Abstract
CD39/ecto-NTPDase 1 (nucleoside triphosphate diphosphohydrolase 1) is an ecto-nucleotidase that influences P2 receptor activation to regulate vascular and immune cell adhesion and signalling events pivotal in inflammation. Whether CD39 interacts with other membrane or cytoplasmic proteins has not been established to date. Using the yeast two-hybrid system, we note that the N-terminus of CD39 binds to RanBPM (Ran binding protein M; also known as RanBP9), a multi-adaptor scaffolding membrane protein originally characterized as a binding protein for the small GTPase Ran. We confirm formation of complexes between CD39 and RanBPM in transfected mammalian cells by co-immunoprecipitation studies. Endogenous CD39 and RanBPM are also found to be co-expressed and abundant in cell membranes of B-lymphocytes. NTPDase activity of recombinant CD39, but not of N-terminus-deleted-CD39 mutant, is substantially diminished by RanBPM co-expression in COS-7 cells. The conserved SPRY [repeats in splA and RyR (ryanodine receptor)] moiety of RanBPM is insufficient alone for complete physical and functional interactions with CD39. We conclude that CD39 associations with RanBPM have the potential to regulate NTPDase catalytic activity. This intermolecular interaction may have important implications for the regulation of extracellular nucleotide-mediated signalling.
Keywords: CD39, ecto-nucleotidase, nucleoside triphosphate diphosphohydrolase 1 (NTPDase1), RanBPM, SPRY domain, yeast two-hybrid system
Abbreviations: ACR, apyrase conserved region; c-Met, an RPTK (receptor protein-tyrosine kinase) for HGF (hepatocyte growth factor); DC, dendritic cells; DMEM, Dulbecco's modified Eagle's medium; EBV, Epstein–Barr virus; FBS, fetal bovine serum; NTPDase1, nucleoside triphosphate diphosphohydrolase 1; P2Y1, P2 (purinergic type-2)-receptor 1; RanBPM, Ran binding protein M; RyR, ryanodine receptor; SD, synthetic dropout; SPRY, repeats in splA and RyR
INTRODUCTION
CD39 was originally identified as an activation marker for B-lymphocytes and subsequently shown to be expressed on subsets of activated NK (natural killer) cells, T-lymphocytes, epidermal Langerhans DC (dendritic cells) and endothelial cells [1]. CD39 acts as an important plasma membrane-bound ecto-nucleotidase with pivotal roles in the hydrolysis of extracellular nucleoside di- and tri-phosphates by the endothelium and leucocytes [2–4]. CD39 has also been shown to modulate vascular inflammation, cellular proliferation and migration [5,6] and to play a crucial role in the regulation of the ADP-purinoreceptor P2Y1 [P2 (purinergic type-2)-receptor 1] function [4,7].
CD39 is an acidic glycoprotein that migrates as a 78 kDa protein. It comprises N- and C-terminal transmembrane-anchoring domains with short cytoplasmic tails, and a large glycosylated extracellular domain, containing five ACR (apyrase conserved region) sequences [1,2,8]. CD39 forms tetramers, and interactions between the transmembrane segments of the CD39 monomers appear to regulate the enzymatic activity of the protein [9]. The ACR motifs are considered critical for enzymatic function of CD39 [7]. CD39 also undergoes several post-translational modifications including proteolytical cleavage, N-glycosylation and constitutive palmitoylation [7,10]. It has been established that palmitoylation greatly stabilizes membrane association, augments NTPDase (nucleoside triphosphate diphosphohydrolase) activity [11], and targets the CD39 molecule, in association with P2Y1 receptors, to specialized membrane microdomains termed caveolae that are crucial in the regulation of signal transduction [10,12–14].
Putative CD39 cellular protein partners that impact upon ectoenzymatic or nucleotide-regulated signalling events are not known. Here, we identify RanBPM (Ran-binding protein M)/RanBP9 as a CD39-interacting protein. RanBPM was originally cloned as a Ran-binding protein and reported to localize both in the nucleus and in the cytoplasm [15,16]. Ectopic expression of RanBPM was subsequently demonstrated to interact with c-Met [an RPTK (receptor protein-tyrosine kinase) for HGF (hepatocyte growth factor)] to regulate Ras activation [17]. RanBPM also has interactions with β1 and β2 families of integrins [18]. RanBPM is well conserved and the endogenous protein is widely expressed in cell lines and tissues [16,18]. RanBPM contains a conserved SPRY [repeats in splA and RyR (ryanodine receptor)] domain of unknown function [19].
In the present study, we demonstrate that CD39 forms a complex with RanBPM. We present results showing that endogenous CD39 and RanBPM co-localize in the cell membranes of B-lymphocytes. Our co-immunoprecipitation studies demonstrate that RanBPM associates with CD39 in B-lymphocytes. We also provide evidence that the enzymatic activity of CD39 is down-regulated specifically, after the binding of RanBPM to the N-terminal intracytoplasmic CD39 domain.
