soluble starch synthase I gene originally identified by our BLAST analysis is indicated by an asterisk. Symbols are as described in Figure 1.
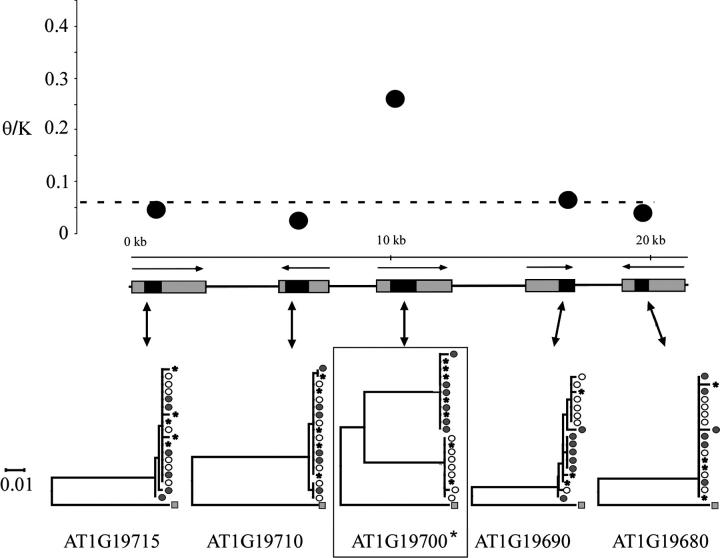
The pattern of nucleotide variation in high-diversity region 3, which includes the homeobox-leucine zipper transcription factor gene (AT1G19700) identified in the genome scan, differs from that observed in the other two regions. We sequenced 3650 nucleotide sites from five genes in this 21.3-kb region, including 1542 silent sites. A total of 80 SNPs were identified by the analysis, including 66 silent-site nucleotide polymorphisms. In high-diversity region 3, only the homeobox-leucine zipper gene has elevated levels of nucleotide diversity (silent site π = 0.05); this increased diversity is still evident in the θ/K ratio for this locus (Figure 3 and Table 2). The four other genes in this region have levels of nucleotide variation ranging from 0.002 to 0.006, which are all lower than the mean for A. thaliana nuclear genes (Figure 3 and Table 2).
Figure 3.—
Nucleotide diversity and neighbor-joining trees of high-diversity region 3. The dashed line indicates the average level of θ/K for A. thaliana. Sequenced sites from each gene are solid. The homeobox-leucine zipper gene originally identified by our BLAST analysis is indicated by an asterisk. Symbols are as described in Figure 1.
Nonsynonymous nucleotide variation across high-diversity gene regions:
Increased levels of nonsynonymous variation can be associated with balanced polymorphisms, and this pattern is exemplified by plant disease resistance and self-incompatibility loci (Bergelson et al. 2001; Charlesworth et al. 2003). Nonsynonymous polymorphism, however, is not generally high across the three high-diversity regions (Table 2). In high-diversity region 1, only the MYB103 gene (AT1G63910) shows elevated levels of nonsynonymous polymorphism; 30 nonsynonymous polymorphisms exist in our sequenced portion of this gene, resulting in nonsynonymous nucleotide diversity of ∼2.7%. Similarly, only one gene in high-diversity region 2, the expressed protein encoding gene MOP9.15, shows elevated nonsynonymous polymorphism with 16 replacement polymorphisms in the sequenced portion of the gene and estimated nonsynonymous nucleotide diversity of 1%. Although increased nonsynonymous variation has been previously identified in genes thought to harbor balanced polymorphisms, the presence of polymorphism at this class of sites is dependent on the mechanism of selection. Patterns of variation at nonsynonymous sites can also be influenced by other evolutionary mechanisms; for example, differing levels of constraint can allow for different patterns of variation at nonsynonymous sites to emerge between genes or regions of genes (Bergelson et al. 2001). Therefore, silent-site polymorphism is likely a more informative tool in delimiting a selected region of the genome and we focus further analyses on this category of sites.
Significant departures from the neutral-equilibrium model and genome-wide variation levels for high-diversity genes:
The HKA test of selection is based on the assumption that under the neutral-equilibrium model, levels of intraspecific diversity and interspecific divergence should be governed by the neutral mutation rate and thus be correlated (Hudson et al. 1987). The multilocus HKA test can be used to compare genes of interest to a neutral set of loci, thereby taking into account levels of neutral variation and divergence from multiple loci in the genome.
Multilocus HKA tests were applied to each of the genes in the three high-diversity regions, with a set of three previously studied genes as neutral reference loci (see Table 3). Significant deviations from neutrality were assessed following Bonferroni correction for multiple testing. The multilocus HKA tests indicate that two genes in high-diversity region 1, the AT1G63910 and AT1G63900 loci, have significantly elevated levels of intraspecific diversity (P < 0.00583 and 0.00525, respectively). For high-diversity region 2, the starch synthase gene (P < 0.001) and two expressed protein genes (P < 0.00025 and 0.00558) are significant. In high-diversity region 3, the homeodomain gene shows significant departures from the expectations of the neutral-equilibrium model (P < 0.00382) (Table 3). In these cases, the significant results arise from both observed high intraspecific diversity and low interspecific divergence compared to expected values (data not shown).
TABLE 3.
