Abstract
Polyploidy, the inheritance of more than two genome copies per cell, has played a major role in the evolution of higher plants. Little is known about the transition from diploidy to polyploidy but in some species, triploids are thought to function as intermediates in this transition. In contrast, in other species triploidy is viewed as a block. We investigated the responses of Arabidopsis thaliana to triploidy. The role of genetic variability was tested by comparing triploids generated from crosses between Col-0, a diploid, and either a natural autotetraploid (Wa-1) or an induced tetraploid of Col-0. In this study, we demonstrate that triploids of A. thaliana are fertile, producing a swarm of different aneuploids. Propagation of the progeny of a triploid for a few generations resulted in diploid and tetraploid cohorts. This demonstrated that, in A. thaliana, triploids can readily form tetraploids and function as bridges between euploid types. Genetic analysis of recombinant inbred lines produced from a triploid identified a locus on chromosome I exhibiting allelic bias in the tetraploid lines but not in the diploid lines. Thus, genetic variation was subject to selection contingent on the final ploidy and possibly acting during the protracted aneuploid phase.
THE genomes of flowering plants, yeast, and vertebrates bear the imprint of multiple ancestral duplications, consistent with sudden and accidental formation of polyploids from diploids followed by gradual diploidization (Wolfe and Shields 1997; Soltis et al. 2003). Considering the importance of polyploidy in evolution, surprisingly little is known about how new polyploids arise and whether gene flow occurs between polyploids and their cognate diploids, a feature that could affect their evolutionary relationship. Particularly, triploids may function as bridges between diploid and tetraploid populations (Ramsey and Schemske 1998).
Triploids are often phenotypically normal plants but are meiotically unstable and therefore transient. Meiosis and chromosome pairing are particularly complicated for triploids in which three sets of chromosomes must be resolved to two poles. This results in frequent chromosome loss and chromosome fragmentation (McClintock 1929). Even if chromosome pairing is successfully resolved, independent assortment produces mostly aneuploid gametes. As a result, the immediate progeny of triploids can be composed of a complex swarm of varied karyotypes, which differ in the number of copies of each chromosome. The negative consequences of aneuploidy on the gametophytes and the progeny, as well as meiotic instability, can contribute to a dramatic reduction in fertility of triploids (Khush 1973). In animals, triploidy almost always results in complete sterility (Benfey 1999; Garnier-Gere et al. 2002). Triploid plants, on the other hand, exhibit various levels of fertility ranging from effective sterility, such as in seedless watermelon, to the severely reduced fertility of Datura (Satina and Blakeslee 1937a,b, 1938) and maize (McClintock 1929) or the mildly reduced fertility of poplar (Johnsson 1942), sugar beet (Levan 1942), or Melandrium (Warmke and Blakeslee 1940).
Among aneuploids, trisomics are by far the best documented (Khush 1973). In the early 1930s, triploids were used as sources of trisomics, which were instrumental in the confirmation of the role of individual chromosomes in development (Blakeslee 1922). Blakeslee (1922) used a triploid to produce all 12 trisomics of Datura, all exhibiting different phenotypes, as predicted. Similar results were obtained later in tobacco (Clausen and Cameron 1944), tomato (Lesley 1928), and maize (McClintock 1929). These studies introduced the idea that triploids could produce viable aneuploids, which were mostly trisomics. Variation for aneuploid production exists. For example, triploids of cherry tomato produce more aneuploids than triploids of the San Marzano variety (Rick and Baron 1953; Rick and Notani 1961). Variation is even greater between species. The triploids of sugar beet (Levan 1942), Melandrium (Warmke and Blakeslee 1940), or poplar (Johnsson 1945) produce other types of viable aneuploids, possessing multiple unbalanced chromosomes. The mechanisms behind the differential response to triploidy and aneuploidy in plants vs. animals, between different plant species, or between varieties of the same species remain poorly understood.
To identify the mechanisms for these differential responses, the consequences of triploidy must be characterized in a genetically tractable system. We chose to investigate the effects of triploidy in A. thaliana. Here, we show that triploids of A. thaliana are fertile, producing a swarm of different aneuploids. The role of genetic variability was tested by comparing triploids generated from crosses between Col-0, a diploid, and either its synthetically produced tetraploid derivative or a natural autotetraploid ecotype (Wa-1). The composition and performance of the swarm was affected by the genotype of the triploid. Furthermore, recombinant inbred lines produced from the progeny of a Col-0 × Wa-1 triploid (Schiff et al. 2001) resolved into diploid and tetraploid cohorts. Thus, Arabidopsis triploids can readily form tetraploids and function as bridges between euploid types. During this process, genetic variation was subject to selection either in aneuploids or in the resulting tetraploids. Genetic analysis of these recombinant inbred lines identified a locus on chromosome I exhibiting allelic bias in the tetraploid lines but not in the diploid lines, consistent with ploidy-dependent selection.
MATERIALS AND METHODS
Plant materials: lines, growth, and crosses:
Most Arabidopsis thaliana ecotypes were obtained from the Arabidopsis Biological Resources Center (ABRC). A few ecotypes were obtained directly from Magnus Nordberg (University of Southern California, Los Angeles) and are now available at the ABRC. The Col-0 × Wa-1 recombinant inbred lines described by Schiff and coworkers (Schiff et al. 2001) were obtained directly from Shauna Somerville (Carnegie Institution, Stanford, CA).
The nomenclature of the different lines used is as follows: Col-0 represents the diploid ecotype Columbia; 4x-Col represents tetraploidized (see below) Col-0; and Wa-1 represents the naturally occurring tetraploid ecotype Warschau. The two triploids were generated as follows: the CCC triploid is the product of a cross between Col-0 as seed parent and 4x-Col as pollen parent while the CWW triploid was generated by crossing Col-0 as seed parent to Wa-1.