EXPERIMENTAL
Reagents
Amino acids, nucleoside phosphates and chemicals were purchased from Sigma. Anti-CD39 and anti-CD39–FITC monoclonal antibodies were purchased from Ancell, and anti-c-Myc monoclonal antibody was from Santa Cruz Biotechnology. Monoclonal antibody to RanBPM was generated as described previously [18].
pCMV-Myc-RanBPM149-729 (N-terminus-truncated RanBPM, amino acids 149–729), pCMV-Myc-RanBPM351-729 (SPRY-truncated RanBPM, amino acids 351–729) and pCMV-Myc-RanBPM149-353 (SPRY moiety of RanBPM, amino acids 149–353) were constructed by inserting the RanBPM coding sequences amplified by PCR respectively into a pCMV-Myc vector (Clontech) by restriction enzyme digestion with SalI and KpnI. The construction of pcDNA3-CD39 and NT-Δ-1–37 mutated CD39 was previously described [2,7]. To construct pGADT7-RanBPM351-729, an NdeI–XhoI PCR product containing the C-terminal 378-amino-acid segment of RanBPM was cloned into the pGADT7 vector (Clontech) containing the Gal4 DNA activation domain. To construct the pGADT7-RanBPM152-353 (SPRY moiety of RanBPM, amino acids 152–353), a DNA fragment encoding the SPRY domain of RanBPM amplified by PCR, was inserted into the pGADT7 vector by digesting with restriction enzymes SmalI/XmaI and XhoI. All sequences amplified by PCR were confirmed by complete sequencing.
Yeast two-hybrid screening
The assay was conducted as described previously [20]. To construct the plasmid (pRW100) expressing the hybrid bait protein, a SalI–BglII DNA fragment encoding the N-terminal 14 amino acids of human CD39, and a FLAG tag (DYKDDDDK) with a two-proline linker added upstream, was cloned into the yeast expression vector containing the Gal4 DNA-binding domain (pGBDU-C1) and the URA3 [this stands for the URA3 gene; the promoter of URA3 marked the DNA-binding domain vector pGBDU-C1; therefore PJ69-4A (which is Ura−) transformed with pGBDU-C1-Bait could survive on uracil-depleted SD (synthetic dropout) plates] marker [20]. The pRW100 plasmid was transformed into the yeast reporter strain PJ69-4A [20]. The resulting strain was subsequently transformed with a pre-transformed human lymphocyte MATCHMAKER cDNA library, and selected, as described by the manufacturer (Clontech).
Cell culture and transient transfection
COS-7 cells were maintained in DMEM (Dulbecco's modified Eagle's medium) with 10% (v/v) FBS (fetal bovine serum), supplemented with L-glutamine (2 mM), penicillin (100 units/ml) and streptomycin (100 μg/ml). Transient transfections were performed using Lipofectamine™ (Invitrogen/Gibco) according to the manufacturer's instructions. Briefly, the cells cultured in 6-well plates were exposed to 1 μg of plasmid DNA and 3 μl of Lipofectamine™ reagent complex for 5 h in Opti-MEM I reduced serum medium (Invitrogen/Gibco), followed by replacement of the medium with fresh culture medium (DMEM/10% FBS). The culture medium was changed every 24 h after transfection and, approx. 48 h post-transfection, the cells were used for analyses. Transfection efficiency was monitored by using the β-galactosidase enzyme assay system with reporter lysis buffer (Promega) (results not shown).
Human EBV (Epstein–Barr virus)-transformed B-lymphocytes (HCC1739 BL, A.T.C.C.) were maintained in RPMI 1640 medium with 2 mM L-glutamine modified by A.T.C.C. to contain 10 mM Hepes, 1 mM sodium pyruvate, 4.5 g/l glucose and 1.5 g/l bicarbonate, supplemented with 10% FBS, penicillin (100 units/ml) and streptomycin (100 μg/ml).
All cells were grown in culture dishes or flasks at 37 °C in a humidified incubator with a 5% CO2 atmosphere.
Cell preparations
CD11b+, B220+ and CD11c+ cells were positively selected from spleens of 8–10-week-old mice through the use of MACS Sort magnetic beads in MACS LS Separation columns (Miltenyi Biotec, Auburn, CA, U.S.A.).
Co-immunoprecipitation and immunofluorescence
Immunoprecipitation was performed using Mouse IgG TrueBlot Set (eBioscience) as suggested by the manufacturer. CD39 was analysed by Western blotting under non-reducing conditions. c-Myc–RanBPM or RanBPM was analysed under reducing conditions.
Human peripheral blood mononuclear cells from healthy blood donor volunteers isolated by Ficoll-Paque™ PLUS (Amersham Biosciences) or cultured B-lymphocytes of peripheral blood, were centrifuged on slides in a cytospin centrifuge and fixed in 2% (w/v) paraformaldehyde. Immunofluorescence was performed as previously described [6,14] following the manufacturer's instructions.
