Abstract
In the fungus Pneumocystis carinii, at least three gene families (PRT1, MSR, and MSG) have the potential to generate high-frequency antigenic variation, which is likely to be a strategy by which this parasitic fungus is able to prolong its survival in the rat lung. Members of these gene families are clustered at chromosome termini, a location that fosters recombination, which has been implicated in selective expression of MSG genes. To gain insight into the architecture, evolution, and regulation of these gene clusters, six telomeric segments of the genome were sequenced. Each of the segments began with one or more unique genes, after which were members of different gene families, arranged in a head-to-tail array. The three-gene repeat PRT1-MSR-MSG was common, suggesting that duplications of these repeats have contributed to expansion of all three families. However, members of a gene family in an array were no more similar to one another than to members in other arrays, indicating rapid divergence after duplication. The intergenic spacers were more conserved than the genes and contained sequence motifs also present in subtelomeres, which in other species have been implicated in gene expression and recombination. Long mononucleotide tracts were present in some MSR genes. These unstable sequences can be expected to suffer frequent frameshift mutations, providing P. carinii with another mechanism to generate antigen variation.
PNEUMOCYSTIS carinii is a parasitic, sometimes pathogenic, yeast-like fungus found in the lungs of laboratory rats (Stringer 2002). Large numbers of P. carinii organisms can be extracted from the lungs of immunosuppressed laboratory rats, which typically develop Pneumocystis pneumonia. This fungus does not proliferate well in culture, although it is phylogenetically related to model ascomycetes such as Schizosaccharomyces pombe (Lee et al. 1993; Sloand et al. 1993; Aliouat et al. 1996; Merali et al. 1999; Cushion et al. 2000). Humans with impaired immune function can also develop Pneumocystis pneumonia, but this disease is caused by a different species called P. jirovecii (Frenkel 1976, 1999; Stringer et al. 2002).
Parasites typically exhibit antigenic variation and P. carinii seems to be no exception. The genome contains three gene families [major surface glycoprotein (MSG), MSG-related (MSR), and protease (PRT1)] that have been implicated as probable contributors to such variation. The members of these gene families are grouped together in clusters located at chromosome ends and encode proteins located on the microbial surface (Stringer et al. 1991; Wada and Nakamura 1994; Lugli et al. 1997, 1999; Stringer and Cushion 1998; Stringer and Keely 2001).
The MSG gene family is known to be expressed in a manner that would create surface variation. Pneumocystis cells carry abundant MSG on their surfaces (Graves et al. 1986; Walzer and Linke 1987; Gigliotti et al. 1988; Kovacs et al. 1988, 1989; Linke and Walzer 1989; Lundgren et al. 1991; Nakamura 1998). Only 1 of the 80 or so open reading frames (ORFs) that encode different isoforms of MSG is expressed at a given time. The expressed MSG ORF resides at a unique site in the genome, called the expression site, which encodes the upstream conserved sequence (UCS), a 365-bp invariant sequence that encodes the signal peptide used to export MSG to the cell surface (Angus et al. 1996; Sunkin et al. 1998; Stringer and Keely 2001; Schaffzin and Stringer 2004). The 5′ ends of messages encoding various MSG proteins were found to begin with the UCS (Wada et al. 1995; Edman et al. 1996). The genomic UCS can be occupied by a wide variety of MSG genes (Sunkin and Stringer 1996, 1997; Keely et al. 2003). Between the UCS and the attached MSG ORF, there is a 25-bp invariant sequence known as the conserved recombination junction element (CRJE), which may be involved in recombination events that install an ORF at the expression site (Wada et al. 1995; Edman et al. 1996; Sunkin and Stringer 1996; Wada and Nakamura 1996b; Stringer and Keely 2001). A copy of the CRJE is also present at the beginning of each non-UCS-linked MSG ORF.
MSR genes bear a strong resemblance to MSG ORFs, but are not dependent on the UCS for expression, lack the CRJE, and are interrupted by a single small intron (Wada and Nakamura 1997; Huang et al. 1999; Schaffzin et al. 1999b). Two size classes of MSR genes have been described. Long MSR genes are similar in size to MSG genes (∼3 kb). Short MSR genes lack a 1-kb segment present in long MSR genes. Expression of MSR genes is not well characterized, but it appears that each gene is transcribed in situ rather than via movement to a unique expression site (Wada and Nakamura 1997; Huang et al. 1999; Schaffzin et al. 1999b). The number of MSR genes expressed at a given time in a single organism is not clear, but transcripts from as many as 13 MSR genes were detected in a population of P. carinii in which 80% of the organisms had the same MSG gene at the expression site (Keely and Stringer 2003). MSR proteins appear to be on the cell surface (Huang et al. 1999).
PRT1 genes are distinct in structure and sequence from MSG and MSR genes and have several small introns (Lugli et al. 1997, 1999; Russian et al. 1999; Wada and Nakamura 1999b). Expression of PRT1 genes is also not completely understood, but studies have suggested that PRT1 proteins can be on the cell surface and that multiple PRT1 genes are expressed in a given organism at a given time (Lugli et al. 1999; Wada and Nakamura 1999a; Keely and Stringer 2003; Ambrose et al. 2004).
The genome of P. carinii contains ∼34 telomeres, each of which probably carries a cluster of surface antigen genes (Cushion 1998; Stringer and Cushion 1998). While the clustering of PRT1, MSR, and MSG genes at telomeres has been established (Underwood et al. 1996; Wada and Nakamura 1996a,b), the sizes and compositions of complete gene arrays, defined as clusters preceded by a unique sequence in the genome and followed by a telomere, were heretofore unclear. To better understand these clusters, large telomeric DNA segments were isolated from a cosmid library and characterized to locate those containing complete gene arrays. Seven arrays, six of which were complete, were sequenced.
MATERIALS AND METHODS
Standard molecular genetic procedures such as preparation of DNA and RNA, cloning, library screening, PCR, restriction mapping, gel electrophoresis, and Southern and Northern blotting were performed using methods described by Sambrook et al. (1989). Southern and Northern blots contained nucleic acids from ∼10 million P. carinii per lane (Schaffzin et al. 1999a,b; Cushion et al. 2001; Schaffzin and Stringer 2004).
A cosmid library was constructed in the vector pWEB (Epicentre Technologies, Madison, WI) by Smulian and colleagues from genomic DNA from a population of P. carinii from a single rat that was infected by the airborne route (Smulian et al. 2001; Keely et al. 2003). The library contained five- to sixfold coverage of the 8 million-bp genome of P. carinii. Cosmids that contained MSG genes were identified by screening 2486 bacterial colonies for hybridization to an MSG DNA probe. Approximately 60 MSG-positive colonies were detected, 90% of which also hybridized to members of the PRT1 and MSR gene families and to a DNA probe specific for subtelomeric P. carinii DNA. A few clones that hybridized to the subtelomere probe did not hybridize to one or more gene family probes, but these clones carried inserts spanning only a few kilobase pairs.
To determine a cosmid sequence, the DNA was fragmented by sonication and end-repaired fragments of ∼2 kb in size were inserted into a pUC plasmid linearized with SmaI. Approximately 1000 plasmids were sequenced per cosmid. The sequences were assembled into one contiguous sequence by methods described previously (Harris and Murphy 2001). Some regions were resequenced to verify the sequence assembled from random fragments. DNA for resequencing was produced by PCR using cosmid DNA as template and primers based on the assembled sequence.
The sequence of each cosmid was tested for accuracy by comparing actual and predicted restriction enzyme sites and fragment sizes. Fragments produced were tested for the presence of gene family members by Southern blot analysis using a battery of radioactive DNA probes specific for MSG, MSR, PRT1, subtelomeres, and telomeres (Keely et al. 2003; Keely and Stringer 2003; Ambrose et al. 2004; Schaffzin and Stringer 2004).
Annotation of the assembled sequences was performed independently in two ways. One research group used the ORF-finder function at the NCBI web site (http://www.ncbi.nlm.nih.gov) to identify ORFs. Each predicted protein was used to query databases using BLASTP. Another group used Artemis software (Berriman and Rutherford 2003), in which case putative genes identified from the output of the software package Genefinder (C. Wilson, L. Hilyer and P. Green, unpublished results) were confirmed manually. Genefinder was trained for Plasmodium species that have a similar base composition to that of P. carinii (low G + C). Functional assignments were based on assessment of FASTA and BLAST searches against public databases. Discrimination between MSR and MSG was confirmed by searching the sequences of interest for the CRJE, which is in MSG genes but not in MSR genes (Stringer and Keely 2001). MSG/MSR/PRT1 annotation was confirmed using the SMART website (http://smart.embl-heidelberg.de). Candidate unique genes (ORFs that did not encode an MSG, a PRT1, or an MSR) were mapped to P. carinii chromosomes by hybridization to Southern blots carrying electrophoretically separated chromosomes prepared as described previously (Hong et al. 1990; Cushion et al. 1993; Cornillot et al. 2002).