Tests of selection for each sampled gene in all three high-diversity regions
| Gene | na | Tajima's D | Fu and Li's D* | Fu and Li's D | No. of haplotypes |
ZnS | HKA test (multilocus)c |
|---|---|---|---|---|---|---|---|
| High-diversity region 1 | |||||||
| AT1G63940 | 16 | 1.6613* | 0.8888 | 1.3556 | 7 | 0.6388* | 0.39277 |
| AT1G63910 | 18 | 0.3111 | 1.5013** | 1.7841** | 6*** | 0.5209* | 0.00583**,d |
| AT1G63900 | 14 | 1.4549 | 0.8756* | 1.0912 | 10 | 0.5480* | 0.00525**,d |
| AT1G63850 | 18 | −0.1764 | 1.1704 | 1.3269 | 5 | 0.4921 | 0.45463 |
| AT1G63840 | 19 | −1.1260 | −0.1052 | −0.6448 | 3 | 1.0000*** | 0.81329 |
| High-diversity region 2 | |||||||
| AT5G24280 | 17 | 0.5368 | 0.5941 | 0.5823 | 6 | 0.1723 | 0.01431* |
| AT5G24290 | 18 | −1.1871 | −1.1379 | −0.8275 | 6*** | 0.4375 | 0.05413 |
| AT5G24300 | 18 | 2.2290** | 1.4535** | 1.9336** | 7*** | 0.9304*** | 0.001***,d |
| AT5G24310 | 18 | 1.4491 | 1.1463 | 0.9705 | 4** | 1.0000*** | 0.15324 |
| AT5G24314 | 17 | 1.5564* | 0.9236 | 1.2390 | 11 | 0.4126 | 0.00025***,d |
| MOP9.15b | 15 | 0.6575 | 0.2994 | 0.9110 | 10 | 0.3536 | 0.00558**,d |
| AT5G24340 | 15 | −0.0269 | 1.0566*** | 0.9025 | 4*** | 0.0452 | 0.01435* |
| AT5G24350 | 16 | −1.0776 | −0.8577 | −0.8408 | 7 | 0.7851** | 0.1492 |
| High-diversity region 3 | |||||||
| AT1G19715 | 18 | −1.9331* | −2.3336 | −1.3439* | 8*** | NA | 0.63494 |
| AT1G19710 | 18 | −0.9452 | −0.3590 | 0.1611 | 5 | 0.3500 | 0.14928 |
| AT1G19700 | 20 | 2.9594*** | 1.3742* | 1.5507* | 5*** | 0.9613*** | 0.00382**,d |
| AT1G19690 | 17 | −1.0650 | −1.7787* | −2.0333* | 10* | 0.2862 | 0.80176 |
| AT1G19680 | 17 | −1.9433* | −2.6319*** | −2.7601* | 6*** | NA | 0.57568 |
NA, no results were obtainable for this test with this sample. ***P < 0.001, **P < 0.01, *P < 0.05.
Number of samples.
BAC locus annotation for a predicted gene with EST support.
Probability estimates based on silent sites for multilocus HKA test.
Significant after Bonferroni correction.
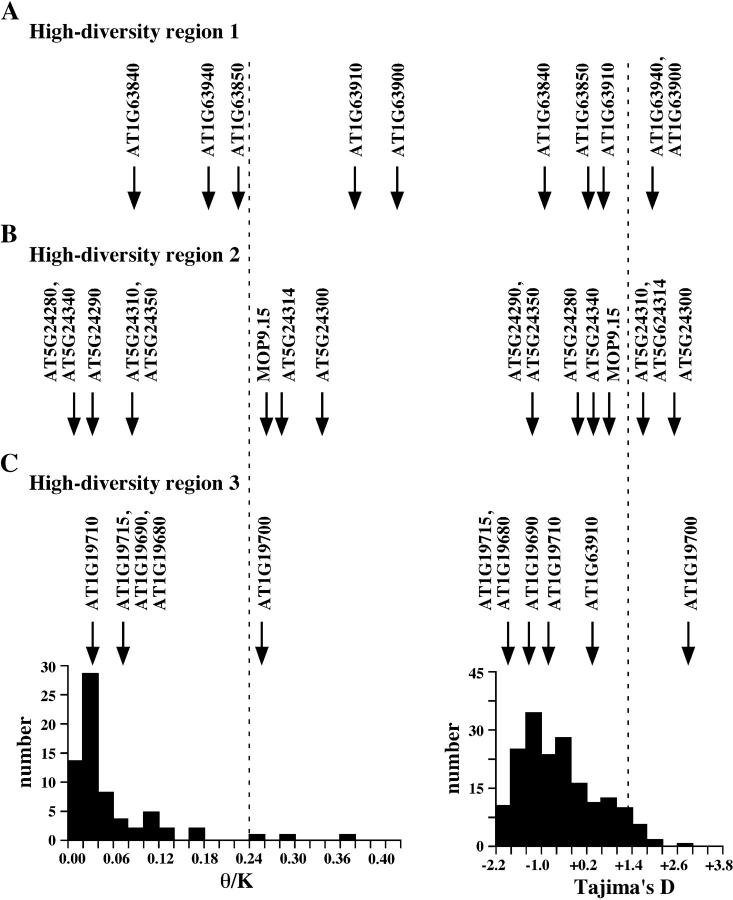
The multilocus HKA tests indicated departures from a neutral-equilibrium model. We also determined where these genes fell within the distribution of θ/K studied in a genome-wide survey of variation in the A. thaliana genome (Schmid et al. 2005). This genome-wide survey examined nucleotide variation in 195 unlinked, randomly selected gene fragments of ∼400 bp in length from 12 ecotypes in Schmid et al. (2005). Of these 195 gene fragments, 68 fragments had both intraspecific nucleotide diversity estimates in A. thaliana and interspecific nucleotide divergence between A. thaliana and A. lyrata, and this subset formed the basis of our θ/K distribution (see Figure 4). All genes identified as departing from neutral-equilibrium expectations in the multilocus HKA test were also found to be in the extreme tail of the genome-wide distribution (top 5%) of θ/K ratios for the genome-wide distribution (see Figure 4), indicating that these genes have exceptionally high levels of nucleotide variation compared to other loci in the A. thaliana genome.