Tetraploid A. thaliana, ecotype Columbia (4x-Col) was generated as follows: Col-0 Arabidopsis seeds were surface sterilized, sown on 0.5× MS agar medium (Murashige and Skoog 1962) supplemented with 0.5% sucrose, stratified for 4 days at 4° in the dark, moved to a 16:8 hr light:dark regime, and grown for 2 weeks. Seedlings on the plate were submerged for 2 hr in 0.1% (w/v) colchicine. Seedlings were gently removed, washed with copious amounts of fresh water, and transplanted to soil (Sunshine Professional Peat-Lite mix 4; SunGro Horticulture, Vancouver, BC). Seeds were collected from individual plants and sown on moist soil, stratified for 4 days, and grown for 4 weeks. Nuclear DNA content from these plants was estimated by flow cytometry and one tetraploid individual was selfed to generate the 4x-Col line used in this study.
All plants were grown in a growth room lit by fluorescent lamps (model TL80; Phillips, Sunnyvale, CA) at 22 ± 3° with a 16:8 hr light:dark photoperiod or in a greenhouse at similar temperatures and light regimes, with supplemental light provided by sodium lamp illumination, as required.
Phenotypic analysis of reproductive traits:
Measurements of pollen counts and viability were estimated using Alexander's stain (Alexander 1969). Pollen grains were counted by collecting five freshly opened flowers from a plant into a microcentrifuge tube containing 70 μl of Alexander's stain (Alexander 1969). The tubes were vortexed and left at room temperature for 30 min. The stain solution and pollen were transferred to a new tube and the volume was adjusted to 100 μl. The pollen grains were resuspended by vortexing and 0.7 μl was pipetted onto a microscope slide and covered with a coverslip. Six aliquots were counted from each sample, and each plant was sampled six independent times.
Pollen viability was tested by placing a flower corolla-side-down in a drop of Alexander stain (Alexander 1969) on a microscope slide for 5 min. The flower was removed and the droplet was covered with a coverslip. The slide was placed on a heated plate (50°) for at least 1 hr. The numbers of viable (purple) and nonviable (blue/green) pollen grains were recorded using a compound microscope. For each plant, estimates of pollen viability were determined from two to seven individual flowers. All flowers were collected in the morning.
For the measurement of seed traits, single siliques were harvested in individual tubes and all seeds were counted using a dissecting microscope. Seeds were characterized as being either “plump” (if they contained a visible embryo structure at least 20% the size of wild-type seed) or “shriveled” (if they did not). Seed germination rates were determined by counting the number of seedlings at 15 days and comparing it to the total number of seeds sown.
For all traits, mean values for each of the five genotypes were compared to each other on a pairwise basis using Student's t-test (α = 0.05).
Flow cytometric determination of genome content:
Approximately 300 mg of leaf tissue or three clusters of unopened buds from inflorescences were harvested from an individual plant and laid in a round petri dish sitting on a bed of ice and containing 1.5 ml of ice-cold chopping buffer (15 mm HEPES, 1 mm EDTA, 80 mm KCl, 20 mm NaCl, 300 mm sucrose, 0.20% Triton-X, 0.5 mm spermine, and 0.10% freshly added β-mercaptoethanol, pH 6.1). The tissue was chopped using a fresh carbon-steel razor blade (VWR) for ∼1 min. The fluid was transferred to a 5-cc luer-loc syringe (Becton-Dickinson, Franklin Lakes, NJ) fitted with a 13-mm filter holder (Millipore, Bedford, MA) containing a 30-μm nylon mesh filter (Small Parts, Miami Lakes, FL; CMN-30 monofilament cloth). The fluid was gently passed from the syringe through the filter and into a 1.7-ml microcentrifuge tube and stored on ice. The tubes were centrifuged at 500 × g for 7 min at 4° and the supernatant was discarded. Pelleted nuclei were resuspended in 0.5 ml of ice-cold staining solution (640 μl of chopping buffer supplemented with 0.31 μl of 100 mg/ml RNAse and 12.8 μl of 5 mg/ml propidium iodide). The fluid was transferred into another syringe fitted with a 30-μm nylon mesh filter and gently forced through the filter into a 5-ml tube (Falcon, Lincoln Park, NJ). The tubes were capped with parafilm and stored in the dark and on ice for at least 2 hr prior to analysis to allow the binding of propidium iodide and DNA to approach equilibrium.
All samples were analyzed with a Becton-Dickinson FACScan. Data from a minimum of 10,000 events were collected from each sample. For each sample, the mean fluorescence intensity of the basal somatic DNA content peak produced by the CellQuest software was saved for further analysis (the same conclusions were drawn when the median of the fluorescence intensity peak was used instead). To allow comparison of different samples and data from different days, the fluorescence intensity of each sample was normalized to that of an external standard as follows: for each cytometry run (all samples prepared and run on the same day), at least two control samples containing Col-0 nuclei were run at least twice each. For each day, a scaling factor, A, was determined by averaging the fluorescence intensities of the first peak (e.g., the 2C nuclei of a diploid) of Col-0 samples and dividing this number by two. The fluorescence intensities of control samples were normalized by dividing them, for each sample, by the corresponding day's A-value. Regression analysis of data from controls of known genome content was used to determine the relationship between the normalized fluorescence intensity (FI/A) and genome content. The controls included diploid Col-0 samples [genome content (GC) of 2.0], samples from triploids of the CCC and CWW genotypes (GC of 3.0), and tetraploid 4x-Col and Wa-1 samples (GC of 4.0). These controls were analyzed on multiple days and originated from either the same plant or different plants of the same genotype. The regression was significant and the data were well fit by the model (P < 0.0001; R2 = 0.989). The genome contents of the unknown samples were calculated using the regression model GC = 0.358685 + 1.6719 × FI/A (supplementary Figure S1 at http://www.genetics.org/supplemental/).
