Abstract
The nonrandom use of synonymous codons (codon bias) is a well-established phenomenon in Drosophila. Recent reports suggest that levels of codon bias differ among genes that are differentially expressed between the sexes, with male-expressed genes showing less codon bias than female-expressed genes. To examine the relationship between sex-biased gene expression and level of codon bias on a genomic scale, we surveyed synonymous codon usage in 7276 D. melanogaster genes that were classified as male-, female-, or non-sex-biased in their expression in microarray experiments. We found that male-biased genes have significantly less codon bias than both female- and non-sex-biased genes. This pattern holds for both germline and somatically expressed genes. Furthermore, we find a significantly negative correlation between level of codon bias and degree of sex-biased expression for male-biased genes. In contrast, female-biased genes do not differ from non-sex-biased genes in their level of codon bias and show a significantly positive correlation between codon bias and degree of sex-biased expression. These observations cannot be explained by differences in chromosomal distribution, mutational processes, recombinational environment, gene length, or absolute expression level among genes of the different expression classes. We propose that the observed codon bias differences result from differences in selection at synonymous and/or linked nonsynonymous sites between genes with male- and female-biased expression.
FOR many years, nucleotide changes that did not alter the encoded amino acid (synonymous substitutions) were believed to reflect neutral variation (Kimura 1968, 1977). However, the phenomenon of codon usage bias, which is the preferential use of certain codons over their synonymous equivalents, has been shown to be widespread across many unicellular and multicellular organisms. In Drosophila melanogaster, as well as several other organisms, codon bias is strongest among the most highly expressed genes (Grantham et al. 1981; Bennetzen and Hall 1982; Gouy and Gautier 1982; Grosjean and Fiers 1982; Duret and Mouchiroud 1999; Coghlan and Wolfe 2000; Akashi 2003). Further, in the species that have been studied, the favored codons correspond to the most abundant isoaccepting tRNAs (Ikemura 1981, 1982; Moriyama and Powell 1997; Kanaya et al. 1999; Duret 2000). Thus, codon bias is largely thought to be due to weak selection favoring the use of codons that are most efficiently and accurately translated (Akashi 1994, 1995; Carlini and Stephan 2003). Selection intensity for codon usage bias, therefore, is expected to vary among genes. Presumably, highly expressed genes have more codon bias because selection for translational efficiency and accuracy is stronger in these genes.
Synonymous codon usage, however, is expected to be under weak selection in D. melanogaster, with the product of the effective population size and the selection coefficient, Nes, being <1 (Akashi 1995; McVean and Vieira 2001). Thus, other processes, either neutral or selective, may influence patterns of synonymous codon usage. For example, Marais et al. (2003) proposed that biased gene conversion favoring G and C nucleotides could result in variation in codon bias among genes and could explain the previously observed positive correlation between codon bias and local recombination rate (Kliman and Hey 1993; Comeron et al. 1999; Marais et al. 2001; Hey and Kliman 2002). This correlation also could have a selective explanation: population genetic theory predicts that selection for synonymous codon usage should be more effective in regions of higher recombination because linkage among selected sites, known as Hill-Robertson interference, is reduced (Hill and Robertson 1966; Kliman and Hey 1993, 2003; Comeron et al. 1999; Hey and Kliman 2002).
Hill-Robertson interference among sites may affect levels of codon bias in other ways as well. Betancourt and Presgraves (2002) examined levels of codon bias in 255 genes, including 153 male-specific accessory protein (Acp) genes, and found a significantly negative correlation between level of codon bias and nonsynonymous divergence between D. melanogaster and D. simulans. The correlation could result from Hill-Robertson interference between linked synonymous and nonsynonymous sites, with the fixation of strongly beneficial amino acid replacements driving the fixation of linked, slightly deleterious synonymous substitutions (Akashi 1996; Betancourt and Presgraves 2002; Kim 2004). This interpretation is supported by the observation that Acp genes evolve significantly faster than non-Acp genes (Swanson et al. 2001) and that several well-studied Acp genes show patterns of molecular evolution consistent with a history of positive selection (Tsaur and Wu 1997; Tsaur et al. 1998; Aguadé 1998, 1999; Begun et al. 2000). In addition to Acp's, a number of other Drosophila reproductive genes showing increased rates of molecular evolution and evidence for positive selection have been reported (Nurminsky et al. 1998; Ting et al. 1998; Parsch et al. 2001; Betrán and Long 2003). These findings suggest that there may be a general pattern of increased evolutionary rates in sex- and reproduction-related genes, presumably as the result of sexual selection (Civetta and Singh 1999; Singh and Kulathinal 2000; Swanson and Vacquier 2002).
In a recent study, Zhang et al. (2004) used expression data from microarray experiments to classify D. melanogaster genes as male-, female-, or non-sex-biased in their expression and compared the evolutionary rates of these genes among Drosophila species. They found that male-biased genes had significantly higher rates of evolution (measured as the ratio of nonsynonymous/synonymous substitution rates) than both female- and non-sex-biased genes. Female-biased genes, in contrast, showed evolutionary rates less than or equal to those of non-sex-biased genes. Population genetic data suggested that these differences were caused by increased positive selection acting on genes with male-biased expression. Zhang et al. (2004) also compared levels of codon bias in sex-biased genes for which interspecific divergence data were available. These results inversely reflected the evolutionary rate comparisons: male-biased genes had significantly less codon bias than both female- and non-sex-biased genes, while female-biased genes had levels of codon bias greater than or equal to those of non-sex-biased genes. The above results, however, were based on a relatively small sample of genes with <100 male- and female-biased genes in each comparison. In this article, we extend the codon bias analyses to >7000 D. melanogaster genes for which annotated coding sequences and microarray data on sex-biased expression were available. We confirm that male-biased genes have significantly less codon bias than female- and non-sex-biased genes, while the latter two groups of genes have relatively equal levels of codon bias. Furthermore, the new data allow us to examine the relationship between codon bias and degree of sex-biased expression. We find that as the level of sex-biased expression increases, male-biased genes show less codon bias, whereas female-biased genes show more codon bias. Differences in chromosomal locations, mutational processes, recombination rates, gene lengths, or absolute expression levels cannot account for the codon bias differences between male- and female-biased genes, suggesting that natural selection influences synonymous codon usage in sex-biased genes.