Figure 4.—
Levels and patterns of nucleotide variation in high-diversity regions compared to a genome-wide distribution. Data for the distributions are from Schmid et al. (2005). Data from 12 A. thaliana accessions were chosen to generate these distributions due to their use as mapping populations (Col-0, Cvi-0, Ler, Nd-0, and Ws-0) and to obtain a maximum average genetic distance between surveyed accessions (Ei-2, CS22491, Gu-0, Lz-0, Wei-0, Ws-0, and Yo-0). The top 5% limits are indicated by dashed lines. A distribution of θ/K ratios for 68 randomly chosen unlinked gene fragments for which estimates of both intraspecific diversity within A. thaliana and interspecific divergence between A. thaliana and A. lyrata are available is shown on the bottom left. A distribution of Tajima's D for 195 randomly chosen unlinked gene fragments for which estimates of intraspecific diversity in A. thaliana only are available is shown on the bottom right. The location of the estimates within these distributions for genes in (A) high-diversity genomic region 1, (B) region 2, and (C) region 3 is shown by arrows.
Haplotype di- and trimorphism of high-diversity genes:
The number of haplotypes in several genes in each of these high-diversity regions is significantly lower than expected under a conservative neutral-equilibrium model of no recombination (see Table 3); many of these are significant even with Bonferroni correction for multiple tests. This arises, in part, because all high-diversity genes identified in this study are organized into two or three distinct allele groups; these are referred to as di- and trimorphic haplotype structuring, respectively (Figures 1–3). Moreover, at least one gene, the myb-like gene (AT1G63910) in high-diversity region 1, also exhibits trans-specific polymorphism, with one of three allele classes (represented by two sampled A. thaliana accessions) more similar to the sequenced A. lyrata allele than to either of the other two A. thaliana allele classes.
Given the close linkage among the genes in these high-diversity regions and the reduced effective recombination rate in A. thaliana as a result of selfing, we would expect gene genealogies across these regions to be strongly correlated. Neighbor-joining trees show that phylogenetic relationships among adjacent genes are not perfectly correlated (Figures 1–3), indicating that intergenic recombination has occurred in these high-diversity regions to limit associations among di- or trimorphic haplogroups across loci. Patterns of linkage disequilibrium (LD; supplementary Figure S1 at http://www.genetics.org/supplemental/) observed across the investigated high-diversity regions, however, indicate that correlations between nucleotide polymorphisms exist among linked genes and are quite strong in high-diversity regions 1 and 2.
Excess of intermediate-frequency alleles in the high-diversity regions:
Tajima's and Fu and Li's tests of selection were applied for all sequenced genes in each of the three high-diversity regions; these tests examine the frequency distribution of nucleotide polymorphisms along branches of a gene tree. Although these tests are often used to infer selection, they are also sensitive to population structure and demographic changes. Given the possible recent population size expansion, ancestral population structure, and selfing nature of A. thaliana, the results of these tests should be interpreted with caution. Despite these concerns, these tests prove useful in comparing patterns of polymorphism among A. thaliana genes and may also be indicative of the types of nonneutral forces acting at specific loci.
All three high-diversity-region genes with elevated levels of nucleotide polymorphism are accompanied by positive Tajima's D and/or Fu and Li's D/D*, and multiple genes in high-diversity regions 1 and 2 show this pattern (Table 3). This is not surprising, given that the three focal genes identified in this study were initially chosen to have highly positive Tajima's D values. In high-diversity region 1, three of five genes have positive Tajima's D, while four have positive Fu and Li's D or D*. In high-diversity region 2, five of eight genes have positive Tajima's D, and six of eight have positive Fu and Li's D/D*. In high-diversity region 3, only the homeodomain-encoding gene has a positive value for these test statistics. Several of the genes with positive Tajima's D in these genomic regions are also in the top 5% tail of the distribution of this test statistic in a recent genome-wide survey of 195 unlinked gene fragments for which intraspecific data in A. thaliana are available (see Figure 4) (Schmid et al. 2005). The latter result indicates that the numbers of intermediate-frequency polymorphisms in several of these high-diversity regions are exceptionally high compared to genes in the rest of the genome.
Linkage disequilibrium within and between genes:
LD is a measure of genetic association at sites both within and between genes and is affected by a wide range of genetic, demographic, and selective factors (Nordborg and Tavare 2002; Gaut and Long 2003). ZnS, the standardized intragenic linkage disequilibrium averaged over all pairwise comparisons, is a test of selection that is expected to be significantly elevated if alleles at a locus are under balancing selection (Kelly 1997). We calculated ZnS for each gene in all three high-diversity genomic regions, and significantly high ZnS values were detected for genes in all three regions. In high-diversity region 1, all loci except for the PRLI-interacting-factor-related gene have significantly high ZnS values (Table 3). Three genes in high-diversity region 2, the soluble starch synthase (AT5G24300) and two expressed protein genes (AT5G24310 and AT5G24350), also have significantly high ZnS values. In contrast, only the homeobox-leucine zipper gene (AT1G19700) reveals a significantly elevated value of ZnS in high-diversity region 3. For regions 2 and 3, the genes initially identified as having elevated levels of nucleotide variation [the soluble starch synthase gene (AT5G24300) in region 2 and the homeodomain gene (AT1G19700) in region 3] also have significantly high ZnS estimates even after Bonferroni correction.