Nucleotide sequences were aligned using DNAMAN software (Lynnon BioSoft, Vaudreuil, Quebec, Canada) set for dynamic full alignment with a gap open penalty of 10, a gap extension of 5, a DNA transition weight of 0.5, and a delay divergent sequence percentage of 30. The alignments were optimized by introducing a limited number of gaps. Ambiguous regions in the alignment were not scored. Relatedness of pairs of aligned nucleotides was calculated by MEGA 2.1 software utilizing p-distance and pairwise deletion programs (Kumar et al. 2001). Synonymous and nonsynonymous p-distances (pS or pN) were calculated by the Nei-Gojobori method (Nei and Gojobori 1986). The number of synonymous differences (Sd) was normalized using the possible number of synonymous sites (S). Nonsynonymous p-distances (pN) were determined with a similar computation (Nei and Gojobori 1986). A neighbor-joining tree was constructed for MSG genes with MEGA 2.1 (Kumar et al. 2001), utilizing the Nei-Gojobori synonymous p-distance (pS) method (Nei and Gojobori 1986). The strength of tree branches was assessed by 1000 bootstrap replications.
The sequences of the MSG expression site and of the ste3 locus carry accession nos. D82031 and AF309805, respectively. The accession numbers for the cosmid sequences are as follows: PCC3G5, AL592382; PCC11A11, CR716157; PCC18A9, CR716158; PCC17D7, CR730243; PCC22C8, CR716159; PCC11H12 + 1B2, CR717231; and PCC21H1, CR717240.
RESULTS
General structure of sequenced telomeric gene arrays:
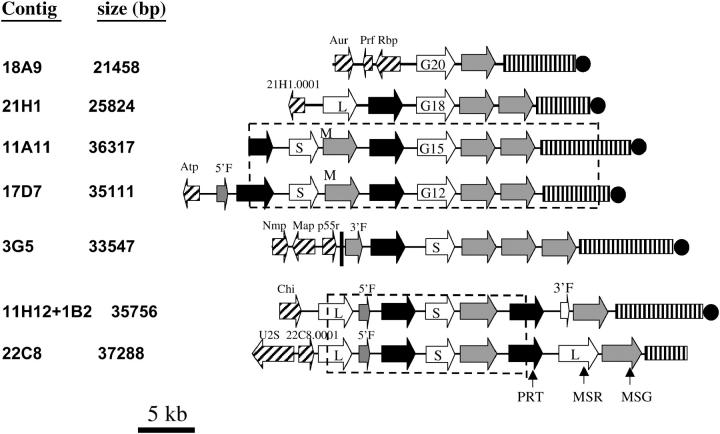
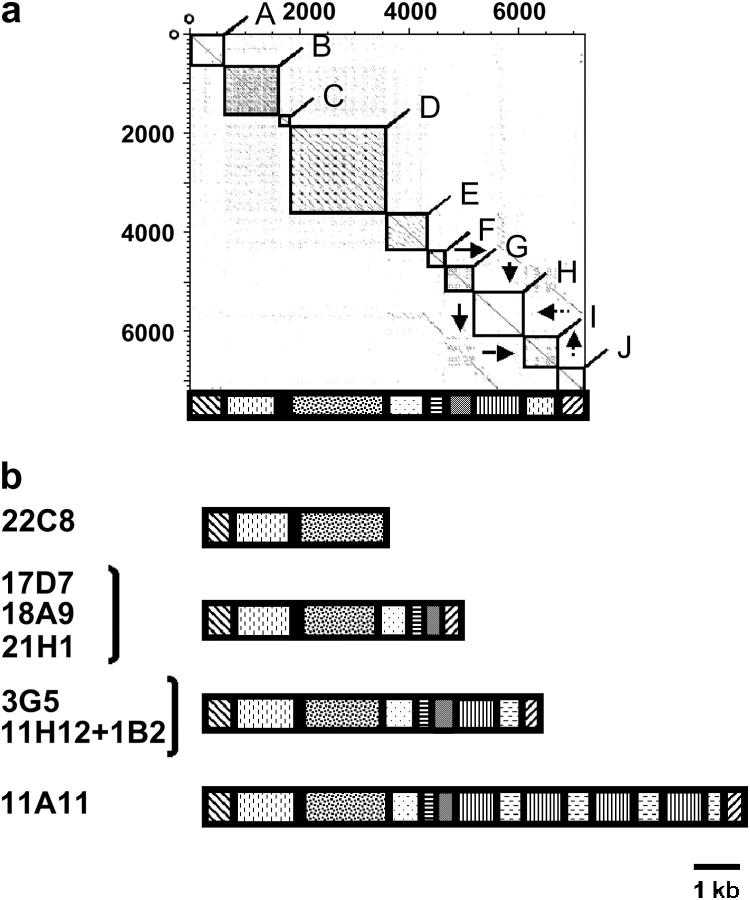
Figure 1 shows maps indicating the locations of the various repeated (solid arrows) and unique genes (cross-hatched arrows) and other sequences in the inserts. At least one putative unique gene was found at the beginning of six of the seven inserts. Eight of these genes were mapped by hybridization to a single Southern-blotted P. carinii chromosome separated by pulsed-field gel electrophoresis (Figure 2). The genes within each repeated-gene array are all in head-to-tail orientation with 3′ ends pointed toward the telomere (Figure 1). All of the gene arrays end with an MSG gene, which is followed by a subtelomere (Figure 1).
Figure 1.—
Maps of gene clusters. Arrows represent ORFs and point in the direction of transcription. Nonhatched arrows represent members of the PRT1 (solid arrows), MSR (open arrows), and MSG (shaded arrows) gene families. Rectangles with vertical lines represent subtelomeres. Solid circles represent telomeres. All features except the telomere are drawn to scale. M indicates the MSG gene that contained two point mutations. 5′F and 3′F indicate ORFs corresponding to the 5′ and 3′ ends, respectively, of either an MSG (shaded arrow) or an MSR (open arrow) gene. G12, G15, G18, and G20 indicate the MSR genes that have a poly(G) mononucleotide tract of the size denoted by the numeral. S and L indicate short and long MSR genes. The dashed-line boxes enclose regions that were at least 99% identical. The hatched arrows represent unique ORFs that had the following presumed functions, BLAST hits, and FASTA E-values: Aur1 (inositol phosphoryl-ceramide synthase, EMBL accession AF076692, 2.9 e-67), Prf (prefoldin-related, NCBI REFSEQ accession XM_331039.1, 3 e-21), Rbp (RNA binding, UniProt YAS9 SCHPO_Q10145, 7.6 e-8), 21H1.0001 (unknown function, no hits), Atp (P-type cation-pumping ATPase, NCBI accession NP_595246, 1 e-6), Nmp (nuclear migration, NCBI accession EAK84394, 2 e-61), Map (microtubule associated protein, NCBI accession XP_323888, 3 e-17), P55 (peptide similar to the p55 antigen of P. carinii, NCBI accession AAQ06671, 4 e-8), Chi (chitin synthesis, NCBI accession NP_013434, 0.084), U2S (U2 snRNP, NCBI accession NP_594538, 0), and 22C8.0001 (unknown function, NCBI accession Q09895, 6.2 e-49). All of the hatched ORFS (except Prf, Rbp, and 22C8.0001, which were not analyzed) hybridized to a single chromosome. Aur1 and Chi mapped to a 440-kb chromosome. Nmp, Map, and p55 mapped to a 290-kb chromosome. Genes 21H1.0001, Atp, and U2S mapped to chromosomes of 680, 620, and 550 kb, respectively.