MATERIALS AND METHODS
Identification of sex-biased genes:
To classify genes as male, female, or non-sex biased in their expression (here referred to as male-, female-, and non-sex-biased genes), we used published data from two independent studies that compared male and female gene expression by competitive microarray hybridization (Parisi et al. 2003; Ranz et al. 2003) following the approach of Zhang et al. (2004). That is, genes with twofold or greater expression in males than in females were classified as male biased, genes with twofold or greater expression in females than in males were classified as female biased, and genes having less than a twofold expression difference between the sexes were classified as non-sex biased. In cases of sex-bias conflict in which a gene was sex biased in one microarray data set and non-sex biased in the other (7% of all genes), the gene was considered sex biased. However, eliminating these genes did not affect our results. In cases of sex-bias conflict in which a gene was male-biased in one microarray data set and female biased in the other (0.08% of all genes), the gene was eliminated from further analysis. Because the twofold cutoff is an arbitrary standard chosen to allow comparison of microarray results across studies, we analyzed the sensitivity of our results to the choice of the cutoff value. Using cutoffs of 1.5- or 3-fold did not alter the qualitative pattern or the statistical significance of our results. Unless noted otherwise, all results presented here use the twofold cutoff for sex-bias classification.
Somatic sex-biased genes were identified using the microarray data of Parisi et al. (2004), which compared gene expression between gonadectomized males and females. Germline sex-biased genes were identified using the testes/ovaries ratio from Parisi et al. (2003). In total, 282 somatic genes (145 male biased and 137 female biased) and 1959 germline genes (1083 male biased and 876 female biased) were used (see supplementary Table S1 at http://www.genetics.org/supplemental/).
Genomic data:
Complete coding sequences (CDS) corresponding to all annotated genes in the D. melanogaster genome (release 3.2) were downloaded from FlyBase (http://www.flybase.org). As a quality control step, we eliminated any CDS that did not begin with an ATG start codon, did not have a length that was a multiple of three, or that contained an internal stop codon (<0.5% of all sequences). For genes with multiple transcripts, we selected only the one with the longest CDS. The final sequence collection contained 13,464 CDSs, each corresponding to a unique gene in the Drosophila genome. Of these genes, 7276 had matches in the combined microarray expression data set (described above) and were used for analyses of codon bias (see supplementary Table S1 http://www.genetics.org/supplemental/). For comparisons of GC content, complete chromosome arm sequences and annotated intron sequences (D. melanogaster genome release 3.2) were downloaded from FlyBase. Estimates of genomic recombination rates (Hey and Kliman 2002) were downloaded from the authors' website (http://lifesci.rutgers.edu/~heylab). The five different recombination estimators described by Hey and Kliman (2002) produced nearly identical results in our analyses. For simplicity, we present only results using the recombination estimator, R, which is based on a comparison of the genetic and physical map locations of 493 genes.
To estimate the absolute expression level of the genes in our analyses, we used the microarray data of Gibson et al. (2004), which were downloaded from the authors' website (http://statgen.ncsu.edu/ggibson/SupplInfo/SupplInfo3.htm). These experiments used microarrays of oligonucleotide probes of a standard length (60 nt) synthesized directly on glass slides and measured the fluorescent intensity of each spot relative to all other spots on the same array. Thus, these data are better suited for comparing expression levels among genes than data from arrays constructed of probes spotted with PCR products of various lengths that measured the male/female fluorescence ratio for each spot (Parisi et al. 2003; Ranz et al. 2003). For each gene, the least-squares mean expression level reported by Gibson et al. (2004) was averaged over both sexes and over both the 2b and Oregon-R strains. In cases where a gene was represented by multiple probes, the expression level was averaged over all probes.
Synonymous codon usage analyses:
Codon usage bias was estimated using two measures: the effective number of codons (ENC; Wright 1990) and the frequency of optimal codons (FOP; Ikemura 1981). For ENC, lower values indicate stronger synonymous codon usage bias, while for FOP higher values indicate stronger bias. Both measures were calculated for all genes using the CodonW program (http://bioweb.pasteur.fr/seqanal/interfaces/codonw.html). Differences in levels of codon bias among male-, female-, and non-sex-biased genes were tested using the nonparametric Mann-Whitney test. To evaluate whether the degree of synonymous codon usage bias in individual genes was correlated with their relative level of sex-biased expression, linear regression and Spearman rank-correlation tests were performed using both FOP and ENC. The two measures of codon bias gave similar results; only those for FOP are reported here. To remove the influence of factors known to correlate with codon usage bias (local GC content, recombination rate, gene length, and expression level) from our comparisons among male-, female-, and non-sex-biased genes, we regressed each of these factors on FOP and calculated the residuals. In all cases, the residuals were not correlated with the original factor, indicating that we had successfully removed its effect on codon bias. We then compared the residual values among male-, female-, and non-sex-biased genes using Mann-Whitney tests.
RESULTS
Levels of codon bias in sex-biased genes:
To investigate levels of codon bias in genes with sex-biased expression, we analyzed synonymous codon usage in 7276 D. melanogaster genes for which we had complete coding sequences and microarray data comparing relative levels of male vs. female gene expression. As can be seen in Table 1, there are significant differences in levels of codon bias among male-, female-, and non-sex-biased genes. Male-biased genes consistently show significantly less codon bias than female- and non-sex-biased genes by both ENC and FOP. As the definition of male-biased genes (i.e., the overexpression cutoff level) becomes more stringent, the degree of significance for these differences increases for both measures of codon bias (Table 1). In contrast, there appears to be little difference in codon bias between female- and non-sex-biased genes. Using the conventional twofold cutoff to define genes as sex biased, there is not a significant difference between female- and non-sex-biased genes by either measure of codon bias (Table 1). When the cutoff is lowered to 1.5-fold, there is less codon bias in the female-biased genes when measured by ENC, but not when measured by FOP (Table 1). Using the more stringent 3-fold cutoff, female-biased genes have more codon usage bias than non-sex-biased genes. This difference is significant for FOP (P = 0.002) and marginally significant for ENC (P = 0.087). The conflicting results that are sometimes observed between FOP and ENC likely are caused by differences in the way that the two methods estimate codon bias. FOP is based on the frequency of a set of species-specific “optimal” codons, while ENC is based on the observed number of codons used for each amino acid. Thus it is possible for the two methods to give different estimates of codon bias. For example, a gene with high AT content at third positions will have very low codon bias when measured by FOP (since the optimal codons for D. melanogaster tend to end in G or C), but can show relatively high codon usage bias when measured by ENC. Indeed, we find that the largest discrepancy between the two measures occurs in such cases when FOP is very low (<35%; data not shown).