Linkage disequilibrium among informative polymorphic sites was also examined across each of the three high-diversity genomic regions using r2, with significantly high levels of LD determined using Fisher's exact test. Statistically significant pairwise LD across each of the three regions is depicted in supplementary Figure S1 at http://www.genetics.org/supplemental/. Significantly high levels of intragenic LD are observed in each of the three high-diversity regions, which conform to the high ZnS estimates. Significantly high linkage disequilibrium between genes is also observed among genes with related neighbor-joining trees (see above) in high-diversity regions 1 and 2, which confirms previous findings of extensive LD in A. thaliana (Nordborg et al. 2002; Shepard and Purugganan 2003).
Local rates of recombination and patterns of variation in high-diversity regions:
The extent of LD and the widths of the peaks of diversity observed in our three high-diversity regions should be affected by local recombination rates. We estimated local recombination rates for each of our three regions using information on genetic and physical map positions of markers flanking our genomic regions. The recombination rate in A. thaliana appears to range from 1 to 14 cM/Mb (Zhang and Gaut 2003) and the genome-wide average was previously shown to be 4.8 cM/Mb (Copenhaver et al. 1999; Zhang and Gaut 2003).
The local recombination rate for high-diversity region 1 was estimated to be 1.75 cM/Mb, which is low in comparison to average chromosome and genome-wide estimates. This low recombination rate is consistent with the observation of a broad peak of nucleotide diversity and significant intergenic LD in high-diversity region 1 (see Figure 1). In contrast, high-diversity region 2 has an estimated recombination rate of 11.50 cM/Mb, which is high compared to average genome-wide estimates. Although elevated levels of nucleotide diversity in this region are observed among several linked genes, the peak is narrower than observed in genomic region 1 (see Figure 2).
The pattern observed in high-diversity region 3, however, appears anomalous. The local recombination rate for high-diversity region 3 is estimated at 2.17 cM/Mb, which is slightly lower than average estimates in this species. As such, we might expect to observe a broad peak of diversity across this region; what we observe, however, is a narrow peak that is centered on only one gene. This departure from expectation may result in higher recombination at smaller scales in this region. Alternatively, differences in peak breadths may result from other factors, including the age of alleles.
DISCUSSION
High-diversity genes represent an important class of genes in organismal genomes. Population genetic theory predicts that high-diversity genes may contain balanced polymorphisms (Hudson and Kaplan 1988), which underlie adaptive variation within species. These balanced polymorphisms are maintained over long evolutionary periods and are characterized by elevated levels of nucleotide diversity in silent sites linked to the selective target (Hudson and Kaplan 1988). This prediction has been substantiated by studies of several genes known to be subject to balancing selection or local adaptation, such as plant disease resistance (Tian et al. 2002) and self-incompatibility loci (Takebayashi et al. 2003). Hunting for high-diversity genes could thus form the basis of an adaptive-trait-locus-mapping approach for scanning genomes for selectively maintained alleles.
Comparison of the A. thaliana Col-0 whole-genome sequence with 4300 short sequence fragments from a genomic shotgun sequence of the Ler ecotype initially identified 60 functionally annotated sequences with 2–10% divergence between the two ecotypes. Further analysis in a secondary screen with five to six other A. thaliana ecotypes confirmed that three of these gene fragments—the myb-like transcription factor gene MYB103 (AT1G63910), a putative soluble starch synthase I gene (AT5G24300), and a locus encoding a homeodomain-leucine zipper protein (AT1G19700)—have elevated nucleotide diversity levels and are high-diversity genes that may represent loci that have or are linked to balanced polymorphisms. The presence of high levels of nucleotide diversity, however, is only one characteristic of the signature of a balanced polymorphism. Additional characteristics can include: (i) a symmetrical peak of nucleotide diversity surrounding the selective target, (ii) maintenance of intermediate-frequency alleles, (iii) a reduction in the number of haplotypes, (iv) high levels of linkage disequilibrium, and (v) the presence of trans-specific polymorphism. The case for balanced polymorphisms is strengthened if the genes that harbor elevated levels of nucleotide diversity also display these other characteristics of selection, although it is worthwhile to note that it is unlikely for every expectation to be fulfilled by every empirical data set. Moreover, although many of these features are not totally independent of each other, they do represent different facets of an underlying pattern of sequence variation associated with selection.
Analysis of the high-diversity genes identified in the genome scan, as well as the loci flanking these genes, reveals that high-diversity region 1 displays all of the characteristic signatures of balanced polymorphisms. This region is characterized by elevated nucleotide variation spanning a local region of the genome, significant levels of intermediate-frequency polymorphisms, intergenic linkage disequilibrium, a significant deficiency in the number of haplotypes among highly variable genes, and trans-specific polymorphism at the MYB103 locus (AT1G63910) in the region. High-diversity region 2 displays all of the characteristics of high-diversity region 1, with the exception of trans-specific polymorphism. This region also contains several genes with high levels of nucleotide diversity, among which the expressed protein gene AT5G24314 has significantly elevated nucleotide polymorphism levels as well as significantly positive Tajima's D. These features are consistent with the hypothesis that both high-diversity regions 1 and 2 harbor balanced polymorphisms.