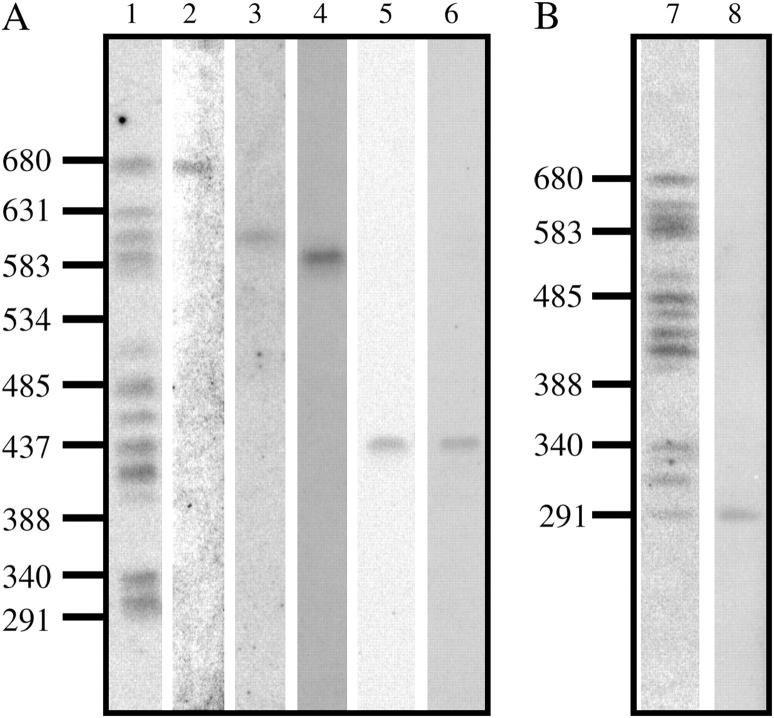
Figure 2.—
Terminal sequence at the end of a cosmid array maps to a single PFGE band. (A) A Southern blot was made from a CHEF gel performed under standard conditions (Cushion et al. 1993). Hybridization probes were as follows: lane 1, CRJE; lane 2, ORF 21H1.0001; lane 3, 17D7 Atp; lane 4, 22C8 U2S; lane 5, 18A9 Aur; lane 6, 11H12 + 1B2 Chi. (B) Southern blot was made from a CHEF gel performed under conditions optimized to resolve the lower four chromosome bands (Cornillot et al. 2002). Hybridization probes were as follows: lane 7, CRJE; lane 8, 3G5 p55-like ORF (p55). Sizes of DNA markers in kilobase pairs are indicated to the left of each part.
Multiple copies of the inserts in cosmids 17D7, 3G5, 22C8, and 1B2 were identified by restriction mapping. The presence of more than one cosmid with a given insert was common, suggesting that certain telomeric segment clusters were more readily cloned than others. The competing hypothesis to explain the high frequency of certain genome segments is that there are far fewer different gene clusters than telomeres. However, this possibility seems remote because 78 different MSG genes and 44 different PRT1 genes have been identified to date (Keely and Stringer 2003; Keely et al. 2003; Ambrose et al. 2004).
Regions enclosed in boxes in Figure 1 were related. The gene array in 11A11 was 99.99% the same as that in 17D7 (four differences in 30 kb), showing that the cloning and sequencing methods were very accurate. The inserts in cosmids 22C8 and 11H12 + 1B2 shared a 15.4-kb central segment, but regions flanking this shared segment were quite different. In fact, the unique genes in the two inserts mapped to different chromosomes. Therefore, this example of genes shared between cosmids was not due to cloning the same genome segment multiple times. It appears that the shared segment was instead present at two different loci.
The structure PRT1-MSR-MSG occurred six times (not counting 11A11 and the region shared between 22C8 and 11H12 + 1B2). In addition, the 11H12 + 1B2 contig had what appears to be a degenerate PRT1-MSR-MSG repeat because there was a fragment of an MSR gene (Figure 1, 3′F) between the terminal PRT1 and MSG genes. Apparent fragments of MSG/MSR genes also appeared in other arrays (17D7 and 3G5). These data suggest that the three-gene structure, PRT1-MSR-MSG, may be the unit that expanded in number to generate all three gene families.
Gene families:
MSG genes:
ORFs encoding MSG isoforms are defined by two features, the presence of a 25-bp sequence called CRJE at the 5′ end and the lack of introns (Stringer and Keely 2001; Keely and Stringer 2003). The sequenced arrays contained 16 ORFs encoding full-size MSG isoforms (Figure 1, large shaded arrows). However, some of these genes were present in more than one cosmid insert (Figure 1, boxed regions). Therefore, the cosmid set contained 12 different MSG ORFs.
One of the MSG ORFs (the first MSG in cosmids 17D7 and 11A11) had a variant form of the CRJE that encodes the peptide sequence MARL instead of the canonical MARP, which is the only peptide sequence encoded by all of the MSG cDNAs examined so far (Wada et al. 1995; Edman et al. 1996). In addition to having a noncanonical CRJE, a frameshift occurred near the end of the MSG-encoding ORF. This mutation would cause a peptide starting with MARL to lack the last 37 amino acids encoded by typical MSG ORFs. The two differences exhibited by the MARL ORF were not artifacts because they were present in both cosmids 17D7 and 11A11. Another example of a divergent CRJE occurred in the array contained in contigs 11H12 + 1B2 and 22C8, where a fragment was encoding MERP instead of MARP. Thus, variant CRJEs were seen only in sequences that differ from typical MSG ORFs in other ways, suggesting that variation in the CRJE may be linked to degeneration of an MSG gene or vice versa.
Pairwise comparisons of the 12 MSG ORFs (including the one encoding MARL) showed that they are between 5 and 19% divergent (at all nucleotide sites). Average divergence is 15%. The MARL ORF exhibited average divergence. The average distance at nonsynonymous sites (16%) was greater than that at synonymous nucleotide sites (15%). The high divergence at nonsynonymous sites caused the average amino acid distance (27%) to be almost twofold greater than the distance at synonymous nucleotide sites. These data suggest that MSG genes are evolving under positive selection for protein variation (Woelk et al. 2002).
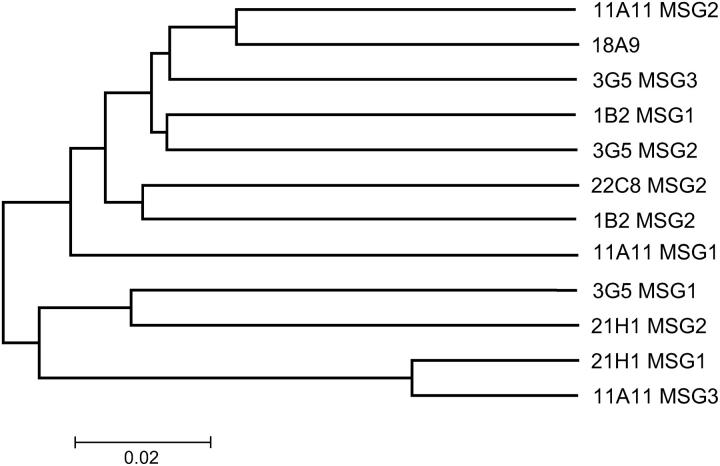
MSG ORFs in the same array were no more identical to each other than to ORFs in other arrays. Figure 3 shows a tree constructed from the synonymous-site data. (The ORF encoding MARL is labeled “11A11 MSG1” in Figure 3.) The lack of close kinship between linked MSG genes suggests that recombination has moved MSG genes from their place of origin and installed them in other arrays (see discussion).
Figure 3.—
Linked MSG genes were not more similar. The tree displays the synonymous p-distances (pS) computed from alignments of synonymous nucleotide sites of the MSG genes in cosmid clones. When a gene was in more than one clone, such as the gene in the region completely shared by contigs 22C8 and 11H12 + 1B2, only one of the two copies of this gene was included in the analysis. Tree branches are labeled to indicate the cosmid and the MSG gene, reading Figure 1 left to right. For example, 11A11 MSG3 refers to the MSG gene that is most proximal to the telomere in cosmid 11A11. Trees produced from either all nucleotide sites or nonsynonymous sites had the same structure as the tree shown. Bar indicates synonymous p-distance (pS).
MSR genes:
The seven sequenced arrays contained 13 genes encoding MSR isoforms (MSR genes are represented by open arrows in Figure 1). However, 2 of these genes are in the region shared completely by contigs 22C8 and 11H12 + 1B2, and 4 are in the region shared by 11A11 and 17D7. Hence, there were 10 different MSR genes.
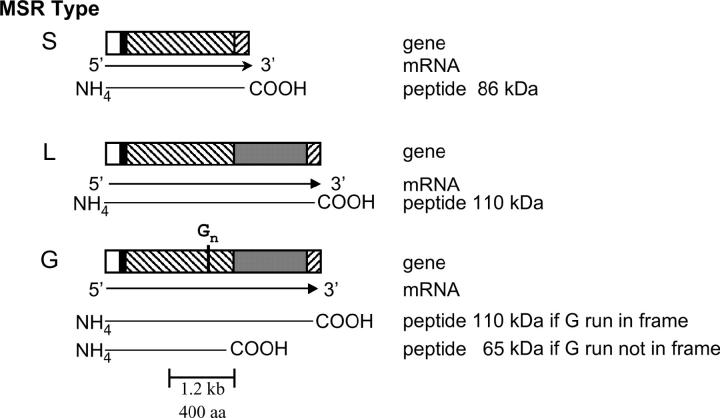
Comparison of these 10 MSR genes to each other and to published cDNAs showed that all begin with a small first exon that encodes ∼28 amino acids. The genes varied, however, with respect to the structure of the second exon and could be divided into three classes (L, G, and S) based on second exon structure. Figure 4 illustrates the differences among the three classes. Class L genes have a second exon of ∼2.4 kb. Class S genes lack a 1-kb region present in class L genes. Class G genes are similar to class L but have a poly(G) tract in the middle of the second exon. The poly(G) tracts disrupt the reading frames of all three of the class G genes in the cosmids, causing translation to stop at a TAA codon located 13 codons downstream of the poly(G) tract. In each gene, however, the sequence downstream of the nonsense codon can be translated in an alternative frame to produce a peptide corresponding to the last half of the peptide encoded by the single ORF in second exons of class L MSR genes. Therefore a frameshift mutation in the poly(G) tract would lead to the production of a full-length class L MSR protein. Although all three of these MSR genes were in the wrong frame to allow translation of all of exon 2, three other MSR genes with poly(G) tracts can be inferred to exist from cDNA data, and all of these have a 2.4-kb exon 2 ORF (Huang et al. 1999). The presence of mononucleotide repeats that can shift the reading frame during translation implies that these motifs may be involved in generating variation (see discussion).