Quantitative TaqMan real-time PCR
Total RNA was extracted and purified from cells or tissues using an RNeasy Mini kit (Qiagen). Reverse transcription was done on 1 μg of RNA using ABI Prism TaqMan reverse transcription reagents (Applied Biosystems, Foster City, CA, U.S.A.). The ABI PRISM 7900HT Sequence Detection system was used for real-time PCR analysis. Primer-probe sets and TaqMan Universal PCR Master mix were purchased from Applied Biosystems. A comparative CT (threshold cycle) was used to determine gene expression and analysed against the endogenous genes of murine glyceraldehyde-3-phosphate dehydrogenase.
NTPDase activity
COS-7 cells were cultured in 24-well plates. At 48 h post-transfection, adherent cells were directly assayed for ecto-nucleotidase activity with phosphate generation as the readout, as previously described [21]. A standard curve was constructed using 0–20 μM KH2PO4. Intact, adherent cells in 24-well plates were washed three times in incubation buffer (20 mM Hepes, pH 7.5, 10 mM glucose, 5 mM KCl, 120 mM NaCl, 2 mM CaCl2 and 5 mM tetramisole) and incubated in 200 μl of the same buffer containing 3 mM nucleoside phosphates for 10 min at 37 °C. Reactions were stopped by taking 100 μl of supernatant from each well into an Eppendorf tube with the addition of 200 μl of 7.5% (w/v) trichloroacetic acid to a final concentration of 5%, vortex-mixing, and immediately putting on ice. Then for each sample, in an ELISA plate, 20 μl of the stopped reaction supernatants and 60 μl of distilled water were incubated together with 200 μl of Malachite Green reagent [2 vol. of distilled water, 2 vol. of 0.0812% Malachite Green base, 1 vol. of 2.32% (w/v) poly-(vinyl alcohol) and 1 vol. of 5.7% (w/v) ammonium molybdate in 6 M HCl] for 20 min. Absorbance (A) at 610 nm was measured by a spectrophotometer (Ultra Microplate Reader Elx 808; Bio-tek Instruments) and generated phosphate levels were determined from standard curves. The residual supernatants in the wells were removed by suction. Adherent cells were then lysed with 100 μl of 1 M NaOH. Protein concentrations were measured with Bio-Rad DC protein assay reagent (Bio-Rad Laboratories, Hercules, CA, U.S.A.) (to normalize ecto-enzymatic activity).
RESULTS
RanBPM is a CD39-binding protein
Using the human CD39 N-terminal moiety (amino acids 1–14) as ‘bait’, an EBV-transformed human B-lymphocyte cDNA library was screened for binding proteins. To decrease the incidence of false positive clone selections, a unique host strain PJ69-4A was used [20]. Three positive clones were identified after screening 2.4×107 clones. DNA sequencing revealed that the three positive clones are identical and each of them encoded the same partial sequence of RanBPM (lacking amino acids 1–148), a protein originally cloned as a Ran-binding protein [15]. Further two-hybrid assays using independent co-transformation of the bait construct with candidate clones, followed by growth selection and β-galactosidase assay, confirmed specific interactions between CD39 and RanBPM (Table 1).
Table 1. Results of yeast two-hybrid screening using the N-terminal CD39 moiety (amino acids 1–14) and the human B-lymphocyte cDNA library.
The symbol ‘+++’ indicates the growth of yeast cells in ‘drop-out media’ as well as high activity of β-galactosidase in the LacZ assay column. The symbol ‘−’ indicates the lack of growth of yeast cells in ‘drop-out media’, or undetectable β-galactosidase activity. ‘pGBDU-C1’ is the expression vector containing the Gal4 DNA-binding domain. ‘Bait’ represents the construct encoding the fusion of the Gal4 DNA-binding domain with the N-terminal 14-amino-acid segment of CD39. ‘RanBPM’ represents the construct encoding the fusion of the Gal4 DNA activation domain with the fragment of RanBPM. ‘Bait+RanBPM’ represents the construct encoding the fusion of the Gal4 DNA activation domain with the fragment of RanBPM in addition to the bait construct.
| Strains | Growth on Ade− | Growth on His− | LacZ assay |
|---|---|---|---|
| Empty vector (PGBDU-C1) | − | − | − |
| Bait (pGBDU-C1-CD39N1-14) | − | − | − |
| RanBPM (pACT2-RanBPM149-729) | − | − | − |
| Bait+RanBPM (pGBDU-C1-CD39N1-14+pACT2-RanBPM149-729) | +++ | +++ | +++ |
These experiments suggest that intact CD39 and RanBPM might interact via the N-terminal moiety of CD39.