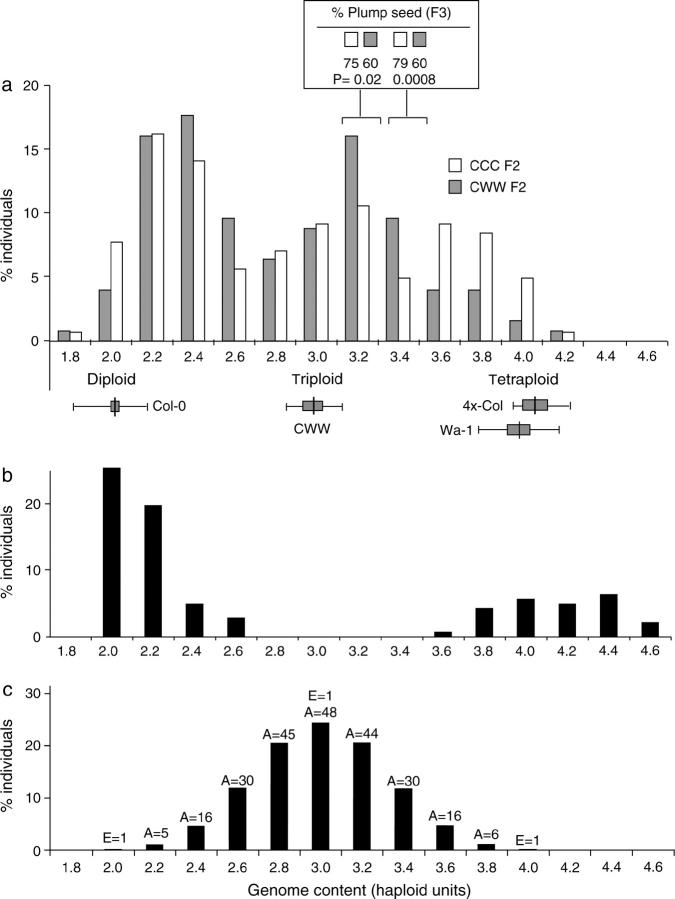
All genome contents were expressed as a multiple of the haploid genome content of Col-0, because we were interested in estimating the genome content of polyploids and aneuploids of the same species relative to diploid individuals of the same species. According to this scale, 2.0 corresponds to diploid individuals and 4.0 corresponds to tetraploid individuals. The range of error obtained for the control samples is summarized in Figure 2a and supplementary Figure S1.
Figure 2.—
(a) Histogram summarizing the distribution of genome contents (expressed as number of haploid Col-0 units) in the aneuploid swarms produced by the CCC and CWW triploids. The height of each bar corresponds to the frequency for each genome content class in the F2 populations derived from CCC (open bars) and CWW (shaded bars). The mean (vertical solid lines), standard deviation (shaded boxes), and range (solid horizontal lines) of genome content measurements obtained for control plants (Col-0, Wa-1, triploid CWW, and 4x-Col) are indicated below the histogram. The percentage of plump seed was different for the CCC and CWW plants in two genome content classes: 3.2 and 3.4. The values obtained for these classes as well as the corresponding t-test P-values are indicated in the inset immediately above the top histogram. (b) Histogram summarizing the distribution of genome contents in the 89 RILs. (c) Expected distribution of genome sizes in the F2 progeny from a triploid assuming random distribution of the chromosomes during meiosis and the absence of selection. Above each bar are indicated the number of euploid (E) and aneuploid (A) types contained in each genome content class.
For the initial characterization of the recombinant inbred lines (RILs), the fluorescence intensity measurements for each RIL were determined by scaling each measurement to Col-0 diploid controls for that day. GC was then estimated by using the regression model calculated above.
Statistical analysis of the F2 genome content distributions:
The expected distribution of genome content in the progeny of a triploid was calculated as follows. The frequency of each aneuploid class was calculated as expected from the random segregation of three sets of chromosomes without selection. On the basis of DNA sequence length (TIGR5.0 release at www.arabidopsis.org) the following genome size percentages were assigned to chromosomes I–V, respectively: 24.5, 15, 20, 18.5, and 22. For each aneuploid class, genome content was calculated using these values. The value obtained was divided by 100 (haploid genome content) and rounded to the nearest decimal point. The different aneuploid types were then binned into the same genome content classes used for the progeny of the CCC and CWW triploids. To compare the observed distributions of genome size (obtained by flow cytometry) to this expected genome size distribution, individuals with the lowest and highest genome contents were pooled in the following categories to ensure enough individuals in every category: 1.8–2.4, 2.6, 2.8, 3.0, 3.2, 3.4, and 3.6–4.2. The theoretical distribution was treated in a similar way and the two distributions were compared using the chi-square test. Similar analyses using different binning patterns were performed to compare the two observed distributions with each other. To compare the overall CCC and CWW F2 distributions, the following genome content classes were used: 1.8–2.0, 2.2, 2.4, 2.6, 2.8, 3.0, 3.2, 3.4, 3.6, and 3.8–4.2. To compare the upper half of the F2 distributions, the following genome content classes were used: 3.2–3.4 and 3.6–4.2. In both cases, the resulting distributions were compared using chi-square tests.
Genetic analyses:
The 94 RILs derived from a CWW triploid were in the F6 or F8 generation following a Col-0 (seed parent) × Wa-1 (pollen parent) cross when genotyped by Schiff et al. (2001). In this prior study, a few seeds from each of 120 F2's were planted and a fertile individual was selected randomly to continue the lineage to the F8 or F6 generation, depending on the plant (Schiff et al. 2001 and personal communication). We obtained these lines directly from Shauna Sommerville (Carnegie Institution) and one individual from each RIL was grown alongside the original parents (Col-0 and Wa-1). Leaf material was harvested both for the measurement of genome content by flow cytometry (see above) and for DNA extraction. Genomic DNA was extracted using the Q-Biogene (Carlsbad, CA) Fast DNA kit according to the manufacturer's recommendations and markers were amplified by PCR. PCR products were resolved on 5% polyacrylamide gels.
The original data set obtained by Schiff et al. is available at http://carnegiedpb.stanford.edu/research/shauna/host_resistance.html. We updated the genotyping data for the following markers: ATEAT1, nga63, AthZFPG, T27K12, nga280, nga111, and nga692 on chromosome I; nga1145, nga1126, and nga168 on chromosome II; AthDET1, SC5, and nga1107 on chromosome IV; and nga106 and nga129 on chromosome V. Data for an additional 23 markers (Table 1) were added to this data set (see supplementary Table S1 at http://www.genetics.org/supplemental/ for complete genotype and genome content data). Five lines, corresponding to RILs 28, 31, 34, 66, and 108 in the original Schiff population, consistently exhibited allele polymorphisms distinct from both Wa-1 and Col-0, suggesting that they had either been contaminated by outcrossing to another ecotype or represented seed contaminants. These lines were excluded from the analysis. To evaluate marker orders, a genetic map from the RIL population was created using an implementation of the Morgan mapping function in Map Manager QTX version b20 (Manly et al. 2001) (supplementary Figure S2 at http://www.genetics.org/supplemental/).