Figure 4.—
MSR genes vary in structure. Depicted are gene, message, and peptide structures for the three classes (S, L, and G) of MSR genes. Open boxes, exon 1; solid boxes, intron; hatched boxes, regions common to all three classes; shaded boxes, region missing in class S genes; Gn, poly(G) tract.
The MSR genes are ∼20% divergent when all coding nucleotide sites are scored. By contrast, the introns in the MSR genes are only 2% divergent. The reason for the very strong conservation of the intron sequence is not known, but this conservation is suggestive of selection against change in this element. The average distance at nonsynonymous sites (20%) was nearly as great as that at synonymous nucleotide sites (21%). The high divergence at nonsynonymous sites caused the average amino acid distance (35%) to be almost twofold greater than the distance at synonymous nucleotide sites. These data suggest that MSR genes are evolving under positive selection for protein variation (Woelk et al. 2002).
MSR genes that were in the same array were no more identical to each other than to MSR genes in other arrays. The first and third MSR genes in cosmid 22C8 are both class L genes, but are only 62% identical. The second MSR gene in that cosmid is a class S gene. The two MSR genes in 17D7 are also of different lengths. Excluding the 1 kb that is not present in the class S gene, the two genes are 82% identical. The two MSRs in cosmid 21H1 are 72% identical. By contrast, the second MSR gene in cosmid 22C8 is 99% identical to the first MSR gene in cosmid 17D7. However, this very high identity did not extend into the upstream PRT1 genes, which are only 89% identical. Nor did it extend downstream into the MSG genes, where cosmid 17D7 has MARL and 22C8 has the canonical MARP. This very high similarity between these two unlinked genes suggests that recombination has placed one or the other or both of these genes in their current locations.
The MSR genes are only 30–60% identical to the MSG genes. However, regions as long as 300 bp with 85% identity were observed (data not shown). Given this level of sequence identity, MSR and MSG genes might be expected to recombine. If such an event were to occur in a reciprocal fashion, it would produce a hybrid gene, e.g., a gene that has the 5′ end of an MSR gene and the 3′ end of an MSG gene. The “MSR” in 18A9 may be an example of such an event because the 5′ end matches MSRs but the 3′ end resembles that of MSGs (data not shown).
PRT1 genes:
There are nine full-length PRT1 genes (solid arrows) on the maps in Figure 1. (Cosmid 11A11 began within a tenth PRT1 gene). Two of these genes are in the region shared by cosmids 11A11 and 17D7. Another two are in the region shared by contigs 22C8 and 11H12 + 1B2. Hence, the cosmid set contains seven different full-size PRT1 genes.
Pairwise comparisons of these seven PRT1 genes showed that they are ∼15% divergent when all coding nucleotide sites are scored. By contrast, the introns in these genes are only 4% divergent. The average distance at nonsynonymous sites (15%) was greater than that at synonymous nucleotide sites (13%). The high divergence at nonsynonymous sites caused the average amino acid distance (26%) to be almost twofold greater than the distance at synonymous nucleotide sites. These data suggest that PRT1 genes are evolving under positive selection for protein variation (Woelk et al. 2002). PRT1 genes that are in the same array are no more identical to each other than to genes in other arrays (data not shown).
Noncoding sequence families:
Subtelomeres:
Excluding cosmid 22C8, subtelomeres ranged between 4.7 and 11.4 kb in length. (The insert in cosmid 22C8 did not end with copies of the telomere repeat, suggesting that the subtelomere was truncated during cloning.) These lengths bracket that reported for the end of a cloned MSG expression site (6.3 kb) (Wada and Nakamura 1996b). The subtelomeres in the cosmids were similar in sequence (75% average identity).
Dotter-plot analysis (Sonnhammer and Durbin 1995) (Figure 5a) revealed 10 blocks (A–J) of internally repetitious sequence. Block A corresponds to the region immediately adjacent to the last MSG gene. Block J corresponds to the terminal telomeric repeats (Trpts). Most of these blocks were composed of multiple copies of one or more short sequence motifs, the presence of which causes rectangular regions in Figure 5a to be filled with points. Blocks B, D, E, F, and G correspond to regions I–IV in the previously published sequence of a P. carinii subtelomere (Wada and Nakamura 1996b). The diagram under the Dotter plot (Figure 5a) represents the block structure of the 3G5 subtelomere as a series of rectangles filled in various ways. Figure 5b illustrates how subtelomere length differences were primarily due to differences in the number of blocks. Subtelomeres in other species are known to vary in similar fashion (Pryde et al. 1997).
Figure 5.—
Subtelomere structures. (a) Dotter plot made by comparing the last 7 kb at the end of the insert in cosmid 3G5 to itself. The DNA compared starts at the nucleotide immediately downstream of the stop codon of the last MSG gene (see Figure 1). The boxes enclose 10 blocks that either contained multiple copies of one or more short sequence motifs or contained copies of sequences found in other blocks. Cases of the second form of repetition are indicated by arrows. For example, sequences in blocks I and J were also present in blocks G and H, respectively. Below the Dotter plot is a diagram that depicts the structure of the 3G5 subtelomere. (b) Comparison of subtelomere structures in different cosmid inserts. Rectangles represent blocks as in a. In cases where a single diagram represents the subtelomeres from multiple cosmids, such as 17D7, 18A9, and 21H1, the width of a rectangle represents the average length of the block it represents.
Terminal telomeric repeats:
All but one of the cloned arrays end with copies of the repeat sequence TTAGGG (Trpt), which has been previously shown to cap the ends of P. carinii chromosomes (Underwood et al. 1996; Wada and Nakamura 1996b). The average number of terminal copies of the Trpt was 12. Cosmid 17D7 had the highest number of terminal Trpts with 15.
Intergenic regions:
The PRT, MSR, and MSG genes in arrays were separated by regions that did not encode recognizable peptides. Four types of intergenic regions (PRT–MSR, MSR–MSG, MSG–PRT, and MSG–MSG) occurred more than once.
The regions upstream of MSG genes (MSR-MSG and MSG-MSG spacers) were relatively short and uniform in length and sequence (280–320 bp, 86% identity). The regions upstream of PRT1 genes (MSG-PRT spacers) were longer and more conserved (1095–1164 bp, 93% identity). The regions upstream of MSR genes (PRT-MSR spacers) were still longer and conserved (1340–1570 bp, 94% identity). All PRT-MSR spacers contained a 600-bp element 90% identical to the 5′ end of the P. carinii thioredoxin reductase gene. The functional copy of the thioredoxin reductase gene is located downstream of a PRT1 gene (Kutty et al. 2003). These data indicate that the evolution of the PRT1 gene family involved coamplification of a part of the thioredoxin reductase gene along with the upstream PRT1 gene.
All of the intergenic spacers were more conserved than the genes they flank, which may serve to facilitate homologous recombination. In addition, subtelomere and telomere repeats can act as cis-acting control regions that function in a domain-wide fashion to modulate expression of a cluster of neighboring genes (Pryde and Louis 1999; Fourel et al. 1999; Verona et al. 2003). Because subtelomeric repeat motifs have these effects in other organisms, it was of interest to determine if repeated DNA sequence motifs found at P. carinii subtelomeres and telomeres were also located upstream of members of the various gene families.