CD39 resides in a complex with RanBPM in transfected cell lysates
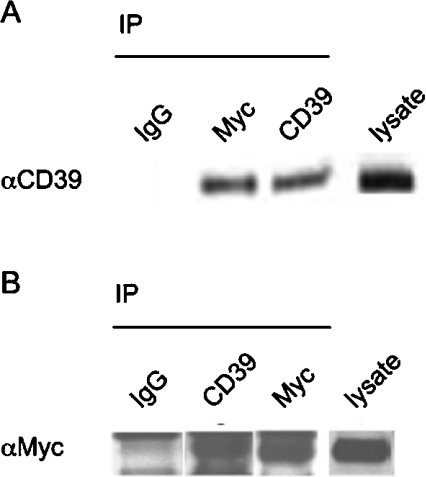
To probe for CD39–RanBPM complex formation in mammalian cells, extracts were prepared from transfected COS-7 cells expressing both recombinant c-Myc-tagged RanBPM and CD39 (Figure 1). We tested the ability of either c-Myc- or CD39-specific antibody to capture the putative protein partner. In the total extract from cells co-expressing CD39 and c-Myc–RanBPM, c-Myc–RanBPM was immunoprecipitated by CD39-specific antibodies, but not by control mouse IgG (Figure 1A). Consistent with this finding, CD39 was found in the immunoprecipitates obtained with c-Myc-specific antibodies, but not in that of control mouse IgG (Figure 1B). Therefore we conclude that the ectopically expressed native CD39 in COS-7 cells exists in complexes with ectopic RanBPM.
Figure 1. Co-immunoprecipitation of recombinant CD39 with c-Myc-tagged RanBPM.
(A, B) Cell lysates (500 μg) from COS-7 cells expressing recombinant CD39 protein and c-Myc–RanBPM were subjected to immunoprecipitation (IP) with CD39-specific antibody or c-Myc-specific antibody or control mouse isotype-matched IgG1, as indicated on the top of the panels. The antibody-captured proteins were analysed by Western blotting (antibody specificity is identified to the left of the panels). CD39 blots were analysed under non-reducing conditions. c-Myc–RanBPM blots were analysed under reducing conditions. The lane on the far right shows the appropriate positive control Western-blot analysis of total lysates. αCD39, anti-CD39; αMyc, anti-Myc.
Endogenous RanBPM associates with CD39 in B-lymphocytes
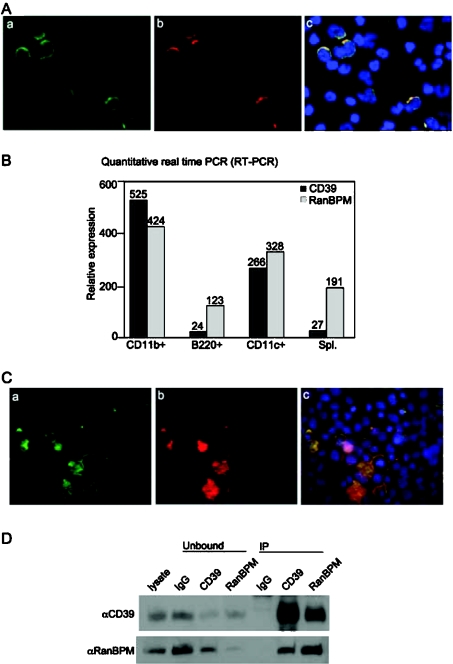
In order to investigate cellular localization of RanBPM and CD39, we stained human peripheral blood mononuclear cells with FITC-labelled monoclonal anti-CD39 antibody and anti-RanBPM monoclonal antibody followed by chicken anti-mouse IgG conjugated with Alexa Fluor® 594. Triple immunofluorescent staining of human peripheral blood mononuclear cells revealed obvious endogenous localization of both proteins and indicated co-localization in the cell membranes (Figure 2A).
Figure 2. Endogenous associations of CD39 and RanBPM.
(A) Triple immunofluorescent analysis for the endogenous co-localization of CD39 (a, green) and RanBPM (b, red) in human peripheral blood mononuclear cells. CD39 staining was performed using FITC-labelled monoclonal anti-CD39 antibody. RanBPM staining was performed using monoclonal anti-RanBPM antibody followed by chicken anti-mouse IgG conjugated with Alexa Fluor® 594. Nuclear staining was performed using Hoechst 33258 (c, blue). Co-localization of the two proteins in the cell membranes of peripheral blood mononuclear cells was observed (c, yellow, overlay of a and b). (B) Quantitative real-time PCR analysis of CD39 and RanBPM mRNA co-expression in murine CD11b+ cells (macrophages), B220+ cells (B-lymphocytes), CD11c+ cells (DC) and total splenocytes (Spl.). These cells all co-expressed CD39 and RanBPM at mRNA level. (C) Triple immunofluorescent staining was performed in EBV-transformed human B-lymphocytes, as described above. Endogenous CD39 (a, green) co-localized with endogenous RanBPM (b, red) in the cell membranes of B-lymphocytes (c, yellow, overlay of a and b). (D) RanBPM interacts with CD39 in B-lymphocytes. EBV-transformed B-lymphocyte lysates were immunoprecipitated with the indicated antibodies. Immunoprecipitates (IP) were resolved on a 4–15% gradient SDS/PAGE, transferred on to a PVDF membrane, and probed with antibodies as indicated to the left of the panels. The far-left lane shows control Western-blot analysis of total lysates (10% input). The residual non-immunoprecipitated proteins within the lysates are indicated as ‘Unbound’ at the top of the panel. CD39 was analysed under non-reducing conditions; RanBPM was analysed under reducing conditions, as before.