TABLE 1.
PCR markers used to complete the Col-0 × Wa-1 genotyping
| Marker name | Chromosome | Sequence primer 1 | Sequence primer 2 |
|---|---|---|---|
| F21M12 | I | GGCTTTCTCGAAATCTGTCC | TTACTTTTTGCCTCTTGTCATTG |
| MSAT1.3 | I | GGAACTGTTGTCTGGGTAAG | CGATTGCACTAAAAGCTCTC |
| MN1.6 | I | TAGTGGAAAGCTGTGCGATG | CTCACCATGTTGTCCGAATG |
| MN1.7 | I | GCAAATCGCCTGTTTTCTTG | CATCTGCGACTGAGAGTTCAA |
| CIW1 | I | ACATTTTCTCAATCCTTACTC | GAGAGCTTCTTTATTTGTGAT |
| MN1.2 | I | TCAATCTCAACATCGGATCAA | TGCTTCCAACAAGTGACAATG |
| NK19K23 | I | GAATTCTGTAACATCCCATTTCC | GGTCTAATTGCCGTTGTTGC |
| F5I14 | I | CTGCCTGAAATTGTCGAAAC | GGCATCACAGTTCTGATTCC |
| MSAT1.5 | I | GCATCGCTCTTAAACAACCAT | CGTTGCAAAACCGTATCAGAA |
| MSAT2.28 | II | AATAGAAATGGAGTTCGACG | TGAACTTGTTGTGAGCTTTG |
| MN2.1 | II | AGGACAAGAGAAATATCAAGAAACAATATC | CTTTTGCAAATTTGTAGGTACTTGTTATAG |
| MSAT2.33 | II | GCTATGCTTTTTCCTGATCT | CATTGCCAGAATCTCGAC |
| MN3.1 | III | AAAAAGGTTTTGATGTTTTACATGAGTAAG | GTTCACAAGAAGAGGGATTAAAGAGTC |
| MSAT4.8 | IV | GTTGGGTTTAGTTGGTAACA | CGGGTAAAGACAGAGCAT |
| MN4.2 | IV | TAAGGTCAGACTATATGTTTACGTTTCATT | GTCATCCTCGTTTAAGTTACGATTG |
| MSAT4.25 | IV | GAATGGTTGTTGATAGTTGA | AAATTTCAGGAGGTGATAGA |
| MSAT4.18 | IV | TGTAAATATCGGCTTCTAAG | CTGAAACAAATCGCATTA |
| MN4.1 | IV | TATAAACTCCTAACATATTGACTTGACCAG | TCGAGAAATTTACTGTGAAAATGAATC |
| nga249 | V | GGATCCCTAACTGTAAAATCCC | TACCGTCAATTTCATCGCC |
| MSAT5.14 | V | AACAACCCTATCTTCTTCTG | TGTGACCCCTTACTCAATA |
| MN5.1 | V | GAAAACCATGTCTATTAACAACAACAAC | AGCTCTAACACGTTTCCCAAGTATAA |
| MSAT5.25 | V | GCTTAATTTGGGTTAAAT | GCACGCAAGTGACT |
| MSAT5.19 | V | AACGCATTTGCTGTTTCCCA | ATGGTTATCTCATCTGGTCT |
Marker associations were evaluated using Fisher's exact test to compare the proportion of individuals homozygous for the Col-0 and the Wa-1 alleles in the diploid and tetraploid populations. This test is more conservative than the chi-square test and is valid for populations with fewer than five individuals in a class. Only homozygous individuals for each marker were considered because our genotyping method cannot unambiguously discriminate between the different polyploid heterozygote types. A Bonferroni correction was applied corresponding to a total of 11 independent tests from ∼550 cM on 10 chromosome arms. The number of tetraploid homozygous individuals determined the observed sample size. Expected values were calculated from the frequencies of Wa-1 and Col-0 homozygotes in the diploid population.
RESULTS
Natural autotetraploid accessions of A. thaliana:
The analysis of natural variation in a genetic trait can provide important information. We undertook a survey of A. thaliana ecotypes to determine whether polyploids are part of natural populations of this model plant. Flow cytometric analyses were used to estimate the nuclear DNA contents of 161 ecotypes of A. thaliana. These included the most widely used laboratory strains, the ecotypes selected for linkage-disequilibrium mapping and single-nucleotide polymorphism discovery by Magnus Nordberg (ABRC number between CS22491 and CS22613), and a few additional types (listed in supplementary Table S2 at http://www.genetics.org/supplemental/). Of these, only two tetraploids were found. These were Wa-1 (from Warsaw; ABRC no. CS6885) and M3385S (from Stockholm; ABRC no. CS3111), both of which were identified in a much smaller sampling of ecotypes reported earlier (Schmuths et al. 2004). Notably, another ecotype called Stockhom (St-0) is diploid although previously reported to be tetraploid (Heslop-Harrison and Maluszynska 1994). Of these two autotetraploids, Wa-1 was chosen for further analysis because, like Col-0, Wa-1 is early flowering while M3385S is late flowering (data not shown).
Fertility of triploid A. thaliana:
We investigated triploids of A. thaliana, which had been reported to be fertile (Steinitz-Sears 1962). We crossed the standard diploid Columbia-0 (Col-0) to both the natural autotetraploid Wa-1 and a synthetic autotetraploid derivative of Col-0 (4x-Col) to make the CWW and CCC triploids (C and W represent Col-0 and Wa-1 genomes, respectively). We sought to determine if triploids with chromosomes from a natural tetraploid accession were more fit than those with only Col-0 chromosomes.
Diploid, triploid, and tetraploid A. thaliana were phenotypically very similar, the most obvious difference being flower size, which increased with ploidy. Comparison of the two tetraploid parents revealed that Wa-1 did not outperform 4x-Col for most fertility-related traits (Figure 1), suggesting that if any adaptations to polyploidy have been selected in this accession, they are not dramatically visible in the seeds and pollen of tetraploids. Neither the CCC nor the CWW triploids performed as well as their diploid or tetraploid parents for these traits (Figure 1). Pollen viability and yield were reduced similarly in both triploids as compared to that of the diploid and tetraploid parents (t-test; all P-values <0.003). The triploids produced between 65% (CCC) and 75% (CWW) plump seeds (i.e., well formed), which was less than the diploid and tetraploid parents produced (t-test; all P-values <0.0001). CWW siliques contained a higher percentage of plump seeds than CCC siliques (t-test; P-value <0.0001). Thus, the genotype of the triploid affected the viability of its progeny.