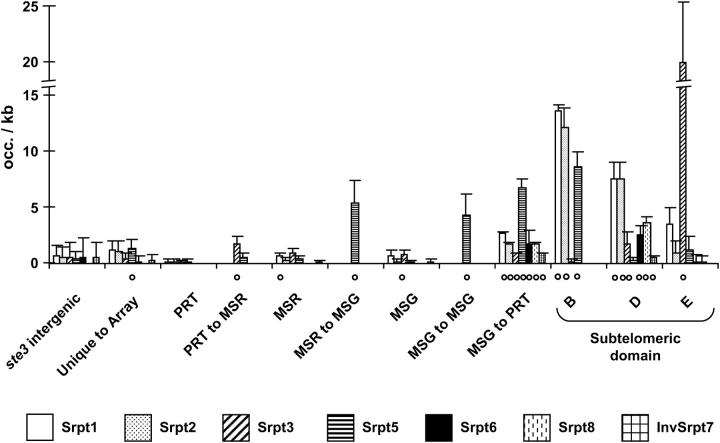
As shown in Figure 6, subtelomeric sequence motifs [subtelomeric repeat (Srpt)] resided in regions between genes. Seven motifs were present in the spacers between MSG and PRT genes (Figure 6, MSG to PRT), a location that demarcates sets of tandem PRT-MSR-MSG arrays. Five of these seven elements were present only in MSG-PRT spacers. Srpt5 was present in all four spacers and in the regions between unique and repeated genes. Four of the borders between unique and repeated DNA also featured a copy of Trpt (data not shown). The presence of subtelomere motifs between genes suggests that genes might have been inserted into one or more subtelomeres.
Figure 6.—
Occurrence of subtelomere short repeats. Normalized occurrences [average occurrence/kilobase pair (+ standard deviation)] of various short subtelomeric repeated sequence motifs (“Srpt”) are shown in various chromosomal regions. The “o” indicates motifs that were present at least once in all members of the region considered. Results for genic regions at the ste3 locus were similar to the intergenic spacers ones (data not shown). Srpt1, T3–4A5–8; Srpt2, T3A5W; Srpt3, GA1–2(GA)2; Srpt5, GT3AT; Srpt6, T4MT2A4; Srpt8, TRAT4KYATYR2; and InvSrpt7, BTGYBA2MWA.
A caveat to such inferences is that the motifs in question are short and/or degenerate and might be found at random in the genome because they are as such. To test this hypothesis, we analyzed the ste3 locus, which contains no PRT, MSR, or MSG genes and is not adjacent to a telomere. Whereas all but one (Srpt8) of the subtelomere sequence motifs occurred between genes in the ste3 locus, they tended to occur at a lower frequency per kilobase (Figure 6).
DISCUSSION
Detailed knowledge of telomeric gene array structures provides clues about the gene families that compose them and their evolution, stability, regulation, and function. The average complete gene array contained 2.2 MSG genes, 1.8 MSR genes, and 1.2 PRT1 genes. There are ∼34 telomeres in P. carinii. Therefore, the cosmid data suggest that there are ∼75 MSG genes, 61 MSR genes, and 41 PRT1 genes in the P. carinii genome. The estimates derived from the cosmids fit well with other data (Stringer et al. 1991; Wada et al. 1993; Sunkin and Stringer 1996; Lugli et al. 1997; Stringer and Cushion 1998; Huang et al. 1999; Schaffzin et al. 1999b; Stringer and Keely 2001; Ambrose et al. 2004).
All of the repeated genes in the cloned arrays were pointed in the same direction, toward the telomere. This is strikingly different from the surface antigen arrays observed in other eukaryotic parasites such as Plasmodium falciparum (Gardner et al. 2002). Members of the three gene families tended to be interdigitated, with PRT1-MSR-MSG a predominant structure. This structure suggests that all three families can grow via duplication of this set of genes. In addition, the tandem repeat structure makes it possible for readthrough transcription to produce mRNAs from all three genes in a PRT1-MSR-MSG set. However, such transcripts have not been detected. In addition, readthrough transcripts would presumably require processing to produce messenger RNAs. Such processing occurs in organisms in the order Kinetoplastida, such as Trypanosomes, which use trans-splicing to attach the same leader RNA to all mRNAs (Borst 1986). However, trans-splicing of single-leader sequence to all transcripts does not occur in P. carinii (Sunkin and Stringer 1997). Furthermore, recent studies have shown that many different PRT1 and MSR transcripts can be present in populations of P. carinii that are dominated by organisms that have one particular MSG gene at the expression site (Wada et al. 1993; Keely and Stringer 2003; Ambrose et al. 2004). At this point, therefore, coordinated transcription of MSG, PRT1, and MSR genes via readthrough seems unlikely.
Expression of an MSG gene appears to require that it be linked to a unique expression site (Wada et al. 1995; Edman et al. 1996; Sunkin and Stringer 1997; Schaffzin and Stringer 2004). Restricting transcription to the expression-site-linked MSG alone allows individual P. carinii organisms to express a single MSG isoform at a time, and changing the gene that is at the expression site produces an organism that has a different MSG on its surface. The mechanism of such changes is not known. One possibility is that a site-specific recombinase might catalyze exchange between a pair of CRJEs, which are invariant sequences located at the beginning of MSG genes, including the one at the expression site. Reciprocal exchange between the CRJE at the expression site and a CRJE in a donor gene would replace the MSG gene at the UCS. In the process of making this change, the genes downstream of the MSG genes would also become linked to the expression site. This movement might simultaneously activate all of the genes in the translocated array. Although transcriptional readthrough seems unlikely to occur, coactivation could occur nevertheless. For example, telomeric genes that are not at the expression site may be silenced (Gottschling et al. 1990; Ai et al. 2002). Other species, such as Trypanosoma brucei and Candida glabrata, have clusters of telomeric surface antigen genes that are transcriptionally silent (Stringer and Keely 2001; Borst 2002; Barry et al. 2003; De Las Penas et al. 2003). One mechanism of silencing is via the actions of cis-acting control regions that function in a domain-wide fashion to modulate chromatin structure (for a review, see Verona et al. 2003). In Saccharomyces cerevisiae interstitial telomeric repeats have been implicated both in silencing of the adjacent chromosomal domain and in insulating a region from the spread of repressive chromatin (Fourel et al. 1999; Pryde and Louis 1999). Such subtelomeric elements may also modulate association between chromosome termini and hence recombination between genes located at telomeres, as is thought to occur in the var genes of P. falciparum (Freitas-Junior et al. 2000).
As an alternative to site-specific recombination, homologous recombination may contribute to changing the expression-site-linked MSG gene sequence. Mitotic yeast cells have been observed to efficiently recombine two identical chromosomal sequence tracts that are 250 bp long (Jinks-Robertson et al. 1993). Pneumocystis gene family members share similar short, highly related sequence tracts. Thus, if the homologous recombination system of P. carinii has requirements similar to those of the system in mitotic yeast, then sequence identity between regions of arrays appears to be sufficient to foster homologous exchanges. In addition, the telomeric location of arrays might increase the frequency of recombination above what it would be on the basis of DNA sequence identity alone. Numerous reports on other species, including the fungus Kluyveromyces lactis and protozoan parasites, have suggested that telomeric genes undergo mitotic recombination more frequently and that these events often involve sequences that are neither on homologous chromosomes nor on sister chromatids (Freitas-Junior et al. 2000; Cornforth and Eberle 2001; McEachern and Iyer 2001; Natarajan and McEachern 2002). Furthermore, the telomeric locations of MSG genes and the expression site would allow reciprocal (i.e., crossing over) recombination to be utilized without causing major genome rearrangement. For the same reasons, the telomeric location of genes may foster evolution via enhanced recombination to form new alleles. In this case, recombination can occur between any two gene family members. Recombination at the ends of chromosomes may contribute to the chromosome length polymorphism seen among P. carinii strains (Cushion et al. 1993). Changes within subtelomeres probably also contribute to such variation. Indeed, the subtelomeres studied here exhibited substantial differences in length that were associated with differences in the number of copies of short repeated sequence motifs. These data suggest that subtelomeres in P. carinii tend to change size due to their repetitive structures, as is the case in other species (Pryde et al. 1997; Mefford and Trask 2002).
While reciprocal exchanges are possible, there is no reason to exclude the possibility of nonreciprocal homologous recombination (commonly referred to as gene conversion) as a means by which to change the MSG gene at the expression site. In mitotic yeast, nonreciprocal recombination is at least as common as crossing over (Prudden et al. 2003). The length of DNA replaced can vary greatly. Conversion tracts can be as short as a few base pairs and as long as many kilobases (Pays et al. 1983; Myler et al. 1988; Nickoloff et al. 1989; Scholler et al. 1989; Weng et al. 1996; Elliott et al. 1998). In principle, the entire end of a chromosome could be changed by gene conversion. The presence in the gene clusters of what appear to be fragments of MSG and MSR genes raises the possibility that pseudogenes contribute to gene family diversity via gene conversion events, as is known to be the case in other species (Howell-Adams and Seifert 2000; Del Portillo et al. 2001; Berriman et al. 2002; Kanti et al. 2004).