TABLE 1.
Levels of codon bias in genes with male-, female-, and non-sex-biased expression
| Cutoffa | Male | Female | Non-sex | PMFb | PMNb | PFNb | |
|---|---|---|---|---|---|---|---|
| N | 1.5 | 1963 | 2290 | 2987 | |||
| ENC | 1.5 | 48.50 | 48.21 | 47.85 | 1.6 × 10−9 | 2.3 × 10−20 | 0.005 |
| FOP | 1.5 | 0.519 | 0.541 | 0.537 | 7.1 × 10−11 | 3.9 × 10−11 | 0.645 |
| N | 2.0 | 1293 | 1443 | 4535 | |||
| ENC | 2.0 | 50.32 | 48.04 | 47.99 | 7.7 × 10−7 | 1.7 × 10−32 | 0.186 |
| FOP | 2.0 | 0.507 | 0.544 | 0.537 | 3.5 × 10−18 | 3.1 × 10−23 | 0.299 |
| N | 3.0 | 860 | 564 | 5851 | |||
| ENC | 3.0 | 51.11 | 47.18 | 48.13 | 7.2 × 10−21 | 2.1 × 10−39 | 0.087 |
| FOP | 3.0 | 0.496 | 0.558 | 0.536 | 2.7 × 10−22 | 5.0 × 10−32 | 0.002 |
N, number of genes; ENC, effective number of codons (Wright 1990); FOP, frequency of optimal codons (Ikemura 1981). Numbers represent the mean value for each category.
Fold expression cutoff used to classify sex-biased genes.
P-value of two-tailed Mann-Whitney test for comparisons among male- (M), female- (F), and non-sex-biased (N) genes.
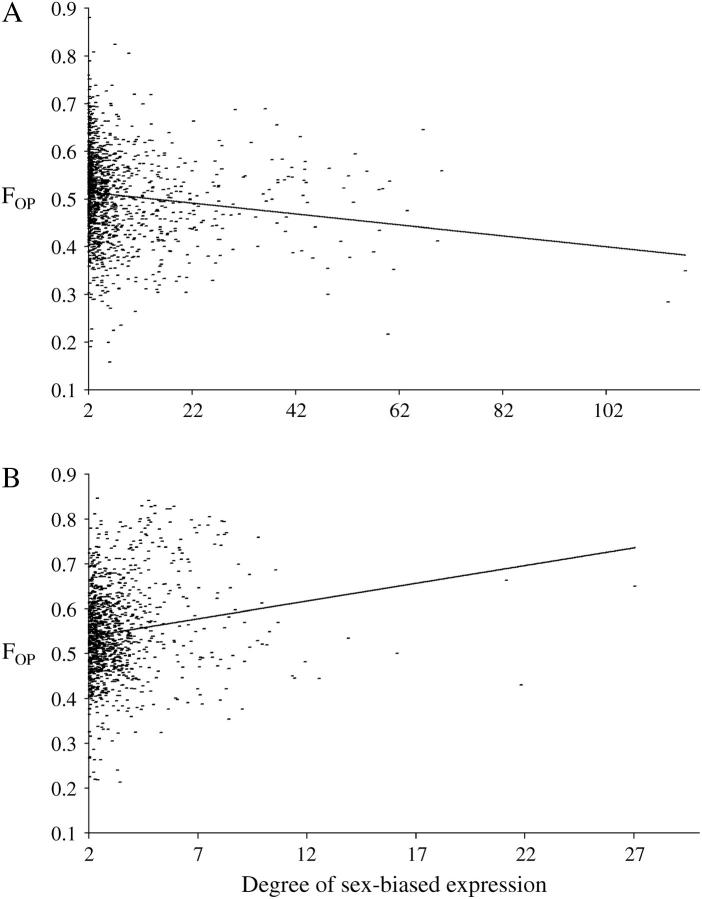
The observation that male-biased genes show less codon bias as the cutoff for defining sex-biased expression becomes more stringent suggests that there may be a negative correlation between codon bias and degree of male-biased expression. Indeed, such a relationship is seen for the male-biased genes (Figure 1A). Although weak, this correlation is significant using both a linear regression (R = −0.13, P = 2.0 × 10−7) and a rank-correlation test (R = −0.19, P = 6.1 × 10−12), indicating that it is not caused by a small number of genes with very highly male-biased expression and low levels of codon bias. Indeed, even after removing the 468 genes with a male/female expression ratio >5, we observe a significantly negative correlation between FOP and degree of sex-biased expression by both linear (R = −0.17, P = 1.0 × 10−5) and rank-order (R = −0.16, P = 5.9 × 10−5) correlation tests. In contrast to the male-biased genes, female-biased genes show a weak but significantly positive correlation between codon bias and degree of female-biased expression by both linear regression (R = 0.23, P = 7.6 × 10−12) and rank correlation (R = 0.16, P = 7.2 × 10−8; Figure 1B). Within the non-sex-biased genes, there was not a significant correlation between codon bias and the male/female expression ratio by either linear regression (R = 2.0 × 10−5, P > 0.1) or rank correlation (R = −0.013, P > 0.1). This is probably due to the fact that male/female expression ratios are constrained to a narrow window (between 0.5 and 2.0). Furthermore, these are the genes for which sex-bias is most likely to be misclassified due to the experimental error inherent in microarray hybridizations. That is, these are the genes in which the male and female fluorescence intensities are closest to each other. Thus the expression ratio can easily be shifted from male biased to female biased (or vice versa) by small intensity variations caused by experimental noise.