To identify further the subsets of peripheral blood mononuclear cells that co-express CD39 and RanBPM, quantitative real-time PCR was performed in different cell preparations from mouse spleen. mRNA co-expression of CD39 and RanBPM was observed in macrophages (CD11b+), B-lymphocytes (B220+; low level of CD39), DC (CD11c+) and splenocytes (Spl.) (Figure 2B). Triple immunofluorescent analyses were performed on human EBV-transformed B-lymphocytes that express high levels of CD39. Endogenous co-localization of CD39 and RanBPM in the cell membranes was observed (Figure 2C).
The partial co-localization of endogenous RanBPM and CD39 in the cell membranes prompted an investigation of the interaction between the two proteins in vivo. Co-immunoprecipitation studies were performed in the B-lymphocyte lysates. We found that anti-RanBPM antibodies co-precipitate associated CD39, and in a reciprocal manner anti-CD39 antibodies co-precipitate endogenous RanBPM (Figure 2D).
These results confirm structural associations between RanBPM and CD39 in non-transfected cells.
RanBPM decreases enzymatic activity of membrane-expressed CD39
Given that CD39 and RanBPM interact structurally in vitro and in vivo and exist as complexes, it is feasible that RanBPM might alter the ecto-nucleotidase activity of CD39. The functional consequences of the CD39–RanBPM interaction were therefore investigated. We studied the effects of ectopic RanBPM on CD39 or NT-Δ-1–37 mutated-CD39 enzymatic activity using phosphohydrolysis activity assays in COS-7 cells expressing ectopic RanBPM and/or CD39, and/or NT-Δ-1–37. COS-7 cells were seeded into 24-well plates and transfected with pcDNA3-CD39 or NT-Δ-1-37 mutant alone and together with pCMV-c-Myc-RanBPM. Transfection experiments with pCMV-c-Myc and pcDNA3 vectors served as negative controls. Cell-associated NTPDase activity was measured 48 h after transfection.
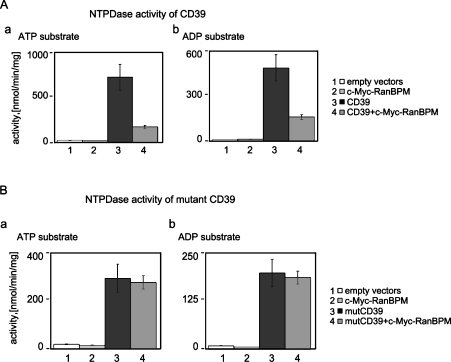
RanBPM did not alter the level of CD39 expression (results not shown), but did down-regulate ecto-NTPDase activity on intact cells (Figure 3A). No NTPDase activity was observed when transfections were performed with control vectors or with c-Myc–RanBPM alone (Figure 3A, panel a, lanes 1 and 2, and panel b, lanes 1 and 2). As expected, pcDNA3-CD39-transfected cells exhibited very high levels of NTPDase activity when using ATP or ADP as substrates, as compared with control vector-transfected cells (Figure 3A, panel a, lane 3, and panel b, lane 3). We observed marked decreases in CD39 enzymatic activity in COS-7 cells expressing ectopic CD39 together with RanBPM (Figure 3A, panel a, lane 4, and panel b, lane 4). RanBPM inhibited CD39 NTPDase activity independently of the substrate of the reaction. Both ATP hydrolysis and ADP hydrolysis were decreased in the presence of RanBPM (Figure 3A, panels a and b). Similar effects were also observed for other extracellular nucleotides (results not shown).
Figure 3. RanBPM inhibits the NTPDase activity of CD39 by interacting with the N-terminus of CD39.
(A) ATP and ADP (as indicated at the top of the panels) are used as substrates of the NTPDase activity in intact transfected COS-7 cells overexpressing CD39 and/or c-Myc–RanBPM. COS-7 cells transfected with empty vectors were used as a negative control. Samples are: bar 1, control reaction with COS-7 cells transfected with empty plasmid vectors; bar 2, cells overexpressing c-Myc–RanBPM; bar 3, cells with ectopic CD39; bar 4, cells expressing both CD39 and c-Myc–RanBPM proteins. Three independent experiments produced consistent results (S.D.=±7%). (B) The same as described in (A) except for NT-Δ-1–37 CD39 mutant (mutCD39) instead of native CD39.
As described previously [7], the NT-Δ-1–37 mutant has decreased NTPDase activity when contrasted with native CD39 (Figure 3B, panel a, lane 3, and panel b, lane 3). RanBPM did not further inhibit the biochemical activity of the NT-Δ-1–37 CD39 mutant, unlike the native CD39 (Figure 3B). No changes in NTPDase activity in COS-7 cells expressing the ectopic NT-Δ-1–37 CD39 mutant together with RanBPM were observed (Figure 3B, panel a, lane 4, and panel b, lane 4).