One gene in high-diversity region 3, the homeodomain-leucine zipper gene (AT1G19700), shows elevated nucleotide diversity and positive Tajima's D and Fu and Li's D/D*. In addition, this gene also displays significant LD and a significant deficiency in haplotype number. Interpretation of these results is complicated, however, by the fact that high nucleotide polymorphism in this region is confined to one gene and does not show the gradual symmetric decline with distance; the four loci flanking the homeobox-leucine zipper gene have levels of nucleotide diversity at or below the mean for A. thaliana nuclear genes (Table 2 and Figure 3). Alternative explanations for the mechanism and/or origin of the two divergent haplogroups observed in this gene may help explain inconsistencies in these observations. Interestingly, a transposable element was identified in the intergenic region 3′ of the homeobox-leucine zipper gene in complete association with the Ler haplogroup (J. M. Cork and M. D. Purugganan, unpublished observations). Potential functional consequences of this insertion and its effect on the molecular evolution of this region are currently being explored.
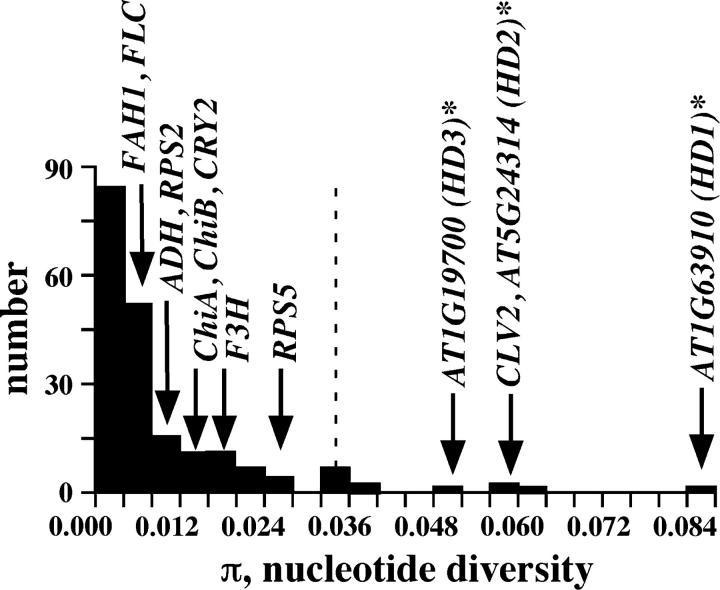
The levels and patterns of nucleotide polymorphisms, particularly those in high-diversity genomic regions 1 and 2, do not conform to the neutral-equilibrium model and are at the extremes for the genome-wide distributions, consistent with the selective maintenance of differentiated alleles. Other alternative possibilities, however, need to be considered. The first possibility is that these differentiated alleles represent ancestral and/or contemporary structure in A. thaliana. Recent studies suggest some isolation by distance as well as evidence for genetic differentiation associated with possible Pleistocene refugia in A. thaliana (Sharbel et al. 2000; Schmuths et al. 2004). Differentiation among dimorphic alleles attributed to population structure, however, is generally modest in other surveys of variation (Kawabe et al. 1997; Kuittinen and Aguade 2000) and does not explain the strong divergence of allelic classes in these high-diversity genomic regions. As a comparison, only 1 of 10 previously reported dimorphic genes in A. thaliana (CLV2) has nucleotide diversity levels in the top 5% of the genome-wide distribution. In contrast, the dimorphic genes in each high-diversity genomic region studied here all show exceptionally high variation levels (see Figure 5).
Figure 5.—
Comparison of nucleotide diversity levels for dimorphic genes compared to a genome-wide distribution. Data for the distribution are from Schmid et al. (2005). The top 5% limit is indicated by a dashed line. Nucleotide diversity estimates (π) for 10 genes previously described as dimorphic are indicated by their names (Miyashita et al. 1996; Kawabe and Miyashita 1999; Aguade 2001; Tian et al. 2002; Mauricio et al. 2003; Caicedo et al. 2004; Olsen et al. 2004). One dimorphic gene from each of the three high-diversity regions is also shown.
The second possibility is that the elevated diversity observed at these genomic regions could arise from gene duplications that result in artifactual comparisons of paralogous rather than allelic sequences. This duplication scenario also requires the loss of alternate duplicates such that A. thaliana ecotypes possess only one or the other duplicate copy. Although this may not be common, such a pattern is indeed possible; at the MAM locus of A. thaliana, alternate duplicate copies in a tandem array are lost in different ecotypes (Kroymann et al. 2003). However, this pattern, like the geographic structuring scenario, is improbable, given the elevated nucleotide diversity observed across multiple linked, unrelated loci in high-diversity regions 1 and 2. In contrast, this scenario cannot be ruled out for high-diversity region 3, where only one gene has elevated nucleotide polymorphism levels.