Recombination appears to have played a role in the evolution of the sequenced gene arrays. This role can be inferred from comparison of linked and unlinked members of a gene family. Gene families grow through duplication events, whereby an ancestral gene gives rise to two identical genes that later diverge. In the absence of recombination, genes that arose from a common ancestor will tend to be more similar to one another than to other copies of the gene family. Even if selection for variation occurs, vestiges of the duplication event should remain at synonymous nucleotide sites, because the bases at these sites can change without changing the sequence of the encoded peptide. However, analysis of the synonymous sites of linked P. carinii gene family members showed that these genes are as divergent from each other as from nonlinked gene family members. Recombination between arrays can explain this observation.
In addition, the cloned gene arrays contain structures that appear to be the result of homologous recombination. Arrays in cosmids 22C8 and contig 11H12 + 1B2 share a 15.4-kb region. This structure could have been generated by one recombination event that implanted the 15.4-kb region present at one locus, say that represented by the array in 22C8, into the other locus, in this case, that represented by array 11H12 + 1B2. Such an event could occur either via two reciprocal simultaneous crossing-over events or via gene conversion. Alternatively, two separate crossovers can produce the same outcome. Both schemes employ homologous recombination, which would have been facilitated by the high identities of gene family members and the spaces between them. MSR genes lie at one end of the shared region, and PRT1 genes lie at the other.
A second example of homologous recombination is suggested by the presence of the nearly identical MSR genes in two different arrays (the second MSR gene in cosmid 22C8 and the first one in cosmid 17D7). The twin MSR genes are flanked by divergent sequences. The region of very high identity is relatively short, suggesting that it may have been generated by a single gene conversion event, although two reciprocal exchanges are also a possible source of these structures.
A third example of recombination is suggested by the structure of the MSR gene in cosmid 18A9, which has the first exon and intron of an MSR gene, but the 3′ end of an MSG gene. Such a gene could have been formed by a reciprocal crossover between an MSR and an MSG gene.
MSG gene expression has been implicated in antigenic variation, but understanding of the roles of PRT1 and MSR genes is less advanced. Nevertheless, the presence of genes encoding multiple isoforms of these proteins provides additional variation potential. Such potential may be exploited by regulation of transcription, as appears to be the case for MSG. However, this is not the only mechanism employed by microbes to generate variation. An example of a pertinent alternative mechanism is seen in the bacterial genus Neisseria. These organisms have a variety of genes that carry sequence motifs that are intrinsically prone to grow and diminish in length due to errors made by the complex that replicates the genome (Burch et al. 1997; Van Belkum et al. 1998; Van Belkum 1999). These errors shift the reading frame of the gene, thus altering the protein produced. Several of the MSR genes in the sequenced arrays contain long poly(G) tracts. Poly(G) tracts and other mononucleotide repeats are more prone to mutation than other sequences because they suffer insertions and deletions at high frequency (Streisinger and Owen 1985; Jonsson et al. 1991). When in an ORF, a change in mononucleotide repeat length causes a frameshift, thus altering the peptide sequence downstream. All of the class G MSR genes are out of frame downstream of the poly(G) tract. Nevertheless, they retain the capacity to produce a protein ∼500 amino acids long. This protein would begin with a signal sequence and therefore should enter the secretory apparatus. Its function, if any, is a matter of speculation at this point, but one possibility is that it is secreted into the extracellular environment. The class G MSR genes also have a latent capacity to produce a full-size MSR protein (∼1000 amino acids). A change in the poly(G) tract length would restore the ORF of exon 2 to its normal size and content. In addition to those described here, three other class G MSR genes have been described. The GenBank database contains three MSR cDNA sequences that have poly(G) tracts between 14 and 16 bp in length (Huang et al. 1999). By contrast with the genes in the cosmids, the poly(G) tracts in the three cDNAs do not disrupt the reading frame. Given the rarity of G:C base pairs in the genome of P. carinii, which is 60% A + T, these poly(G) tracts seem unlikely to have been generated by chance. They are probably used to generate diversity within populations of P. carinii. In bacteria, there are numerous examples of phase variation conferred by changes in mononucleotide repeats (Jonsson et al. 1991; Yogev et al. 1993; Hammerschmidt et al. 1996; Carroll et al. 1997; Zhang and Wise 1997; Chen et al. 1998; Lavitola et al. 1999; Park et al. 2000; Ekins and Niven 2003; Kearns et al. 2004; Segura et al. 2004).
Telomeric gene arrays are a common feature of pathogenic microbes, suggesting that they reflect the needs of a pathogenic lifestyle (Barry et al. 2003). One such need is to limit the ability of the host to eliminate the microbial population. A mechanism that works to this end is high-frequency antigenic variation, whereby one or more individual microbes within the population shed antigenic determinants that are attracting immune attack and replace these with novel determinants. Survival of these individuals and their descendants perpetuates the infection. P. carinii appears to create antigenic variation by regulated expression of gene families, a mechanism that can generate variants at a frequency that is much greater than what can be produced by random mutation of a single antigen-encoding gene. The positioning of gene family members in telomeric clusters facilitates antigenic variation because this location allows more recombination, which has two advantageous effects. Recombination both provides a mechanism to change antigen expression and fosters expansion and evolution of gene copies.
Acknowledgments
We thank Ed Louis for stimulating discussions and continuous support and Kim Rutherford for advice in programming. This work was supported by grants R01AI36701 and R01AI44651 from the National Institutes of Health and TW01200-02 from Acquired Immune Deficiency Syndrome/Fogerty International Research Collaboration Award (AIDS/FIRCA) program and by a grant from The Wellcome Trust.
References
- Ai, W., P. G. Bertram, C. K. Tsang, T. F. Chan and X. F. Zheng, 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell 10: 1295–1305. [DOI] [PubMed] [Google Scholar]
- Aliouat, E. M., E. Dei-Cas, L. Dujardin, J. P. Tissier, P. Billaut et al., 1996. High infectivity of Pneumocystis carinii cultivated on L2 rat alveolar epithelial cells. J. Eukaryot. Microbiol. 43: 22S. [DOI] [PubMed] [Google Scholar]
- Ambrose, H. E., S. P. Keely, E. M. Aliouat, E. Dei-Cas, A. E. Wakefield et al., 2004. Expression and complexity of the PRT1 multigene family of Pneumocystis carinii. Microbiology 150: 293–300. [DOI] [PubMed] [Google Scholar]
- Angus, C. W., A. Tu, P. Vogel, M. Qin and J. A. Kovacs, 1996. Expression of variants of the major surface glycoprotein of Pneumocystis carinii. J. Exp. Med. 183: 1229–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, J. D., M. L. Ginger, P. Burton and R. McCulloch, 2003. Why are parasite contingency genes often associated with telomeres? Int. J. Parasitol. 33: 29–45. [DOI] [PubMed] [Google Scholar]
- Berriman, M., and K. Rutherford, 2003. Viewing and annotating sequence data with Artemis. Brief. Bioinform. 4: 124–132. [DOI] [PubMed] [Google Scholar]
- Berriman, M., N. Hall, K. Sheader, F. Bringaud, B. Tiwari et al., 2002. The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 122: 131–140. [DOI] [PubMed] [Google Scholar]
- Borst, P., 1986. Discontinuous transcription and antigenic variation in trypanosomes. Annu. Rev. Biochem. 55: 701–732. [DOI] [PubMed] [Google Scholar]
- Borst, P., 2002. Antigenic variation and allelic exclusion. Cell 109: 5–8. [DOI] [PubMed] [Google Scholar]
- Burch, C. L., R. J. Danaher and D. C. Stein, 1997. Antigenic variation in Neisseria gonorrhoeae: production of multiple lipooligosaccharides. J. Bacteriol. 179: 982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, P. A., K. T. Tashima, M. B. Rogers, V. J. DiRita and S. B. Calderwood, 1997. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol. Microbiol. 25: 1099–1111. [DOI] [PubMed] [Google Scholar]