These results demonstrate that RanBPM can modulate CD39 enzymatic function and this requires specific interactions mediated by the N-terminal moiety of CD39.
The SPRY domain of RanBPM, in isolation, is insufficient for full functional interactions with CD39
RanBPM is known to contain an SPRY domain that is located at the N-terminus [19]. Several lines of evidence suggest that the SPRY domain is involved in the mediation of protein–protein interactions (including RanBPM complex formation with c-Met) [17,22]. Hence, this domain might also be involved in the interaction between RanBPM and CD39. To test this hypothesis, we performed two assays.
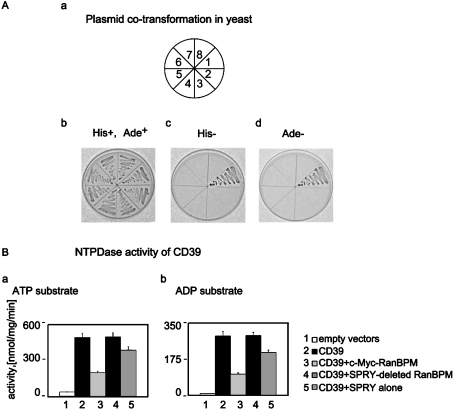
Plasmid constructs for ectopic cellular expression of mutant SPRY-truncated RanBPM protein, as well as the SPRY domain alone, were used in yeast two-hybrid assays (Figure 4A). Yeast PJ69-4A cells were co-transformed with bait expression vector (pGBDU-C1-CD39N1-14), with the SPRY expression plasmid (pGADT7-RanBPM152-353), or the SPRY-deleted RanBPM (pGADT7-RanBPM351-729), as well as RanBPM mutants containing intact SPRY (pGADT7-RanBPM149-729). These experiments were done in several combinations (Figure 4A, panel a). Only the RanBPM variant with intact SPRY could support yeast growth on minimal complete uracil-, leucine-, histidine- and adenine-depleted SD plates (Figure 4A, panel b, lane 1, panel c, lane 1, and panel d, lane 1). No growth was observed in yeasts that received SPRY domain-deleted-RanBPM or the SPRY domain alone (Figure 4A, panel c, lanes 2 and 3, and panel d, lanes 2 and 3).
Figure 4. SPRY domains of RanBPM and interactions with the N-termini of CD39.
(A) Probing for SPRY–CD39 interactions by yeast two-hybrid assays. Yeast PJ69-4A cells were co-transformed with the CD39 N-terminal ‘bait’ (pGBDU-C1-CD39N1-14) and either the originally deleted clone lacking N-terminal amino acids 1–148 (pACT2-RanBPM149-729) (position 1), or the SPRY-truncated RanBPM expression plasmid (pGADT7-RanBPM351-729) (position 2) and the SPRY domain alone (pGADT7-RanBPM149-353) (position 3). All the remaining co-transformations served as controls. These were conducted with: the ‘bait-less’ empty vector pGBDU-C1 and the empty control AD vector pGADT7 (position 4), ‘bait’ and pGADT7 (position 5), empty pGBDU-C1 and pACT2-RanBPM149-729 (position 6), pGBDU-C1 and pGADT7-RanBPM351-729 (position 7), and finally pGBDU-C1 and pGADT7-RanBPM149-353 (position 8). His+ and Ade+ indicate uracil- and leucine-deficient plates. His− plates lack uracil, leucine and histidine. Ade− plates lack uracil, leucine and adenine. Plates with the co-transformed yeast cells were incubated at 30 °C for 3 days. (B) SPRY domains, in isolation, are not sufficient for full functional interaction between RanBPM and CD39. NTPDase activity assay with ATP and ADP as substrates (indicated at the top of the panels) in COS-7 cells with CD39 alone (bar 2); CD39 and intact RanBPM (bar 3); CD39 and SPRY-truncated RanBPM variant (bar 4); CD39 and RanBPM with isolated SPRY moiety (bar 5). As a control COS-7 cells transfected with empty vectors lacking the protein inserts were used (bar 1). Three independent experiments produced consistent results (variance=±7%).
In the second assay, we analysed the NTPDase activity of CD39 in COS-7 cells overexpressing the SPRY moiety alone or SPRY-deleted mutant of RanBPM (Figure 4B). The specific enzymatic activity of CD39 was not altered by co-expression of SPRY-deleted RanBPM (Figure 4B, panel a, lane 4, and panel b, lane 4). Cells expressing CD39 together with isolated SPRY moiety, did show a slight decrease in NTPDase activity when compared with cells expressing truncated RanBPM (Figure 4B, panel a, lane 5, and panel b, lane 5).
Our results suggest that the SPRY domain in isolation is insufficient for RanBPM–CD39 binding and complete inhibition of CD39 NTPDase activity. SPRY domains may play a role in modulating this interaction, but the association requires further components of RanBPM to exert full inhibitory effects on NTPDase activity.