It should be noted that both the geographic structure and the duplication scenarios are not mutually exclusive from a selection hypothesis. The former two scenarios relate to the origins of allelic differentiation, but selection can still be invoked to explain the intraspecific maintenance of these differentiated alleles. For example, geographic structure could explain the divergence in CRY2 (Olsen et al. 2004) and FLC dimorphic haplotypes (Caicedo et al. 2004) and duplication accounts for differences in gene content and apparent nucleotide polymorphisms among genes at the MAM locus (Kroymann et al. 2003). In these cases, however, these different alleles are associated with trait variation in flowering time in the case of CRY2 (Olsen et al. 2004) and FLC (A. L. Caicedo, J. R. Stinchmobe, K. M. Olsen, J. Schmitt and M. D. Purugganan, unpublished results) and with glucosinilate levels for the MAM locus (Kroymann et al. 2003), which suggests that maintenance of differentiated allele classes could result from selection of these ecologically relevant phenotypes. In each case, the precise mechanistic origins of alleles do not preclude the resultant phenotypic consequences that may lead to selective maintenance of alternate alleles.
While the genome-scan approach appears able to identify high-diversity gene regions that may contain candidate balanced polymorphisms, identifying the specific polymorphism(s) and associated phenotype(s) that are possible targets of selection requires further work. In high-diversity region 1, the region of high diversity flanks tandemly repeated genes that belong to the TIR-NBS-LRR family of disease resistance loci. Disease resistance loci are known to be subject to various selective forces, including diversifying selection (Bergelson et al. 2001). The presence/absence of deletion alleles, for example, is the basis for balancing selection at the disease resistance gene RPS5 (Tian et al. 2002). This previous knowledge and the pattern of variation observed in this region make the TIR-NBS-LRR genes the most likely putative target of selection in high-diversity region 1. Interestingly, PCR amplifications consistently fail to amplify the second TIR-NBS-LRR duplicate copy in this region in 9 of 19 A. thaliana ecotypes. Although this, as well as problems in designing copy-specific primers, makes it difficult to obtain completed sequence data sets for these putative disease resistance loci, these results suggest that this locus may segregate for the presence or absence of the second duplicate copy in Arabidopsis ecotypes. This finding also demonstrates the possible utility of the adaptive-trait-locus-mapping approach in exploiting the relationship between linkage disequilibrium and selection in identifying genes of potential adaptive significance when their direct sampling cannot be easily achieved.
The potential target of selection in high-diversity region 2 remains uncertain. One candidate is an expressed protein gene of unknown function (AT5G24314) that segregates for three distinct haplotype groups, yields a significant multilocus HKA test, and has the highest level of nucleotide polymorphism in the region. The precise function of this gene is unknown, but a T-DNA insertion mutant at this locus displays aberrant seed pigmentation associated with a defective embryo (Budziszewski et al. 2001). Another candidate, however, is the putative starch synthase I locus, which is characterized by two highly divergent allele classes that show almost no polymorphism. This gene also has elevated levels of nucleotide diversity and highly positive Tajima's D estimates (see Figures 2 and 4).
Finally, only the homeodomain-leucine zipper transcription factor gene in high-diversity region 3 has high-diversity and intermediate-frequency alleles, although whether this is due to selection remains ambiguous, given that this gene does not show other key characteristics of a balanced polymorphism. At present, the precise function of this gene is unknown. Detailed functional reverse genetic studies are currently underway to determine the functions and the precise phenotypic consequences of the alternatively maintained alleles for all these candidate adaptive trait genes.
It is unclear how common these high-diversity genomic regions are in the genome. Moreover, not all balanced polymorphisms may have the extreme levels of diversity observed in this study, and thus this approach is inherently conservative. It remains important, however, to continue to identify and study high-diversity genes and genomic regions, and their possible contribution to adaptive variation. A. thaliana is particularly suited for these studies, given that the genomic resources (Jander et al. 2002) and predominantly selfing nature of this species make it easier to identify high-diversity regions associated with balanced polymorphisms (Nordborg et al. 1996; Tian et al. 2002; Shepard and Purugganan 2003). Detailed analysis of these genes may shed light on the extent of selection, as well as other evolutionary and genetic forces that act on these loci, and on their contribution to genome evolution.
Acknowledgments
We thank members of the Purugganan laboratory for a critical reading of this manuscript. We also thank M. Barrier for programming assistance and K. Shepard, R. C. Moore, P. Awadalla, M. Uyenonyama, and T. Mitchell-Olds for helpful discussions. This work was supported in part by National Science Foundation Integrated Research Challenges in Environmental Biology and Frontiers in Integrative Biological Research grants to M.D.P. and a National Institutes of Health Training Grant graduate fellowship to J.M.C.