Figure 1.—
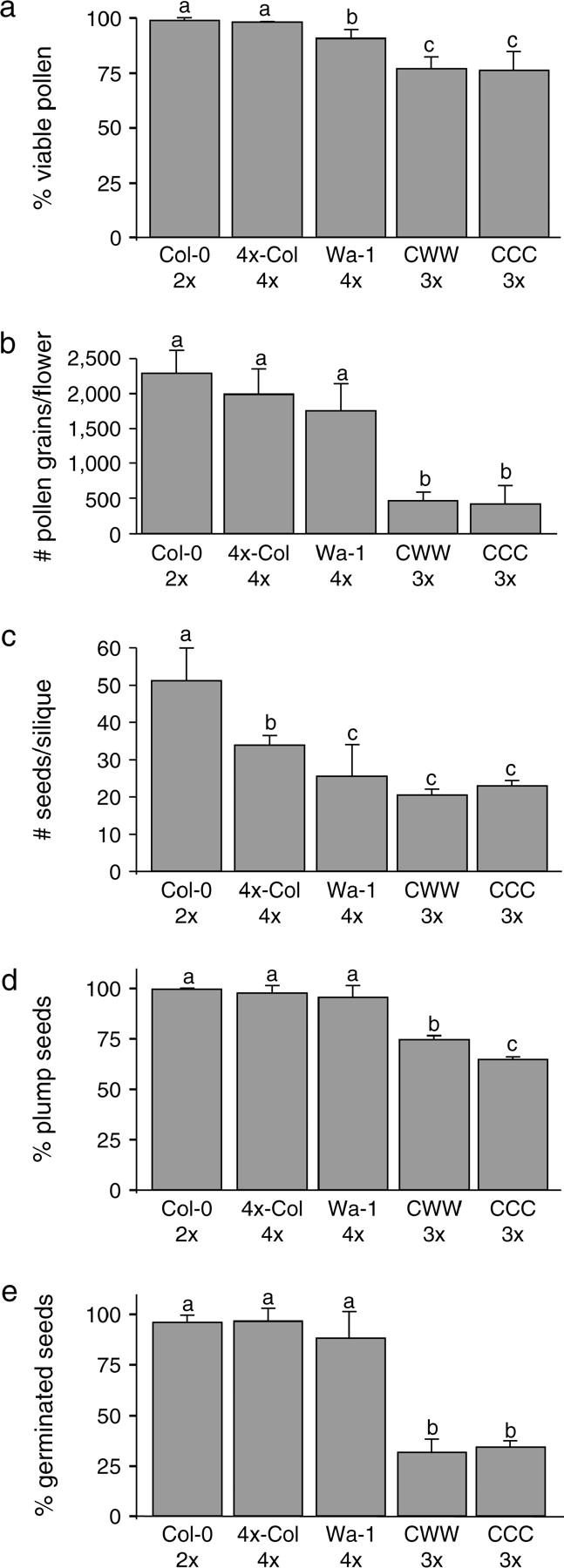
Effect of genotype and ploidy on fertility-associated phenotypes. The mean percentage of viable pollen grains per flower (a), mean number of pollen grains per flower (b), mean number of seeds per silique (c), mean percentage of plump seeds (as a measure of seed viability) per silique (d), and mean percentage of germinated seeds per silique (e) were measured in diploid Columbia (Col-0), the CCC and CWW triploids, and the tetraploids Wa-1 and 4x-Col. The mean (indicated by the height of each column) and standard deviation (indicated by the error bars) are displayed. Means were compared by Student's t-test. Different letters above two columns indicate significantly different means for these two measurements (P-value <0.05).
Triploids of Arabidopsis produce an aneuploid swarm:
The triploids were allowed to self-pollinate and the genome contents of 124 CWW F2 plants and 141 CCC F2 plants were estimated by flow cytometric analysis of nuclear DNA. Both triploids produced a swarm of aneuploids including trisomics and other aneuploids spanning a wide range of genome contents (Figure 2a). The distributions of genome content in these swarms deviated from the theoretical distribution of genome contents (see materials and methods) expected from random chromosome segregation and an absence of selection (Figure 2, a and c; χ2 = 345.3, d.f. = 6, and P-value <0.0001 for the CCC F2 and χ2 = 269.5, d.f. = 6, and P-value <0.0001 for the CWW F2). The proportion of euploid F2 plants was greater than predicted, suggesting a strong selection against aneuploids or meiotic mechanisms favoring euploid gamete formation.
The distributions of the two F2 families were different (χ2 = 24.6, d.f. = 9, and P-value = 0.0035). Most of the difference between the two distributions stemmed from the upper half of the distributions. CWW produced more individuals in genome content classes 3.4 and 3.6 while CCC produced more individuals in genome content classes 3.6–4.2 (χ2 = 18.17, d.f. = 1, and P-value <0.0001). Thus, either possessing Wa-1 alleles influences the effect of different types of aneuploidy or the CWW and CCC triploids produce different types of gametes in different proportions. Unfortunately, flow cytometric measurement of genome content does not distinguish between different aneuploid types of similar chromosome number or genome content. It is also possible that some of the F2 individuals contain chromosome fragments due to chromosome breaks during meiosis. Finally, the relative size of the different chromosome types may not be exactly the same in Col-0 as in Wa-1. Therefore, we cannot rule out the possibility that within each genome content class, the types of aneuploids produced by the CCC and CWW triploids differ as well.
Phenotype and fertility of the aneuploid swarm:
The progeny of both triploids contained plants exhibiting various phenotypes, such as altered leaf color and shape, dwarfism ranging from subtle to extreme, and changes in flowering time (Figure 3, a–h). The phenotypes of the F3 seeds were also variable: individuals in some families aborted much earlier in their development than others (Figure 3, i–p; data not shown). With the possible exception of the two extreme classes, each genome content class can contain several (up to 50) different types of aneuploids (Figure 2c). Therefore, each genome content class contained individuals exhibiting various phenotypes and no phenotype was representative of a particular genome class.