DISCUSSION
Global deletion of cd39 in mice results in disordered purinergic signalling that perturbs vascular thromboregulation and impacts upon immune responses [4]. These findings are associated with the loss of the phosphohydrolytic properties of CD39 and consequent perturbation of P2-receptors. These mice also show abnormalities in growth factor-mediated cellular responses, e.g. seen in recovery from liver injury [23] and angiogenesis [6], that are difficult to fully explain on the basis of secondary abnormalities in P2-receptor signalling alone. Hence, CD39 may have functions distinct from ecto-enzymatic activity. Properties that are shared by CD39 and other ecto-nucleotidases include localization in membrane lipid rafts and the regulation of cellular signalling pathways [24]. Interestingly, CD38 interacts with CD31 and has functions independent of enzymatic activities [25,26].
In the present paper, we describe novel structural and functional intracellular interactions between CD39, a prototypic member of the NTPDase family, and RanBPM, a membrane scaffolding protein originally characterized as a binding protein for Ran, a small GTPase. We performed co-immunoprecipitation and co-localization experiments that validated structural interactions between CD39 and RanBPM, as initially noted in the yeast two-hybrid system.
In previous work, Zhong et al. [27] have shown that the yeast ecto-apyrase Ynd1p interacts with the activator subunit Vmal3p of vacuolar H-ATPase. We did not observe any such links between CD39 and vacuolar ATPases. Indeed, our results clearly show that RanBPM interacts solely with the N-terminal intracytoplasmic moiety of CD39. RanBPM (RanBP9) is a Ran-binding protein with a molecular mass of 90 kDa. The protein was first thought to be located in the centrosome, the microtubule-organizing centre [15,16]. Recently, however, RanBPM has been shown to be associated with plasma membranes and to interact with a broad spectrum of transmembrane proteins and receptors [18].
We also demonstrate functional interactions between CD39 and RanBPM with respect to the modulation of the CD39 NTPDase ecto-enzymatic activity (Figure 3). This association could be important in the regulation of NTPDase enzymatic activity by RanBPM [17,28]. The exact mechanism whereby the NTPDase activity of CD39 is inhibited by co-expression with RanBPM remains to be determined. Wang et al. [9] have previously reported that the transmembrane segments of CD39 are involved in the association of monomers to form tetramers that are necessary for expression of full NTPDase activity. Dissociation of the tetramer to monomers by membrane disruption or mutagenesis of the transmembrane domains causes concomitant loss of NTPDase activity [9]. We, and others, have shown that CD39 mutant proteins lacking one or both transmembrane domains have substantially lower enzymatic activity than the intact enzyme, and have ascribed this to disruption of multimeric formation [7,9]. Here we show that RanBPM modulates NTPDase activity of CD39 by specific interactions with the N-terminus of CD39. The biochemical activity of NT-Δ-1–37 CD39 mutant is not inhibited by co-expressed RanBPM. We propose that the interaction of RanBPM with the N-terminus of CD39 might influence multimeric formation of the enzyme in the plasma membrane. This would be predicted to result in decreases of ecto-enzymatic activity of CD39.
RanBPM contains a conserved SPRY domain that has unknown functions [19]. The SPRY domain comprises a 140-amino-acid domain (amino acids 212–333 of RanBPM), a structural motif that was originally identified in a Dictyostelium discoideum dualspecificity kinase termed splA, and RyR [19]. The SPRY domain is found in one or three copies (as in splA and RyR) in eukaryotic nuclear and cytoplasmic proteins, as well as in transmembrane and secreted proteins. Suggested functions for some SPRY domain-containing proteins include RNA binding, cell growth and differentiation. The SPRY domain might also play a role in RNA binding or protein–protein interaction [19]. We investigated whether the RanBPM SPRY domain is involved in the interaction with CD39. Mutants of RanBPM were derived and yeast two-hybrid assays together with NTPDase activity assays were performed. Our results show that the SPRY domain of RanBPM in isolation is insufficient for binding to CD39 and concomitant full inhibition of CD39 NTPDase activity (Figure 4). SPRY domains do appear to play roles in modulating associations between defined proteins. Wang et al. [17] have also reported that RanBPM interacts with c-Met receptor through the SPRY domain and might thereby influence the Ras/ERK/SRE (where ERK is extracellular-signal-regulated kinase and SRE is serum response element) pathway. Whether CD39 has the potential to modulate this pathway via interactions with RanBPM to influence Ras activation remains to be determined.
RanBPM interacts with several signalling molecules [17,18,22,29] and might also play roles as a membrane scaffolding protein by associating with LFA-1 (lymphocyte function-associated antigen-1) integrin protein [18]. Integrins are a family of transmembrane cellular receptors that mediate cell–cell and cell–matrix interactions [30]. Both cellular adhesion to matrix proteins and mitogenic signalling by growth factors are required for cellular proliferation, suggesting that integrin and growth factor receptor signalling pathways might converge on down-stream components of shared signal transduction pathways. Currently, it remains unclear whether, and if so, how precisely, CD39 might influence such interactions by complexation with RanBPM. However, the results shown here suggest that CD39 might regulate signal transduction pathways in a dualistic manner. This could occur by CD39 modulating P2 receptor function following hydrolysis of extracellular nucleotides, and by further interacting with other signalling proteins, potentially using RanBPM as a bridge.