- Chen, C. J., C. Elkins and P. F. Sparling, 1998. Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect. Immun. 66: 987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth, M. N., and R. L. Eberle, 2001. Termini of human chromosomes display elevated rates of mitotic recombination. Mutagenesis 16: 85–89. [DOI] [PubMed] [Google Scholar]
- Cornillot, E., B. Keller, M. T. Cushion, G. Metenier and C. P. Vivares, 2002. Fine analysis of the Pneumocystis carinii f. sp. carinii genome by two-dimensional pulsed-field gel electrophoresis. Gene 293: 87–95. [DOI] [PubMed] [Google Scholar]
- Cushion, M. T., 1998. Genetic heterogeneity of rat-derived Pneumocystis. FEMS Immunol. Med. Microbiol. 22: 51–58. [DOI] [PubMed] [Google Scholar]
- Cushion, M. T., M. Kaselis, S. L. Stringer and J. R. Stringer, 1993. Genetic stability and diversity of Pneumocystis carinii infecting rat colonies. Infect. Immun. 61: 4801–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushion, M. T., M. Collins, B. Hazra and E. S. Kaneshiro, 2000. Effects of atovaquone and diospyrin-based drugs on the cellular ATP of Pneumocystis carinii f. sp. carinii. Antimicrob. Agents Chemother. 44: 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushion, M. T., S. Orr, S. P. Keely and J. R. Stringer, 2001. Time between inoculations and karyotype forms of Pneumocystis carinii f. sp. carinii influence outcome of experimental coinfections in rats. Infect. Immun. 69: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Penas, A., S. J. Pan, I. Castano, J. Alder, R. Cregg et al., 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17: 2245–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Portillo, H. A., C. Fernandez-Becerra, S. Bowman, K. Oliver, M. Preuss et al., 2001. A superfamily of variant genes encoded in the subtelomeric region of Plasmodium vivax. Nature 410: 839–842. [DOI] [PubMed] [Google Scholar]
- Edman, J. C., T. W. Hatton, M. Nam, R. Turner, Q. Mei et al., 1996. A single expression site with a conserved leader sequence regulates variation of expression of the Pneumocystis carinii family of major surface glycoprotein genes. DNA Cell Biol. 15: 989–999. [DOI] [PubMed] [Google Scholar]
- Ekins, A., and D. F. Niven, 2003. Transferrin-dependent expression of TbpA by Histophilus ovis involves a poly G tract within tbpA. FEMS Microbiol. Lett. 220: 95–98. [DOI] [PubMed] [Google Scholar]
- Elliott, B., C. Richardson, J. Winderbaum, J. A. Nickoloff and M. Jasin, 1998. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 18: 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel, G., E. Revardel, C. E. Koering and E. Gilson, 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 18: 2522–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior, L. H., E. Bottius, L. A. Pirrit, K. W. Deitsch, C. Scheidig et al., 2000. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature 407: 1018–1022. [DOI] [PubMed] [Google Scholar]
- Frenkel, J. K., 1976. Pneumocystis jiroveci n. sp. from man: morphology, physiology, and immunology in relation to pathology. Natl. Cancer Inst. Monogr.. 43: 13–30. [PubMed] [Google Scholar]
- Frenkel, J. K., 1999. Pneumocystis pneumonia, an immunodeficiency-dependent disease (IDD): a critical historical overview. J. Eukaryot. Microbiol. 46: 89S–92S. [PubMed] [Google Scholar]
- Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman et al., 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigliotti, F., L. R. Ballou, W. T. Hughes and B. D. Mosley, 1988. Purification and initial characterization of a ferret Pneumocystis carinii surface antigen. J. Infect. Dis. 158: 848–854. [DOI] [PubMed] [Google Scholar]
- Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian, 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Graves, D. C., S. J. McNabb, M. A. Worley, T. D. Downs and M. H. Ivey, 1986. Analyses of rat Pneumocystis carinii antigens recognized by human and rat antibodies by using western immunoblotting. Infect. Immun. 54: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt, S., A. Muller, H. Sillmann, M. Muhlenhoff, R. Borrow et al., 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20: 1211–1220. [DOI] [PubMed] [Google Scholar]
- Harris, D. E., and L. Murphy, 2001. Sequencing bacterial artificial chromosomes. Methods Mol. Biol. 175: 217–234. [DOI] [PubMed] [Google Scholar]
- Hong, S. T., P. E. Steele, M. T. Cushion, P. D. Walzer, S. L. Stringer et al., 1990. Pneumocystis carinii karyotypes. J. Clin. Microbiol. 28: 1785–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell-Adams, B., and H. S. Seifert, 2000. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol. Microbiol. 37: 1146–1158. [DOI] [PubMed] [Google Scholar]
- Huang, S. N., C. W. Angus, R. E. Turner, V. Sorial and J. A. Kovacs, 1999. Identification and characterization of novel variant major surface glycoprotein gene families in rat Pneumocystis carinii. J. Infect. Dis. 179: 192–200. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson, S., M. Michelitch and S. Ramcharan, 1993. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 3937–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson, A. B., G. Nyberg and S. Normark, 1991. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J. 10: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanti, B. M., D. E. Norris and N. Kumar, 2004. Molecular players of homologous recombination in protozoan parasites: implications for generating antigenic variation. Infect. Genet. Evol. 4: 91–98. [DOI] [PubMed] [Google Scholar]
- Kearns, D. B., F. Chu, R. Rudner and R. Losick, 2004. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol. Microbiol. 52: 357–369. [DOI] [PubMed] [Google Scholar]
- Keely, S. P., and J. R. Stringer, 2003. Sequence diversity of transcripts from Pneumocystis carinii gene families MSR and PRT1. J. Eukaryot. Microbiol. 50(Suppl): 627–628. [DOI] [PubMed] [Google Scholar]
- Keely, S. P., M. T. Cushion and J. R. Stringer, 2003. Diversity at the locus associated with transcription of a variable surface antigen of Pneumocystis carinii as an index of population structure and dynamics in infected rats. Infect. Immun. 71: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs, J. A., J. L. Halpern, J. C. Swan, J. Moss, J. E. Parrillo et al., 1988. Identification of antigens and antibodies specific for Pneumocystis carinii. J. Immunol. 140: 2023–2031. [PubMed] [Google Scholar]
- Kovacs, J. A., J. L. Halpern, B. Lundgren, J. C. Swan, J. E. Parrillo et al., 1989. Monoclonal antibodies to Pneumocystis carinii: identification of specific antigens and characterization of antigenic differences between rat and human isolates. J. Infect. Dis. 159: 60–70. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Kutty, G., S. N. Huang and J. A. Kovacs, 2003. Characterization of thioredoxin reductase genes (trr1) from Pneumocystis carinii and Pneumocystis jiroveci. Gene 310: 175–183. [DOI] [PubMed] [Google Scholar]
- Lavitola, A., C. Bucci, P. Salvatore, G. Maresca, C. B. Bruni et al., 1999. Intracistronic transcription termination in polysialyltransferase gene (siaD) affects phase variation in Neisseria meningitidis. Mol. Microbiol. 33: 119–127. [DOI] [PubMed] [Google Scholar]
- Lee, C. H., N. L. Bauer, M. M. Shaw, M. M. Durkin, M. S. Bartlett et al., 1993. Proliferation of rat Pneumocystis carinii on cells sheeted on microcarrier beads in spinner flasks. J. Clin. Microbiol. 31: 1659–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke, M. J., and P. D. Walzer, 1989. Analysis of a surface antigen of Pneumocystis carinii. J. Protozool. 36: 60S–61S. [DOI] [PubMed] [Google Scholar]
- Lugli, E. B., A. G. Allen and A. E. Wakefield, 1997. A Pneumocystis carinii multi-gene family with homology to subtilisin-like serine proteases. Microbiology 143: 2223–2236. [DOI] [PubMed] [Google Scholar]
- Lugli, E. B., E. T. Bampton, D. J. Ferguson and A. E. Wakefield, 1999. Cell surface protease PRT1 identified in the fungal pathogen Pneumocystis carinii. Mol. Microbiol. 31: 1723–1733. [DOI] [PubMed] [Google Scholar]
- Lundgren, B., G. Y. Lipschik and J. A. Kovacs, 1991. Purification and characterization of a major human Pneumocystis carinii surface antigen. J. Clin. Invest. 87: 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern, M. J., and S. Iyer, 2001. Short telomeres in yeast are highly recombinogenic. Mol. Cell 7: 695–704. [DOI] [PubMed] [Google Scholar]
- Mefford, H. C., and B. J. Trask, 2002. The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 3: 91–102. [DOI] [PubMed] [Google Scholar]
- Merali, S., U. Frevert, J. H. Williams, K. Chin, R. Bryan et al., 1999. Continuous axenic cultivation of Pneumocystis carinii. Proc. Natl. Acad. Sci. USA 96: 2402–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myler, P. J., R. F. Aline, Jr., J. K. Scholler and K. D. Stuart, 1988. Multiple events associated with antigenic switching in Trypanosoma brucei. Mol. Biochem. Parasitol. 29: 227–241. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., 1998. The major surface antigen of Pneumocystis carinii. FEMS Immunol. Med. Microbiol. 22: 67–74. [DOI] [PubMed] [Google Scholar]