Figure 1.—
Relationship between codon bias and degree of sex-biased expression. (A) FOP plotted against the male/female expression ratio for 1293 male-biased genes (Spearman rank correlation, R = −0.19, P = 6.1 × 10−12). The line represents the least-squares linear regression (R = −0.13, P = 2.0 × 10−7) with a slope of −0.0011. (B) FOP plotted against the female/male expression ratio for 1443 female-biased genes (Spearman rank correlation, R = 0.16, P = 7.2 × 10−8). The line represents the least-squares linear regression (R = 0.23, P = 7.6 × 10−12) with a slope of 0.0079.
Comparison of X-linked and autosomal genes:
Previous studies have shown that genes with male-biased expression are significantly underrepresented on the X chromosome (Parisi et al. 2003; Ranz et al. 2003). The same pattern is observed for the genes included in our survey: 11% (144/1293) of the male-biased genes were located on the X, while 18% (823/4535) of the non-sex-biased genes and 23% (331/1443) of the female-biased genes were located on the X. Thus it is possible that interchromosomal differences in synonymous codon bias could account for the observed differences among genes of the different expression classes, particularly if X-linked genes tend to show greater codon bias than autosomal genes. To test this possibility, we analyzed levels of codon bias separately for X-linked and autosomal genes. Indeed, there is a general tendency for X-linked genes to show greater codon bias than autosomal genes across all expression classes (Table 2). This difference is significant for ENC within each expression class (P < 0.001), while for FOP it is significant within the female- and non-sex-biased genes (P < 0.001), but not male-biased genes (P = 0.18). These interchromosomal differences, however, cannot explain the observed reduction of codon bias in male-biased genes. If we consider only X-linked genes, male-biased genes still show significantly less codon bias than both female- and non-sex-biased genes (Table 2). The same result is seen when only autosomal genes are considered (Table 2).
TABLE 2.
Codon bias in X-linked and autosomal genes
| Chromosome | Male | Female | Non-sex | PMFa | PMNa | PFNa | |
|---|---|---|---|---|---|---|---|
| N | X | 144 | 331 | 823 | |||
| ENC | X | 48.87 | 45.65 | 46.11 | 7.9 × 10−7 | 1.8 × 10−6 | 0.250 |
| FOP | X | 0.515 | 0.568 | 0.553 | 2.0 × 10−7 | 4.8 × 10−5 | 0.013 |
| N | Auto | 1149 | 1112 | 3712 | |||
| ENC | Auto | 50.50 | 48.75 | 48.40 | 3.8 × 10−8 | 1.7 × 10−32 | 0.003 |
| FOP | Auto | 0.506 | 0.536 | 0.534 | 3.5 × 10−18 | 6.3 × 10−10 | 0.531 |
N, number of genes; ENC, effective number of codons (Wright 1990); FOP, frequency of optimal codons (Ikemura 1981). Numbers represent the mean value for each category.
P-value of two-tailed Mann-Whitney test for comparisons among male- (M), female- (F), and non-sex-biased (N) genes.
Comparison of germline and somatic sex-biased genes:
The vast majority of gene expression differences between the sexes is attributable to genes that are differentially expressed between male and female reproductive tissues (Parisi et al. 2004). Since the microarray data that we used to classify sex-biased genes were based on comparisons of either dissected reproductive tissues or whole flies, our results apply mainly to germline-expressed genes. To investigate whether there were differences between germline and somatic sex-biased genes, we examined levels of codon bias separately in the two groups (Table 3). Germline sex-biased genes were identified from the Parisi et al. (2003) microarray data set that compared gene expression between dissected testes and ovaries, while somatic sex-biased genes were identified from the Parisi et al. (2004) data set that compared gene expression between gonadectomized males and females. For both germline and somatic sex-biased genes, we observed highly significant differences in level of codon bias between male- and female-biased genes (Mann-Whitney test, P < 1.0 × 10−16), with male-biased genes having less codon bias in both cases (Table 3).
TABLE 3.
Codon bias in somatic and germline sex-biased genes
| Expression | Male | Female | Pa | |
|---|---|---|---|---|
| N | Soma | 145 | 137 | |
| ENC | Soma | 52.62 | 43.42 | 3.9 × 10−17 |
| FOP | Soma | 0.455 | 0.598 | 2.1 × 10−17 |
| N | Germ | 1083 | 876 | |
| ENC | Germ | 50.32 | 46.70 | 2.2 × 10−29 |
| FOP | Germ | 0.507 | 0.564 | 1.3 × 10−29 |
N, number of genes; ENC, effective number of codons (Wright 1990); FOP, frequency of optimal codons (Ikemura 1981). Numbers represent the mean value for each category.
P-value of two-tailed Mann-Whitney test comparing male- and female-biased genes.
Interestingly, we observed significant differences in codon bias between somatic and germline tissues for both male- and female-biased genes (Table 3). Male-biased genes that are expressed in the germline show more codon bias than those expressed in somatic tissues (ENC, P = 1.3 × 10−6; FOP, P = 5.9 × 10−9). Female-biased genes, on the other hand, show the opposite pattern. Female-biased genes expressed in germline tissues show significantly less codon bias than those expressed in somatic tissues (ENC, P = 5.1 × 10−5; FOP, P = 1.0 × 10−3).
Comparison of intron base composition, recombination rates, gene lengths, and expression levels:
Because most of the preferred codons in D. melanogaster end in G or C, it is possible that different mutational or gene conversion biases among genes could lead to differences in codon bias (Kliman and Hey 1994; Marais et al. 2003). For example, if female-biased genes were more prone to G or C mutations, then one would expect them to show greater codon bias than male-biased genes. Such biases in mutation (or gene conversion) are expected to affect not only coding regions, but also linked intron sequences. To test this, we examined intron GC content for the male-, female-, and non-sex-biased genes in our data set. We find that intron GC content is remarkably consistent and does not differ significantly among the three expression classes with intron %GC values of 39.5, 39.5, and 40.0 for male-, female-, and non-sex-biased, respectively. As an additional measure to remove the influence of local GC content on codon bias differences among the three groups of genes, we regressed intron %GC on FOP and used the residuals for Mann-Whitney tests among groups. After this correction, male-biased genes still had significantly less codon bias than both female- and non-sex-biased genes (P = 5.6 × 10−9 and 7.4 × 10−7, respectively), while female-biased genes had slightly higher codon bias than non-sex-biased genes (P = 0.02).