Figure 3.—

Phenotypes of CCC and CWW F2 individuals. The estimated genome content of each individual is expressed as in Figure 2. The origin of each plant (progeny of CCC or CWW) is indicated in parentheses. (a–f) Whole aerial portion of the plant, 20 days postgermination. (i–p) All seeds contained in a single silique at maturity. Seeds surrounded by a black box were scored as dead (not plump). a–h are on the same scale (bar, 1 cm). All seed pictures (i–p) are on the same scale (bar, 1 mm).
The viability of the F3 seeds was estimated by the percentage of plump seeds and seed number per F2 silique. Surprisingly, most of the aneuploids produced viable seeds although the number of viable seeds was variable and lower in aneuploids than in euploids. The negative effect of aneuploidy on seed viability was stronger in the F2 plants of CCC than in the F2 plants of CWW. Specifically, CWW F2 plants of genome content consistent with 16 or 17 chromosomes produced a higher percentage of plump seeds than CCC plants of similar genome content (Figure 2a, inset). Thus selection against aneuploid types continued in the F3 and was influenced by genotype similarly in the F2 and F3.
Several generations of selfing resolved the aneuploid swarm into two groups:
Assuming that similar selection influences the distribution of genome content in subsequent generations as well, we sought to investigate the genome content of RILs derived from a triploid. A set of RILs had previously been derived from a CWW triploid to the F6 or F8 generation (Schiff et al. 2001). The ploidy level of Wa-1 being unknown at the time, the authors were unaware of the genome content of their lines. Yet, they did note high lethality and sterility in early filial generations (one-fourth of the lineages were lost to fertility problems; I. W. Wilson, personal communication), a low Col-0 to Wa-1 allelic ratio, and an unusually high level of heterozygosity in the RILs. All these observations are consistent with aneuploidy and polyploidy arising from an interploidy cross.
From this population, we obtained the 89 surviving lines, 80 of which had been propagated to the F8 while the remaining 9 had been propagated only to the F6. We measured the nuclear DNA content of these 89 lines (Figure 2b) and were able to partition the RILs into two classes: near-diploids (n = 63; genome content 1.92–2.60) and near-tetraploids (n = 26; genome content 3.56–4.63). As expected, the near-diploid lines exhibited a lower level of heterozygosity (mean of 2.0 ± 2.3%) than the near-tetraploid lines (mean of 15.9 ± 6.6%).
Within both groups, many plants possessed a genome content that deviated from the euploid parents by at least the approximate size of one chromosome (Figure 2b). It was unclear whether the wide range of genome contents observed within the two RIL classes was due to measurement imprecision or to aneuploidy. To obtain a more precise determination of genome size in some of these lines, multiple individuals from a few of the lines with genome content values furthest from euploidy were grown and their genome size was determined. Aneuploidy in these lines was investigated by examination of genome content in siblings. In 7 of the 15 lines tested, siblings segregated for genome content deviation consistent with an aneuploid parent. Interestingly, these lines included 3 of the 9 lines that were propagated only to the F6 generation while only 4 of the 80 lines that were propagated to the F8 generation were determined to be aneuploid, suggesting that earlier generations are more likely to contain aneuploid individuals.
Genetic analysis of the RILs and evidence for ploidy-dependent selection:
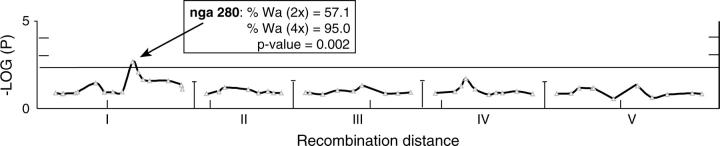
The influence of genotype on the F2 and F3 generations (Figures 1 and 2), along with the gradual production of euploids from this aneuploid swarm, may result in ploidy-dependent selection in the RILs. Selection of alleles in the tetraploid individuals but not in the diploid individuals, or vice versa, should be detectable as a difference in allelic frequency for a given marker in the two ploidy types. To test this, we investigated whether genome content in the RIL population was associated with marker genotype. A linkage model for triploid or aneuploid inheritance is not available. We therefore took a single-locus approach and searched for loci exhibiting segregation distortion as well as loci conditionally selected in one of the two ploidy classes. For the purpose of this analysis, the two euploid classes included lines with near-diploid and near-tetraploid individuals. We compared the allelic ratios in the tetraploid RILs to the allelic ratios in the diploid RILs at each of the 53 markers using Fisher's exact test with the significance threshold corrected for multiple tests (see materials and methods). Marker nga280 and the closely linked MN1.2 marker, both on chromosome I, exhibited a significantly higher ratio of Wa-1 to Col-0 alleles in the tetraploid lines than in the diploid lines (Figure 4). The nga280 Wa-1 allele was not underrepresented in the diploid population (57%) and was present in 95% of the homozygous tetraploid lines. To verify that this was not due to a chromosomal inversion, we determined the marker order around nga280 and found it to be identical to that of the physical map of Col-0 (TIGR5.0 release at www.arabidopsis.org). Our data are therefore consistent with conditional selection of the Wa-1 alleles in the tetraploid lines and not in the diploid lines. The lack of a suitable linkage model hinders the mapping of a QTL. Nevertheless, genotypic differences between Col-0 and Wa-1 in the response to tetraploid formation from a triploid could be localized to a specific chromosome.
Figure 4.—
Analysis of ploidy-dependent selection in the Col-0 × Wa-1 RIL population. Chromosomes I–V are represented across the x-axis. Short vertical lines indicate the positions of the centromeres. The allele frequencies in putative diploid and tetraploid populations were compared using Fisher's exact test. The allele frequency of the diploid population was used as the expected ratio and the observed allelic ratio was that of the putative tetraploid population. The logarithm of the P-value at each marker is shown and the dashed horizontal line indicates a Bonferroni-corrected P-value of 0.05 for 11 independent tests. The inset indicates the percentages of individuals homozygous for the Wa-1 allele as well as the Fisher's exact test P-value for the most significant marker (nga280).