References
- Abbot, R. J., and M. F. Gomes, 1989. Population genetic structure and outcrossing rate of Arabidopsis thaliana (L.) Heynh. Heredity 62: 411–418. [Google Scholar]
- Aguade, M., 2001. Nucleotide sequence variation at two genes of the phenylpropanoid pathway, the FAH1 and F3H genes, in Arabidopsis thaliana. Mol. Biol. Evol. 18: 19. [DOI] [PubMed] [Google Scholar]
- Akey, J. M., G. Zhang, K. Zhang, L. Jin and M. D. Shriver, 2002. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12:1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Bamshad, M., and S. P. Wooding, 2003. Signatures of natural selection in the human genome. Nat. Rev. Genet. 4: 99–111. [DOI] [PubMed] [Google Scholar]
- Barbash, D. A., P. Awadalla and A. M. Tarone, 2004. Functional divergence caused by ancient positive selection of a Drosophila hybrid incompatibility locus. PLoS Biol. 2: 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrier, M., C. D. Bustamante, J. Yu and M. D. Purugganan, 2003. Selection on rapidly evolving proteins in the Arabidopsis genome. Genetics 163: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson, J., M. Kreitman, E. A. Stahl and D. Tian, 2001. Evolutionary dynamics of plant R-genes. Science 292: 2281–2285. [DOI] [PubMed] [Google Scholar]
- Budziszewski, G. J., S. P. Lewis, L. W. Glover, J. Reineke, G. Jones et al., 2001. Arabidopsis genes essential for seedling viability: isolation of insertional mutants and molecular cloning. Genetics 159: 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A. L., J. R. Stinchcombe, K. M. Olsen, J. Schmitt and M. D. Purugganan, 2004. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. USA 101: 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., 2003. Effects of inbreeding on the genetic diversity of populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1051–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., and P. Awadalla, 1998. Flowering plant self-incompatibility: the molecular population genetics of Brassica S-loci. Heredity 81(Pt 1): 1–9. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., C. Bartolome, M. Schierup and B. K. Mable, 2003. Haplotype structure of the stigmatic self-incompatibility gene in natural populations of Arabidopsis lyrata. Mol. Biol. Evol. 20: 1741–1753. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., S. Glanowski, R. Nielsen, P. D. Thomas, A. Kejariwal et al., 2003. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science 302: 1960–1963. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G. N., K. Kuromori, T. Benito, M. I. Kaul, S. Lin et al., 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286: 2468–2474. [DOI] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and L. Hubbard, 1997. The evolution of apical dominance in maize. Nature 386: 485–488. [DOI] [PubMed] [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. C. Wendl and P. Green, 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Fu, Y. X., and W. H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan, D., and P. W. Hedrick, 2003. Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution 57: 1707–1722. [DOI] [PubMed] [Google Scholar]
- Gaut, B. S., and A. D. Long, 2003. The lowdown on linkage disequilibrium. Plant Cell 15: 1502–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad, Y., S. Rosenberg, M. Przeworski, D. Lancet and K. Skorecki, 2002. Evidence for positive selection and population structure at the human MAO-A gene. Proc. Natl. Acad. Sci. USA 99: 862–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel, N., and S. B. Carroll, 2003. Genetic mechanisms and constraints governing the evolution of correlated traits in drosophilid flies. Nature 424: 931–935. [DOI] [PubMed] [Google Scholar]
- Greenberg, A. J., J. R. Moran, J. A. Coyne and C.-I Wu, 2003. Ecological adaptation during incipient speciation revealed by precise gene replacement. Science 302: 1754–1757. [DOI] [PubMed] [Google Scholar]
- Harr, B., M. Kauer and C. Schlotterer, 2002. Hitchhiking mapping: a population-based fine-mapping strategy for adaptive mutations in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99: 12949–12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W. G., and A. Robertson, 1968. The effects of inbreeding at loci with heterozygote advantage. Genetics 60: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1988. The coalescent process in models with selection and recombination. Genetics 120: 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguade, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander, G., S. R. Norris, S. D. Rounsley, D. F. Bush, I. M. Levin et al., 2002. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson, U., J. West, C. Lister, S. Michaels, R. Amasino et al., 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347. [DOI] [PubMed] [Google Scholar]
- Kawabe, A., and N. T. Miyashita, 1999. DNA variation in the basic chitinase locus (ChiB) region of the wild plant Arabidopsis thaliana. Genetics 153: 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe, A., H. Innan, R. Terauchi and N. T. Miyashita, 1997. Nucleotide polymorphism in the acidic chitinase locus (ChiA) region of the wild plant Arabidopsis thaliana. Mol. Biol. Evol. 14: 1303–1315. [DOI] [PubMed] [Google Scholar]
- Kelly, J. K., 1997. A test of neutrality based on interlocus associations. Genetics 146: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn, M. H., H. J. Pelz and R. K. Wayne, 2000. Natural selection mapping of the warfarin-resistance gene. Proc. Natl. Acad. Sci. USA 97: 7911–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn, M. H., H. J. Pelz and R. K. Wayne, 2003. Locus-specific genetic differentiation at Rw among warfarin-resistant rat (Rattus norvegicus) populations. Genetics 164: 1055–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman, M. E., and M. Aguade, 1986. Excess polymorphism at the Adh locus in Drosophila melanogaster. Genetics 114: 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman, M., and R. R. Hudson, 1991. Inferring the evolutionary histories of the Adh and Adh-dup loci in Drosophila melanogaster from patterns of polymorphism and divergence. Genetics 127: 565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann, J., S. Donnerhacke, D. Schnabelrauch and T. Mitchell-Olds, 2003. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc. Natl. Acad. Sci. USA 100(Suppl. 2): 14587–14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuittinen, H., and M. Aguade, 2000. Nucleotide variation at the CHALCONE ISOMERASE locus in Arabidopsis thaliana. Genetics 155: 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Li, S. F., T. Higginson and R. W. Parish, 1999. A novel MYB-related gene from Arabidopsis thaliana expressed in developing anthers. Plant Cell Physiol. 40: 343–347. [DOI] [PubMed] [Google Scholar]