Biased gene conversion also is expected to increase as the local recombination rate increases (Marais et al. 2003). Thus, differences in the recombinational environment among male-, female-, and non-sex-biased genes could lead to differences in codon bias through this process. To examine this possibility, we compared local recombination rate estimates (Hey and Kliman 2002) for the genes in our survey. Average recombination rate estimates, R, for male-, female-, and non-sex-biased genes were 2.45, 2.51, and 2.48, respectively, and did not differ significantly among genes of the three expression classes (Mann-Whitney test, P > 0.40). Four other estimators of recombination rate presented in Hey and Kliman (2002) also showed no significant differences among male-, female-, and non-sex-biased genes (data not shown). To remove the potential influence of local recombination rate on codon bias differences among the three groups of genes, we regressed R on FOP and compared the residuals. After this correction, male-biased genes still had significantly less codon bias than both female- and non-sex-biased genes (P = 1.3 × 10−19 and 5.6 × 10−27, respectively), while there was no difference between female- and non-sex-biased genes (P = 0.48). Although there were no differences in average recombination rate among genes of the three expression classes, there were differences in the strength of the correlation between FOP and recombination rate within each class. This correlation was significant for the female-biased (R = 0.09; P = 0.002) and the non-sex-biased genes (R = 0.05; P = 0.004), but not for the male-biased genes (R = 0.02; P = 0.45).
Comeron (2004) presented evidence for transcription-associated mutational biases (TAMB) in human testes. This process is expected to alter base composition by increasing G (relative to C) and T (relative to A) content on the coding strand (Green et al. 2003). If TAMB were also common in Drosophila testes, then it would be expected to lead to increased %GT in the coding strand of testis-expressed genes and could disproportionately affect synonymous codon usage in male-biased genes. To test whether TAMB could explain the differences in codon bias observed among male-, female-, and non-sex-biased genes, we examined coding-strand GT content of introns occurring in genes of the three expression classes. As with intron GC contents, we find that intron GT contents are remarkably consistent, with %GT values of 50.1, 50.6, and 50.1 for male-, female-, and non-sex-biased genes, respectively. Thus, there is no evidence that TAMB are responsible for the reduced codon bias observed in male-biased genes. Furthermore, TAMB cannot explain the observation that somatic male-biased genes have significantly less codon bias than both somatic female-biased and germline male-biased genes (see above).
Levels of codon bias also have been shown to negatively correlate with CDS length (Powell and Moriyama 1997; Comeron et al. 1999; Duret and Mouchiroud 1999). Thus, if there were an overall trend for male-biased genes to be longer than female-biased genes, we would expect to see reduced levels of codon bias in male-biased genes. The genes in our survey, however, show the opposite trend: male-biased genes tend to be shorter than both female- and non-sex-biased genes. For the male-biased genes, the median CDS length is 1347 bp, while the median CDS lengths of the female- and non-sex-biased genes are 1449 bp and 1386 bp, respectively. The CDS length difference between male- and female-biased genes is marginally significant (Mann-Whitney test, P = 0.04), while all other comparisons are not significant (P > 0.20). To remove the potential influence of CDS length on codon bias differences among the three groups of genes, we regressed the length on FOP and compared the residuals. After this correction, male-biased genes still had significantly less codon bias than both female- and non-sex-biased genes (P = 2.5 × 10−21 and 1.9 × 10−26, respectively), while there was no difference between female- and non-sex-biased genes (P = 0.25).
The microarray experiments that served as the basis for our identification of sex-biased genes measured the ratio of male-to-female expression for each gene (Parisi et al. 2003; Ranz et al. 2003). These ratios, however, do not provide information on the expression level of the genes relative to other genes in the genome. Because codon bias is known to positively correlate with expression level, it may be that differences in absolute expression level among male-, female-, and non-sex-biased genes are responsible for the observed codon bias differences among these groups. To test this possibility, we used the microarray data of Gibson et al. (2004) to estimate the absolute expression level of all genes in our analysis (see materials and methods). In general, we find that sex-biased genes are expressed at higher levels than non-sex-biased genes, with male-biased genes expressed 1.4-fold higher and female-biased expressed 1.5-fold higher than non-sex-biased genes. To remove the potential influence of absolute expression on codon bias differences among the three groups of genes, we regressed expression on FOP and compared the residuals. After this correction, male-biased genes still had significantly less codon bias than both female- and non-sex-biased genes (P = 9.3 × 10−13 and 2.7 × 10−22, respectively), while there was no difference between female- and non-sex-biased genes (P = 0.34).