DISCUSSION
Polyploidy has played a major role in the evolution of flowering plants (Ramsey and Schemske 1998; Soltis et al. 2003) yet little is known about the genetic mechanisms that contribute to the formation and stability of polyploids. In this study, we sought to characterize the response of A. thaliana to triploidy and to identify genetic variability affecting this response as well as investigate the contribution of triploids to the formation of polyploids.
Triploidy occurs relatively frequently in diploid species of animals and plants, resulting either from the fusion of an accidental 2N gamete to a regular 1N gamete, both produced by diploid individuals, or from crosses between diploid and tetraploid individuals. In humans, 10% of spontaneous miscarriages are due to triploidy (Sankaranarayanan 1979), but triploids of fish and amphibians can grow to adults and some species are even fixed in the triploid state (Tock et al. 2002). Plants can be classified in two categories according to their response to triploidy: in some species, most triploid embryos die because of abnormal endosperm development and the few individuals that do develop exhibit highly reduced fertility (Satina and Blakeslee 1938; Woodell and Valentine 1961; Tsuchiya 1967). This phenomenon is called the triploid block. Interestingly, data from triploids of some of these species suggest that they produce only a narrow range of low genome content aneuploids or diploid progeny. In other species, interploidy crosses are successful (Woodell and Valentine 1961) and the resulting triploids undergo virtually normal development until reproduction. These triploids exhibit various levels of fertility and data from triploids of some of these species suggest that they produce progeny with a much wider range of genome content. Such species include sugar beet (Levan 1942), Melandrium (Warmke and Blakeslee 1940), or poplar (Johnsson 1942, 1945). Our study demonstrates that A. thaliana belongs to the latter category, exhibiting only a mild reduction in fertility (Figure 1).
Studies on natural populations support a role for triploids as bridges between ploidy classes with the assumption that triploids directly produce euploids (Ramsey and Schemske 1998; Burton and Husband 2001). Our study supports a role for triploids of Arabidopsis both in polyploidy formation and as vectors for gene flow between diploid and tetraploid populations. Most remarkable, however, is the observation that this triploid bridge acts through at least one and possibly several generations of aneuploidy.
Aneuploidy has severe effects on phenotype and viability probably because of a requirement for the balanced dosage of many factors (Bridges 1922; Birchler et al. 2001). While this adequately describes the behavior of trisomics, its application to higher-order aneuploids is unclear. Trisomics have been described in a variety of species and, in all cases, each trisomic exhibits stereotypical phenotypes dependent on which chromosome was present in excess (Blakeslee 1922; McClintock 1929; Rick and Baron 1953; Steinitz-Sears 1962). Yet, little information is available about the phenotypes of aneuploids involving more than one unbalanced chromosome. The availability of multiple aneuploid types in a model system should facilitate the development of a more comprehensive model for the effects of aneuploidy.
The CWW F2 aneuploid swarm (Figure 2a) likely reflects the composition of the F2 generation used to generate the recombinant inbred lines produced by Schiff and coworkers (Schiff et al. 2001). Analysis of these RILs demonstrated that repeated selfing had resolved the aneuploid swarm found in the F2 into two groups: near-diploids and near-tetraploids. Thus in Arabidopsis severe aneuploidy (i.e., aneuploids with several unbalanced chromosomes) is not always a terminal condition and a majority of the F2 aneuploids were able to produce euploid progeny in a few generations. This is consistent with the factors affecting the distribution of karyotypes in the F2 continuing to influence the subsequent generations, progressively depleting the population of aneuploids and enriching it with euploid individuals. These data suggest a possible role of triploids and aneuploids as bridges between diploid and tetraploid populations. Of course, both the strength and nature of selection acting on karyotype swarms in the wild may be radically different from those acting on plants grown under controlled laboratory conditions.
The genetic analysis of these RILs also identified one marker that exhibited a significantly higher percentage of Wa-1 alleles in the tetraploid lines than in the diploid lines. These data are consistent with the conditional selection of a Wa-1 allele in tetraploids or their progenitors. This bias suggests a role for the linked locus in polyploid formation or establishment. The mechanisms underlying this selection remain unclear.
Indeed, a variety of mechanisms can influence the composition and phenotype of the aneuploid swarm produced by a triploid as well as the composition of the subsequent generations. Mechanisms affecting meiosis could include the production of 2N gametes (Bretagnolle and Thompson 1995; Carputo et al. 2003), preferential pairing of chromosomes, preferential migration of unpaired chromosomes to one pole, bivalent vs. trivalent formation, or chromosome loss (McClintock 1929; Satina and Blakeslee 1937b). Other mechanisms could act at later stages. For example, aneuploidy might be alleviated by the epigenetic silencing of unpaired chromosomes (de la Casa-Esperon and Sapienza 2003; Bean et al. 2004). Imprinted factors required for endosperm development limit the viability of interploidy fertilizations (Vinkenoog et al. 2003) and may similarly affect the development of the endosperm of aneuploid genome content. The collection of data regarding meiosis in Arabidopsis triploids, as well as the development of a statistical model of both triploid inheritance and the consequences of aneupoidy on survival, will be instrumental in documenting the role of each of these mechanisms in triploid inheritance. Elucidation of the mechanisms responsible for the differences between Col-0 and Wa-1 may both inform evolutionary studies on the roles of multiple ploidy types in speciation and identify mechanisms for the species-specific differences in triploidy effects.
Recurring selection of plants for aneuploidy tolerance may play an important evolutionary role and have consequences on their response to dosage regulation. The tolerance of the model organism Arabidopsis to multiple aneuploid states and the connected genetic variation should provide important tools to understand the relationship between ploidy, evolution, and the genetic responses underlying dosage sensitivity.
Acknowledgments
We thank Shauna Sommerville (Carnegie Institution, Stanford, CA) for the Col-0 × Wa-1 recombinant inbred lines. We thank the Cell Analysis Facility (Department of Immunology, University of Washington) and Biology Greenhouse (Biology Department, University of Washington) for material support. This article was greatly improved by discussions with Saunak Sen (University of California, San Francisco) on triploid and aneuploid inheritance. This work was supported by a Fulbright grant-in-aid and a Belgian-American Educational Foundation Fellowship (both to I.M.H.), the National Science Foundation (NSF) Polyploidy Project (NSF no. 0077774 to L.C.), and the U.S. Department of Agriculture National Research Initiative (2003-35300-13248 to B.P.D.).