- Lister, C., and C. Dean, 1993. Recombinant inbred lines for mapping RFLP and phenotypic markers in A. thaliana. Plant J. 4: 745–750. [Google Scholar]
- Luikart, G., P. R. England, D. Tallmon, S. Jordan and P. Taberlet, 2003. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4: 981–994. [DOI] [PubMed] [Google Scholar]
- Mauricio, R., E. A. Stahl, T. Korves, D. Tian, M. Kreitman et al., 2003. Natural selection for polymorphism in the disease resistance gene Rps2 of Arabidopsis thaliana. Genetics 163: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita, N. T., H. Innan and R. Terauchi, 1996. Intra- and interspecific variation of the alcohol dehydrogenase locus region in wild plants Arabis gemmifera and Arabidopsis thaliana. Mol. Biol. Evol. 13: 433–436. [DOI] [PubMed] [Google Scholar]
- Nielsen, R., 2001. Statistical tests of selective neutrality in the age of genomics. Heredity 86: 641–647. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., 1997. Structured coalescent processes on different time scales. Genetics 146: 1501–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., and S. Tavare, 2002. Linkage disequilibrium: what history has to tell us. Trends Genet. 18: 83–90. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., B. Charlesworth and D. Charlesworth, 1996. Increased levels of polymorphism surrounding selectively maintained sites in highly selfing species. Proc. R. Soc. Lond., Ser. B, Biol. Sci. 263: 1033–1039. [Google Scholar]
- Nordborg, M., J. O. Borevitz, J. Bergelson, C. C. Berry, J. Chory et al., 2002. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 30: 190–193. [DOI] [PubMed] [Google Scholar]
- Olsen, K. M., S. S. Halldorsdottir, J. R. Stinchcombe, C. Weinig, J. Schmitt et al., 2004. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167: 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H., and J. Coyne, 1992. The genetics of adaptation: a reassessment. Am. Nat. 140: 725–742. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., 2000. Detecting the form of selection from DNA sequence data. Trends Genet. 16: 526–529. [DOI] [PubMed] [Google Scholar]
- Payseur, B. A., A. D. Cutter and M. W. Nachman, 2002. Searching for evidence of positive selection in the human genome using patterns of microsatellite variability. Mol. Biol. Evol. 19: 1143–1153. [DOI] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-Delbarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Rozen, S., and H. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Schierup, M. H., X. Wekemans and F. B. Christiansen, 1998. Allelic genealogies in sporophytic self-incompatibility systems in plants. Genetics 150: 1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotterer, C., 2002. Towards a molecular characterization of adaptation in local populations. Curr. Opin. Genet. Dev. 12: 683–687. [DOI] [PubMed] [Google Scholar]
- Schmid, K. J., S. Rasmos-Onsins, H. Ringys-Beckstein, B. Weisshaar and T. Mitchell-Olds, 2005. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from the standard neutral model of DNA sequence polymorphism. Genetics 169: 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuths, H., M. H. Hoffmann and K. Bachmann, 2004. Geographic distribution and recombination of genomic fragments on the short arm of chromosome 2 of Arabidopsis thaliana. Plant Biol. 6: 128–139. [DOI] [PubMed] [Google Scholar]
- Schulte, P. M., H. C. Glemet, A. A. Fiebig and D. A. Powers, 2000. Adaptive variation in lactate dehydrogenase-B gene expression: role of a stress-responsive regulatory element. Proc. Natl. Acad. Sci. USA 97: 6597–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbel, T. F., B. Haubold and T. Mitchell-Olds, 2000. Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Mol. Ecol. 9: 2109–2118. [DOI] [PubMed] [Google Scholar]
- Shepard, K. A., and M. D. Purugganan, 2003. Molecular population genetics of the Arabidopsis CLAVATA2 region: the genomic scale of variation and selection in a selfing species. Genetics 163: 1083–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, E. A., G. Dwyer, R. Mauricio, M. Kreitman and J. Bergelson, 1999. Dynamics of disease resistance polymorphism at the Rpm1 disease resistance locus of Arabidopsis. Nature 400: 667–671. [DOI] [PubMed] [Google Scholar]
- Storz, J. F., B. A. Payseur and M. W. Nachman, 2004. Genome scans of DNA variability in humans reveal evidence for selective sweeps outside of Africa. Mol. Biol. Evol. 21: 1800–1811. [DOI] [PubMed] [Google Scholar]
- Strobeck, C., 1983. Expected linkage disequilibrium for a neutral locus linked to a chromosomal arrangement. Genetics 103: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., A. G. Clark, H. M. Waldrip-Dail, M. F. Wolfner and C. F. Aquadro, 2001. a Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98: 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., Z. Yang, M. F. Wolfner and C. F. Aquadro, 2001. b Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl. Acad. Sci. USA 98: 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1993. Statistical analysis of DNA polymorphism. Jpn. J. Genet. 68: 567–595. [DOI] [PubMed] [Google Scholar]
- Takebayashi, N., P. B. Brewer, E. Newbigin and M. K. Uyenoyama, 2003. Patterns of variation within self-incompatibility loci. Mol. Biol. Evol. 20: 1778–1794. [DOI] [PubMed] [Google Scholar]
- Tian, D., H. Araki, E. Stahl, J. Bergelson and M. Kreitman, 2002. Signature of balancing selection in Arabidopsis. Proc Natl Acad Sci U S A 99: 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux, Y., M. McMullen, C. T. Hittinger, K. Houchins, L. Schulz et al., 2002. Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proc. Natl. Acad. Sci. USA 99: 9650–9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Wootton, J. C., X. Feng, M. T. Ferdig, R. A. Cooper, J. Mu et al., 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418: 320–323. [DOI] [PubMed] [Google Scholar]
- Yoshida, K., T. Kamiya, A. Kawabe and N. T. Miyashita, 2003. DNA polymorphism at the ACAULIS5 locus of the wild plant Arabidopsis thaliana. Genes Genet. Syst. 78: 11–21. [DOI] [PubMed] [Google Scholar]
- Zhang, L., and B. Gaut, 2003. Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res. 13: 2533–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]