DISCUSSION
Our survey of synonymous codon usage in sex-biased genes revealed a strong and consistent pattern of reduced codon bias in genes with male-biased expression relative to those with female- and non-sex-biased expression (Table 1). This result is in agreement with that previously reported for a much smaller sample of sex-biased genes (Zhang et al. 2004). The Zhang et al. (2004) study produced conflicting results regarding levels of codon bias in female- vs. non-sex-biased genes: comparison of 78 female-biased and 126 non-sex-biased genes for which D. yakuba EST sequences were available indicated significantly greater levels of codon bias in female-biased genes, while comparison of 92 highly female-biased genes to 99 genes with equal expression between the sexes indicated no significant difference in codon bias. In the present survey, we compared 1443 female-biased and 4535 non-sex-biased genes using a twofold expression cutoff and observed no significant difference in codon bias. The contradictory results seen in the first comparison by Zhang et al. (2004) may be attributable to differences in absolute expression level among the female- and non-sex-biased genes. Because the genes used in this comparison were identified in an EST screen (Domazet-Loso and Tautz 2003), they should be biased toward genes that are highly expressed. Furthermore, since the EST clones were derived from a mixed pool of males and females in unknown proportion, the expression bias could be stronger for sex-biased genes. For example, if females were underrepresented in the original pool of flies, then a female-biased gene would have to show relatively high levels of expression to be represented in the EST library. Indeed, using the microarray data of Gibson et al. (2004) to estimate absolute expression level, we find that the genes in the EST data set show such a bias: the male- and non-sex-biased genes have 4.5-fold higher expression than those in this study, while the female-biased genes have a 7.5-fold higher expression. This can explain why the average values of codon bias are higher for all three groups of genes in the Zhang et al. (2004) data set and why the greatest difference is in the female-biased genes. After correcting for this expression difference by regressing the expression level on FOP and performing a Mann-Whitney test on the residuals, we find no significant difference between the female- and non-sex-biased genes (P = 0.67). In contrast to the EST data set, we observe no effect of absolute expression level on codon bias differences between female- and non-sex-biased genes in this study (see results). Because there appears to be little or no difference in codon bias between female- and non-sex-biased genes, we conclude that reduced codon bias is not a general property of sex-biased genes, but instead is specific to genes with male-biased expression. Thus, an explanation for our findings must be related to differences, either neutral or selective, between male- and female-biased (or non-sex-biased) genes.
Neutral processes, such as mutational or gene conversion biases, are thought to influence patterns of synonymous codon usage differentially throughout the genome (Kliman and Hey 1994; Marais et al. 2003). Therefore, if male-biased genes are subject to different replicational or recombinational conditions than female- and non-sex-biased genes, they might be expected to differ in patterns of codon bias. However, several observations argue against such an explanation. First, differences in the above conditions would need to be irregularly dispersed throughout the genome, because male-, female-, and non-sex-biased genes are found dispersed throughout all chromosome arms. A possible exception is on the X chromosome, where male-biased genes are significantly underrepresented (Parisi et al. 2003; Ranz et al. 2003). However, when X-linked and autosomal genes are considered separately, there is still significantly less codon bias in male-biased genes than in female- or non-sex-biased genes (Table 2), indicating that the X/autosome distribution of genes cannot explain the observed differences. Second, we see no difference in intron GC or coding-strand GT content among male-, female-, and non-sex-biased genes, which would be expected if the three classes of genes experienced different mutational biases. Finally, there is no significant difference in local recombination rate among male-, female-, and non-sex-biased genes, which would be expected if biased gene conversion were responsible for synonymous codon usage differences among the three groups of genes.
Differences in the type and/or strength of natural selection acting on male-, female-, and non-sex-biased genes could affect levels of codon bias in the three groups of genes in a number of ways. One possibility is that synonymous codon usage in male- and female-biased genes is adapted to match the tRNA pools in the tissues where these genes are predominantly expressed. For example, synonymous codon usage in male-biased genes could be adapted to match a testis-specific tRNA pool. There is, however, little support for this hypothesis. First, significant differences in codon bias are observed whether it is measured as FOP or ENC (Table 1), and the latter measure of codon bias makes no assumptions about which codons are favored. Second, an analysis of synonymous codon usage in all genes included in our survey indicates that the same synonymous codons are favored/avoided in male-, female-, and non-sex-biased genes (see supplementary Tables S2 and S3 at http://www.genetics.org/supplemental/). Third, there is a negative correlation between codon bias and degree of sex-biased expression for male-biased genes (Figure 1A). If synonymous codon usage in male-biased genes were adapted to match male-specific tRNA pools, then one would expect codon bias to increase as the degree of male-biased expression increased. The above results could be explained if there are not qualitative, but rather quantitative, differences in tRNA abundance between testes and other tissues. For example, the most abundant tRNAs could be the same in all tissues, but the extent to which the tRNA pool is biased could be less in testes than in other tissues. However, the codon bias differences between male- and female-biased genes are not limited to genes expressed in sex-specific reproductive tissues, but are also seen for genes expressed in nonreproductive tissues (Table 3), which presumably share the same tRNA pools. Furthermore, genes with somatic male-biased expression genes show significantly less codon bias than genes with germline male-biased expression (Table 3). This would not be expected if synonymous codon usage in male-biased genes were predominantly influenced by testes tRNA pools.
Differences in the strength of purifying selection acting at synonymous sites in male-, female-, and non-sex-biased genes could also lead to differences in levels of codon bias. For example, it has been proposed that the strength of selection at synonymous sites in a gene is inversely proportional to gene length (Comeron et al. 1999). Indeed, as is predicted by this model, a negative correlation between codon bias and CDS length has been observed in Drosophila (Powell and Moriyama 1997; Comeron et al. 1999; Duret and Mouchiroud 1999). This model, however, cannot explain the codon bias differences among male-, female-, and non-sex-biased genes seen in our survey, as the male-biased genes tend to be shorter than both female- and non-sex-biased genes. Thus one would expect male-biased genes to show more codon bias, rather than less, than genes of the other two classes.