References
- Alexander, M. P., 1969. Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Bean, C. J., C. E. Schaner and W. G. Kelly, 2004. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet. 36: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey, T. J., 1999. The physiology and behavior of triploid fishes. Rev. Fish. Sci. 7: 39–67. [Google Scholar]
- Birchler, J. A., U. Bhadra, M. P. Bhadra and D. L. Auger, 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol. 234: 275–288. [DOI] [PubMed] [Google Scholar]
- Blakeslee, A., 1922. Variation in Datura due to changes in chromosome number. Am. Nat. 56: 16–31. [Google Scholar]
- Bretagnolle, F., and J. D. Thompson, 1995. Gametes with somatic chromosome number: their mechanisms of formation and role in the evolution of autopolyploid plants. New Phytol. 129: 1–22. [DOI] [PubMed] [Google Scholar]
- Bridges, C. B., 1922. The origins of variation in sexual and sex-limited characters. Am. Nat. 56: 51–63. [Google Scholar]
- Burton, T. L., and B. C. Husband, 2001. Fecundity and offspring ploidy in matings among diploid, triploid and tetraploid Chamerion angustifolium (Onagraceae): consequences for tetraploid establishment. Heredity 87: 573–582. [DOI] [PubMed] [Google Scholar]
- Carputo, D., L. Frusciante and S. J. Peloquin, 2003. The role of 2n gametes and endosperm balance number in the origin and evolution of polyploids in the tuber-bearing solanums. Genetics 163: 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, R. E., and D. R. Cameron, 1944. Inheritance in Nicotiana tabacum. XVIII. Monosomic analysis. Genetics 29: 447–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Casa-Esperon, E., and C. Sapienza, 2003. Natural selection and the evolution of genome imprinting. Annu. Rev. Genet. 37: 349–370. [DOI] [PubMed] [Google Scholar]
- Garnier-Gere, P., Y. Naciri-Graven, S. Bougrier, A. Magoulas, M. Heral et al., 2002. Influences of triploidy, parentage and genetic diversity growth of the Pacific oyster Crassostrea gigas reared in contrasting environments. Mol. Ecol. 11: 1499–1514. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison, J., and J. Maluszynska, 1994 Molecular cytogenetics of Arabidopsis, pp. 63–87 in Arabidopsis, edited by E. Meyerowitz and C. Somerville. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Johnsson, H., 1942. Cytological studies of triploids progenies of Populus tremula. Hereditas 28: 306–312. [Google Scholar]
- Johnsson, H., 1945. The triploid progeny of the cross diploid x tetraploid Populus tremula. Hereditas 31: 411. [DOI] [PubMed] [Google Scholar]
- Khush, G., 1973 Cytogenetics of Aneuploids. Academic Press, New York.
- Lesley, J. W., 1928. A cytological and genetical study of progenies of triploid tomatoes. Genetics 13: 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levan, A., 1942. The effect of chromosomal variation in sugar beets. Hereditas 28: 345–399. [Google Scholar]
- Manly, K. F., R. H. Cudmore, Jr. and J. M. Meer, 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12: 930–932. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1929. A cytological and genetical study of triploid maize. Genetics 14: 180–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and F. Skoog, 1962. A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Ramsey, J., and D. W. Schemske, 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29: 467–501. [Google Scholar]
- Rick, C. M., and D. W. Baron, 1953. Cytological and genetical identification of the primary trisomics of the tomato. Genetics 39: 640–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick, C. M., and N. K. Notani, 1961. The tolerance of extra chromosomes by primitive tomatoes. Genetics 46: 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan, K., 1979. The role of non-disjunction in aneuploidy in man. An overview. Mutat. Res. 61: 1–28. [DOI] [PubMed] [Google Scholar]
- Satina, S., and A. Blakeslee, 1937. a Chromosome behavior in triploids of Datura stramonium. I. The male gametophyte. Am. J. Bot. 24: 518–526. [Google Scholar]
- Satina, S., and A. Blakeslee, 1937. b Chromosome behavior in triploids of Datura stramonium. II. The female gametophyte. Am. J. Bot. 24: 621–627. [Google Scholar]
- Satina, S., and A. Blakeslee, 1938. Chromosome behavior in triploid Datura. III. The seed. Am. J. Bot. 25: 595–602. [Google Scholar]
- Schiff, C. L., I. W. Wilson and S. C. Somerville, 2001. Polygenic powdery mildew disease resistance in Arabidopsis thaliana: quantitative trait analysis of the accession Warschau-1. Plant Pathol. 50: 690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuths, H., A. M. Eister, R. Horres and K. Bachmann, 2004. Genome size variation among accessions of Arabidopsis thaliana. Ann. Bot. 93: 317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis, D., P. Soltis and J. Tate, 2003. Advances in the study of polyploidy since plant speciation. New Phytol. 161: 173–191. [Google Scholar]
- Steinitz-Sears, L., 1962. Chromosome studies in Arabidopsis thaliana. Genetics 48: 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tock, M., D. Lamatsch, C. Steinlein, J. Epplen, W. Grosse et al., 2002. A bisexually reproducing all-triploid vertebrate. Nat. Genet. 30: 325–328. [DOI] [PubMed] [Google Scholar]
- Tsuchiya, T., 1967. The establishment of a trisomic series in a two-rowed cultivated variety of barley. Can. J. Genet. Cytol. 9: 667–682. [Google Scholar]
- Vinkenoog, R., C. Bushell, M. Spielman, S. Adams, H. G. Dickinson et al., 2003. Genomic imprinting and endosperm development in flowering plants. Mol. Biotechnol. 25: 149–184. [DOI] [PubMed] [Google Scholar]
- Warmke, H. E., and A. Blakeslee, 1940. The establishment of a 4N dioecious race in Melandrium. Am. J. Bot. 27: 751–762. [Google Scholar]
- Wolfe, K. H., and D. C. Shields, 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713. [DOI] [PubMed] [Google Scholar]
- Woodell, S., and D. Valentine, 1961. Studies in British primulas IX. Seed incompatibility in diploid-autotetraploid crosses. New Phytol. 60: 282–294. [Google Scholar]