- Natarajan, S., and M. J. McEachern, 2002. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 22: 4512–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., and T. Gojobori, 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Nickoloff, J. A., J. D. Singer, M. F. Hoekstra and F. Heffron, 1989. Double-strand breaks stimulate alternative mechanisms of recombination repair. J. Mol. Biol. 207: 527–541. [DOI] [PubMed] [Google Scholar]
- Park, S. F., D. Purdy and S. Leach, 2000. Localized reversible frameshift mutation in the flhA gene confers phase variability to flagellin gene expression in Campylobacter coli. J. Bacteriol. 182: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays, E., S. Van Assel, M. Laurent, M. Darville, T. Vervoort et al., 1983. Gene conversion as a mechanism for antigenic variation in trypanosomes. Cell 34: 371–381. [DOI] [PubMed] [Google Scholar]
- Prudden, J., J. S. Evans, S. P. Hussey, B. Deans, P. O'Neill et al., 2003. Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J. 22: 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde, F. E., and E. J. Louis, 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18: 2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde, F. E., H. C. Gorham and E. J. Louis, 1997. Chromosome ends: all the same under their caps. Curr. Opin. Genet. Dev. 7: 822–828. [DOI] [PubMed] [Google Scholar]
- Russian, D. A., V. Andrawis-Sorial, M. P. Goheen, J. C. Edman, P. Vogel et al., 1999. Characterization of a multicopy family of genes encoding a surface- expressed serine endoprotease in rat Pneumocystis carinii. Proc. Assoc. Am. Physicians. 111: 347–356. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. F., E. F. Fristch and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schaffzin, J. K., and J. R. Stringer, 2004. Expression of the Pneumocystis carinii major surface glycoprotein epitope is correlated with linkage of the cognate gene to the upstream conserved sequence locus. Microbiology 150: 677–686. [DOI] [PubMed] [Google Scholar]
- Schaffzin, J. K., T. R. Garbe and J. R. Stringer, 1999. a Major surface glycoprotein genes from pneumocystis carinii f. sp. ratti. Fungal Genet. Biol. 28: 214–226. [DOI] [PubMed] [Google Scholar]
- Schaffzin, J. K., S. M. Sunkin and J. R. Stringer, 1999. b A new family of Pneumocystis carinii genes related to those encoding the major surface glycoprotein. Curr. Genet. 35: 134–143. [DOI] [PubMed] [Google Scholar]
- Scholler, J. K., P. J. Myler and K. D. Stuart, 1989. A novel telomeric gene conversion in Trypanosoma brucei. Mol. Biochem. Parasitol. 35: 11–19. [DOI] [PubMed] [Google Scholar]
- Segura, A., A. Hurtado, E. Duque and J. L. Ramos, 2004. Transcriptional phase variation at the flhB gene of Pseudomonas putida DOT-T1E is involved in response to environmental changes and suggests the participation of the flagellar export system in solvent tolerance. J. Bacteriol. 186: 1905–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloand, E., B. Laughon, M. Armstrong, M. S. Bartlett, W. Blumenfeld et al., 1993. The challenge of Pneumocystis carinii culture. J. Eukaryot. Microbiol. 40: 188–195. [DOI] [PubMed] [Google Scholar]
- Smulian, A. G., T. Sesterhenn, R. Tanaka and M. T. Cushion, 2001. The ste3 pheromone receptor gene of Pneumocystis carinii is surrounded by a cluster of signal transduction genes. Genetics 157: 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E. L., and R. Durbin, 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: 1–10. [DOI] [PubMed] [Google Scholar]
- Streisinger, G., and J. Owen, 1985. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics 109: 633–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer, J. R., 2002. Pneumocystis. Int. J. Med. Microbiol. 292: 391–404. [DOI] [PubMed] [Google Scholar]
- Stringer, J. R., and M. T. Cushion, 1998. The genome of Pneumocystis carinii. FEMS Immunol. Med. Microbiol. 22: 15–26. [DOI] [PubMed] [Google Scholar]
- Stringer, J. R., and S. P. Keely, 2001. Genetics of surface antigen expression in Pneumocystis carinii. Infect. Immun. 69: 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer, J. R., C. B. Beard, R. F. Miller and A. E. Wakefield, 2002. A new name (Pneumocystis jiroveci) for Pneumocystis from humans. Emerg. Infect. Dis. 8: 891–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer, S. L., S. T. Hong, D. Giuntoli and J. R. Stringer, 1991. Repeated DNA in Pneumocystis carinii. J. Clin. Microbiol. 29: 1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkin, S. M., and J. R. Stringer, 1996. Translocation of surface antigen genes to a unique telomeric expression site in Pneumocystis carinii. Mol. Microbiol. 19: 283–295. [DOI] [PubMed] [Google Scholar]
- Sunkin, S. M., and J. R. Stringer, 1997. Residence at the expression site is necessary and sufficient for the transcription of surface antigen genes of Pneumocystis carinii. Mol. Microbiol. 25: 147–160. [DOI] [PubMed] [Google Scholar]
- Sunkin, S. M., M. J. Linke, F. X. McCormack, P. D. Walzer and J. R. Stringer, 1998. Identification of a putative precursor to the major surface glycoprotein of Pneumocystis carinii. Infect. Immun. 66: 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood, A. P., E. J. Louis, R. H. Borts, J. R. Stringer and A. E. Wakefield, 1996. Pneumocystis carinii telomere repeats are composed of TTAGGG and the subtelomeric sequence contains a gene encoding the major surface glycoprotein. Mol. Microbiol. 19: 273–281. [DOI] [PubMed] [Google Scholar]
- Van Belkum, A., 1999. Short sequence repeats in microbial pathogenesis and evolution. Cell Mol. Life Sci. 56: 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belkum, A., S. Scherer, L. van Alphen and H. Verbrugh, 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62: 275–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verona, R. I., M. R. Mann and M. S. Bartolomei, 2003. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 19: 237–259. [DOI] [PubMed] [Google Scholar]
- Wada, M., and Y. Nakamura, 1994. MSG gene cluster encoding major cell surface glycoproteins of rat Pneumocystis carinii. DNA Res. 1: 163–168. [DOI] [PubMed] [Google Scholar]
- Wada, M., and Y. Nakamura, 1996. a Antigenic variation by telomeric recombination of major-surface-glycoprotein genes of Pneumocystis carinii. J. Eukaryot. Microbiol. 43: 8S. [DOI] [PubMed] [Google Scholar]
- Wada, M., and Y. Nakamura, 1996. b Unique telomeric expression site of major-surface-glycoprotein genes of Pneumocystis carinii. DNA Res. 3: 55–64. [DOI] [PubMed] [Google Scholar]
- Wada, M., and Y. Nakamura, 1997. cDNA cloning and overexpression of cell surface subtilisin-like proteases (SSP) of Pneumocystis carinii. J. Eukaryot. Microbiol. 44: 54S. [DOI] [PubMed] [Google Scholar]
- Wada, M., and Y. Nakamura, 1999. a Immunological characterization of surface subtilisin-like protease (SSP) of Pneumocystis carinii. J. Eukaryot. Microbiol. 46: 151S–152S. [PubMed] [Google Scholar]
- Wada, M., and Y. Nakamura, 1999. b Type-II major-surface-glycoprotein family of Pneumocystis carinii under the control of novel expression elements. DNA Res. 6: 211–217. [DOI] [PubMed] [Google Scholar]
- Wada, M., K. Kitada, M. Saito, K. Egawa and Y. Nakamura, 1993. cDNA sequence diversity and genomic clusters of major surface glycoprotein genes of Pneumocystis carinii. J. Infect. Dis. 168: 979–985. [DOI] [PubMed] [Google Scholar]
- Wada, M., S. M. Sunkin, J. R. Stringer and Y. Nakamura, 1995. Antigenic variation by positional control of major surface glycoprotein gene expression in Pneumocystis carinii. J. Infect. Dis. 171: 1563–1568. [DOI] [PubMed] [Google Scholar]
- Walzer, P. D., and M. J. Linke, 1987. A comparison of the antigenic characteristics of rat and human Pneumocystis carinii by immunoblotting. J. Immunol. 138: 2257–2265. [PubMed] [Google Scholar]
- Weng, Y. S., J. Whelden, L. Gunn and J. A. Nickoloff, 1996. Double-strand break-induced mitotic gene conversion: examination of tract polarity and products of multiple recombinational repair events. Curr. Genet. 29: 335–343. [DOI] [PubMed] [Google Scholar]
- Woelk, C. H., O. G. Pybus, L. Jin, D. W. Brown and E. C. Holmes, 2002. Increased positive selection pressure in persistent (SSPE) versus acute measles virus infections. J. Gen. Virol. 83: 1419–1430. [DOI] [PubMed] [Google Scholar]
- Yogev, D., R. Rosengarten and K. S. Wise, 1993. Variation and genetic control of surface antigen expression in mycoplasmas: the Vlp system of Mycoplasma hyorhinis. Zentralbl. Bakteriol. 278: 275–286. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., and K. S. Wise, 1997. Localized reversible frameshift mutation in an adhesin gene confers a phase-variable adherence phenotype in mycoplasma. Mol. Microbiol. 25: 859–869. [DOI] [PubMed] [Google Scholar]