Another possibility is that there may be general differences in selective constraint among male-, female-, and non-sex-biased genes. It has been suggested that there is a correlation between the level of constraint on synonymous and nonsynonymous sites (Akashi 1994; Comeron and Kreitman 1998). This could explain the observation that codon bias is negatively correlated with both the synonymous and the nonsynonymous substitution rate. Since it has been shown that male-biased genes have significantly higher rates of nonsynonymous substitution than female- and non-sex-biased genes (Zhang et al. 2004), it may be that male-biased genes are subject to less purifying selection at both synonymous and nonsynonymous sites. A possible cause for this may be that, due to their greater variance in reproductive success, males have a smaller Ne than females. Thus selection is expected to be less effective on male traits. Although all of the genes in our survey are physically present in both sexes (i.e., none are Y-linked), there may be sex-related differences in the degree of selection that they experience. For example, in the extreme case of a gene with male-specific expression, purifying selection against synonymous or nonsynonymous mutations will occur only in males and is expected to be weaker because of their reduced Ne. Patterns of polymorphism in sex-biased genes, however, argue against the above explanation. Zhang et al. (2004) analyzed population genetic data for 55 D. melanogaster genes and found that, in contrast to their elevated ratio of nonsynonymous-to-synonymous divergence, male-biased genes did not have an elevated ratio of nonsynonymous-to-synonymous polymorphism relative to female- and non-sex-biased genes, as would be expected if they were evolving under less selective constraint. However, Zhang et al. (2004) pointed out several caveats to this interpretation. First, it is based on a small number of sex-biased genes (13 male biased and 12 female biased) for which both polymorphism and divergence data were available from the literature. Second, the data were collected by many independent groups that employed different population sampling schemes. Thus, there is no control for demographic factors that might affect observed levels of polymorphism. Finally, there may be a sampling bias toward genes with an a priori expectation of either positive or balancing selection. When all genes showing evidence for selection are removed from the analysis (i.e., genes giving a significant result by the test of McDonald and Kreitman (1991), the male-biased genes still show the highest average nonsynonymous/synonymous divergence ratio and the lowest average nonsynonymous/synonymous polymorphism ratio, although in this case the sample size drops to 7 male-biased and 9 female-biased genes.
The negative correlation between codon bias and the nonsynonymous substitution rate also could be explained by Hill-Robertson interference between synonymous and nonsynonymous mutations within a gene (Akashi 1996; Betancourt and Presgraves 2002; Kim 2004). Under this scenario, the fixation of strongly favored amino acid replacements in adaptively evolving proteins results in the fixation of linked, slightly deleterious synonymous mutations. Thus, if male-biased genes were targets of positive selection more often than female- or non-sex-biased genes, one would expect them to show reduced levels of codon bias. Because interference is reduced in regions of higher recombination, one might expect that the positive correlation between codon bias and local recombination rate would be stronger for male-biased genes than for female- or non-sex-biased genes. However, male-biased genes show the weakest correlation (see results). Such a pattern could be explained by a greater rate of adaptive amino acid substitution in male-biased genes in regions of higher recombination. If positively selected amino acid replacements are more frequent in these regions, then there would be more opportunity for the fixation of linked, slightly deleterious synonymous mutations. This would partially counteract the relaxation of Hill-Robertson interference in the regions of higher recombination described above and weaken the correlation between codon bias and recombination rate. Support for this hypothesis comes from the observation that there is a positive correlation between the nonsynonymous substitution rate and the local recombination rate for Acp genes, which are thought to undergo frequent adaptive evolution in general (Swanson et al. 2001; Betancourt and Presgraves 2002), and for male-biased genes in general (our unpublished results).
Zhang et al. (2004) compared ratios of polymorphism and divergence at synonymous and nonsynonymous sites in male-, female-, and non-sex-biased genes and found evidence for increased adaptive evolution in male-biased genes. These results also suggest that Hill-Robertson interference between strongly selected nonsynonymous mutations and weakly selected synonymous mutations is more common in male-biased genes (although see the caveats mentioned above). In addition to an elevated rate of nonsynonymous substitution, male-biased genes also show increased rates of expression polymorphism and divergence, at least some of which appear to be attributable to positive selection (Meiklejohn et al. 2003; Ranz et al. 2003; Nuzhdin et al. 2004). Given that the majority of expression differences among Drosophila species are caused by differences in cis-regulatory elements (Wittkopp et al. 2004), it is likely that positive selection on linked regulatory sequences also results in Hill-Robertson interference that could reduce codon bias in male-biased genes. We wish to emphasize, however, that not every male-biased gene in our survey must be affected by positive selection to cause the observed reduction in codon bias. We can estimate the minimum number of genes that need to be affected by seeing how many of the male-biased genes with the lowest FOP values need to be removed for the difference between male- and female-biased genes to become insignificant (P > 0.05) by the Mann-Whitney test. In this case, it is 215 genes, or ∼17% of the male-biased genes in our survey. A similar analysis indicates that a higher number (>500) of the male-biased genes are responsible for the negative correlation between FOP and degree of male-biased expression (Figure 1A) and suggests that up to 40% of the male-biased genes may be affected by positive selection. It should be noted, however, that the positive selection and relaxed constraint hypotheses are not mutually exclusive. It is possible that some fraction of the male-biased genes is subject to increased positive selection, while another fraction experiences less selective constraint. It is also possible that this latter fraction is overrepresented among highly male-biased genes. If so, the fraction of male-biased genes affected by positive selection could be much lower than the above estimates.
In this study, we have shown that male-biased genes have significantly less codon bias than female- and non-sex-biased genes on a genomic scale. We propose that this is at least partially attributable to increased positive selection acting on structural and/or regulatory changes in male-biased genes. Further studies of polymorphism and divergence in both sequence and expression are needed to determine the fraction of male-biased genes that are subject to positive selection and the extent to which this selection influences levels of codon bias.
Acknowledgments
We thank H. Akashi, J. Baines, J. Hermisson, C. Meiklejohn, D. Presgraves, J. Ranz, L. Rose, W. Stephan, and N. Stoletzki for critical reading of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft grant PA 903/2-1.
References
- Aguadé, M., 1998. Different forces drive the evolution of the Acp26Aa and Acp26Ab accessory gland genes in the Drosophila melanogaster species complex. Genetics 150: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., 1999. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics 152: 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., 1994. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136: 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., 1995. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics 139: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., 1996. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution and larger proteins in D. melanogaster. Genetics 144: 1297–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., 2003. Translational selection and yeast proteome evolution. Genetics 164: 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., P. Whitley, B. L. Todd, H. M. Waldrip-Dail and A. G. Clark, 2000. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics 156: 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J. L., and B. D. Hall, 1982. Codon selection in yeast. J. Biol. Chem. 257: 3026–3031. [PubMed] [Google Scholar]
- Betancourt, A., and D. C. Presgraves, 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. USA 99: 13616–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán, E., and M. Long, 2003. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics 164: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlini, D. B., and W. Stephan, 2003. In vivo introduction of unpreferred synonymous codons into the Drosophila Adh gene results in reduced levels of ADH protein. Genetics 163: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 1999. Broad-sense sexual selection, sex gene pool evolution, and speciation. Genome 42: 1033–1041. [PubMed] [Google Scholar]
- Coghlan, A., and K. H. Wolfe, 2000. Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast 16: 1131–1145. [DOI] [PubMed] [Google Scholar]
- Comeron, J. M., 2004. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics 167: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J. M., and M. Kreitman, 1998. The correlation between synonymous and nonsynonymous substitutions in Drosophila: Mutation, selection or relaxed constraints? Genetics 150: 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J. M., M. Kreitman and M. Aguadé, 1999. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics 151: 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso, T., and D. Tautz, 2003. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 13: 2213–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret, L., 2000. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16: 287–289. [DOI] [PubMed] [Google Scholar]
- Duret, L., and D. Mouchiroud, 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, G., R. Riley-Berger, L. Harshman, A. Kopp, S. Vacha et al., 2004. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics 167: 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy, M., and C. Gautier, 1982. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 10: 7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham, R., C. Gautier, M. Gouy, M. Jacobzone and R. Mercier, 1981. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 9: r43–r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, P., B. Ewing, W. Miller, P. J. Thomas and E. D. Green, 2003. Transcription-associated mutational asymmetry in mammalian evolution. Nat. Genet. 33: 514–517. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., and W. Fiers, 1982. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene 18: 199–209. [DOI] [PubMed] [Google Scholar]
- Hey, J., and R. M. Kliman, 2002. Interactions between natural selection, recombination and gene diversity in the genes of Drosophila. Genetics 160: 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W. G., and A. Robertson, 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Ikemura, T., 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151: 389–409. [DOI] [PubMed] [Google Scholar]
- Ikemura, T., 1982. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J. Mol. Biol. 158: 573–597. [DOI] [PubMed] [Google Scholar]
- Kanaya, S., Y. Yamada, Y. Kudo and T. Ikemura, 1999. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238: 143–155. [DOI] [PubMed] [Google Scholar]
- Kim, Y., 2004. Effect of strong directional selection on weakly selected mutations at linked sites: implication for synonymous codon usage. Mol. Biol. Evol. 21: 286–294. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1968. Genetic variability maintained in a finite population due to mutational production of neutral and nearly neutral isoalleles. Genet. Res. 11: 247–269. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1977. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 267: 275–276. [DOI] [PubMed] [Google Scholar]
- Kliman, R. M., and J. Hey, 1993. Reduced natural selection associated with low recombination in Drosophila melanogaster. Mol. Biol. Evol. 10: 1239–1258. [DOI] [PubMed] [Google Scholar]
- Kliman, R. M., and J. Hey, 1994. The effects of mutation and natural selection on codon bias in the genes of Drosophila. Genetics 137: 1049–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman, R. M., and J. Hey, 2003. Hill-Robertson interference in Drosophila melanogaster: reply to Marais, Mouchiroud and Duret. Genet. Res. 81: 89–90. [DOI] [PubMed] [Google Scholar]
- Marais, G., D. Mouchiroud and L. Duret, 2001. Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc. Natl. Acad. Sci. USA 98: 5688–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais, G., D. Mouchiroud and L. Duret, 2003. Neutral effect of recombination on base composition in Drosophila. Genet. Res. 81: 79–87. [DOI] [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- McVean, G. A., and J. Vieira, 2001. Inferring parameters of mutation, selection and demography from patterns of synonymous site evolution in Drosophila. Genetics 157: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn, C. D., J. Parsch, J. M. Ranz and D. L. Hartl, 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 100: 9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, E. N., and J. R. Powell, 1997. Codon usage bias and tRNA abundance in Drosophila. J. Mol. Evol. 45: 514–523. [DOI] [PubMed] [Google Scholar]
- Nurminsky, D. I., M. V. Nurminskaya, D. De Aguiar and D. L. Hartl, 1998. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature 396: 572–575. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S. V., M. L. Wayne, K. L. Harmon and L. M. McIntyr, 2004. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol. Biol. Evol. 21: 1308–1317. [DOI] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, D. Naiman, G. Bouffard, J. Malley et al., 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299: 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, P. Edwards, L. Minor, D. Naiman et al., 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch, J., C. D. Meiklejohn and D. L. Hartl, 2001. Patterns of DNA sequence variation suggest the recent action of positive selection in the janus-ocnus region of Drosophila simulans. Genetics 159: 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J., and E. N. Moriyama, 1997. Evolution of codon bias in Drosophila. Proc. Natl. Acad. Sci. USA 94: 7784–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J. M., C. I. Castillo-Davis, C. D. Meiklejohn and D. L. Hartl, 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300: 1742–1745. [DOI] [PubMed] [Google Scholar]
- Singh, R. S., and R. J. Kulathinal, 2000. Sex gene pool evolution and speciation: a new paradigm. Genes Genet. Syst. 75: 119–130. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3: 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., A. G. Clark, H. M. Waldrip-Dail, M. F. Wolfner and C. F. Aquadro, 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98: 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, C. T., S. C. Tsaur, M. L. Wu and C.-I Wu, 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504. [DOI] [PubMed] [Google Scholar]
- Tsaur, S. C., and C.-I Wu, 1997. Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa of Drosophila. Mol. Biol. Evol. 14: 544–549. [DOI] [PubMed] [Google Scholar]
- Tsaur, S. C., C. T. Ting and C.-I Wu, 1998. Positive selection driving the evolution of a gene of male reproduction, Acp26Aa, of Drosophila: II. Divergence versus polymorphism. Mol. Biol. Evol. 15: 1040–1046. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., B. K. Haerum and A. G. Clark, 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. [DOI] [PubMed] [Google Scholar]
- Wright, F., 1990. The ‘effective number of codons’ used in a gene. Gene 87: 23–29. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., T. M. Hambuch and J. Parsch, 2004. Molecular evolution of sex-biased genes in Drosophila. Mol. Biol. Evol. 21: 2130–2139. [DOI] [PubMed] [Google Scholar]