Additionally, the C-terminus of CD39 contains a peptide sequence (KDMV) that could make up a putative PDZ domain. These PDZ domains can assemble with other protein interaction domains [for instance, SH3 domain (Src homology domain), PTB domain (phosphotyrosine-binding domain) and WW domain (protein–protein interaction domain containing two conserved tryptophan residues)] to facilitate signalling responses or determine the localization of receptors [31–34]. It is possible that the C-terminal domain of CD39 may further dictate interactions with other proteins, including P2 receptors. The P2Y1 and P2Y2 receptors have C-terminal sequences ending with DTSL and DIRL respectively that could serve as suitable PDZ-ligands [35].
In conclusion, our studies confirm that CD39 might be a bifunctional molecule with ecto-nucleotidase activity expressed by the large ectodomain, and the ability to bind to RanBPM expressed by the N-terminal intracytoplasmic domain. As RanBPM may in turn modulate the ecto-enzymatic functions of CD39, the functional sequelae of this intermolecular interaction may have important implications for the regulation of extracellular nucleotide signalling pathways [24].
Acknowledgments
We thank X. Zhong and G. Guidotti (Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA, U.S.A.) for providing the yeast strain PJ69-4A and pGBDU-C1 vector, M. Wink (Liver Center, Department of Medicine, Beth Israel Deaconess Medical Center, Harvard Medical School) for her helpful advice concerning the NTPDase activity assay, and E. Csizmadia and S. Vlajkovik (Liver Center, Department of Medicine, Beth Israel Deaconess Medical Center, Harvard Medical School) for the isolation and staining of peripheral blood mononuclear cells. This work was supported by HL57307 (S.C.R.), HL66167 (E.K.), HL63972 (S.C.R.) and HL076540 (project to S.C.R.) from the National Institutes of Health.
References
- 1.Maliszewski C. R., Delespesse G. J., Schoenborn M. A., Armitage R. J., Fanslow W. C., Nakajima T., Baker E., Sutherland G. R., Poindexter K., Birks C. The CD39 lymphoid cell activation antigen. Molecular cloning and structural characterization. J. Immunol. 1994;153:3574–3583. [PubMed] [Google Scholar]
- 2.Kaczmarek E., Koziak K., Sevigny J., Siegel J. B., Anrather J., Beaudoin A. R., Bach F. H., Robson S. C. Identification and characterization of CD39/vascular ATP diphosphohydrolase. J. Biol. Chem. 1996;271:33116–33122. doi: 10.1074/jbc.271.51.33116. [DOI] [PubMed] [Google Scholar]
- 3.Wang T. F., Guidotti G. CD39 is an ecto-(Ca2+,Mg2+)-apyrase. J. Biol. Chem. 1996;271:9898–9901. [PubMed] [Google Scholar]
- 4.Enjyoji K., Sevigny J., Lin Y., Frenette P. S., Christie P. D., Esch J. S., II, Imai M., Edelberg J. M., Rayburn H., Lech M., et al. Targeted disruption of cd39/ATP diphosphohydrolase results in disordered hemostasis and thromboregulation. Nat. Med. 1999;5:1010–1017. doi: 10.1038/12447. [DOI] [PubMed] [Google Scholar]
- 5.Goepfert C., Imai M., Brouard S., Csizmadia E., Kaczmarek E., Robson S. C. CD39 modulates endothelial cell activation and apoptosis. Mol. Med. 2000;6:591–603. [PMC free article] [PubMed] [Google Scholar]
- 6.Goepfert C., Sundberg C., Sevigny J., Enjyoji K., Hoshi T., Csizmadia E., Robson S. Disordered cellular migration and angiogenesis in cd39-null mice. Circulation. 2001;104:3109–3115. doi: 10.1161/hc5001.100663. [DOI] [PubMed] [Google Scholar]
- 7.Schulte am Esch J., II, Sevigny J., Kaczmarek E., Siegel J. B., Imai M., Koziak K., Beaudoin A. R., Robson S. C. Structural elements and limited proteolysis of CD39 influence ATP diphosphohydrolase activity. Biochemistry. 1999;38:2248–2258. doi: 10.1021/bi982426k. [DOI] [PubMed] [Google Scholar]
- 8.Handa M., Guidotti G. Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum) Biochem. Biophys. Res. Commun. 1996;218:916–923. doi: 10.1006/bbrc.1996.0162. [DOI] [PubMed] [Google Scholar]
- 9.Wang T. F., Ou Y., Guidotti G. The transmembrane domains of ectoapyrase (CD39) affect its enzymatic activity and quaternary structure. J. Biol. Chem. 1998;273:24814–24821. doi: 10.1074/jbc.273.38.24814. [DOI] [PubMed] [Google Scholar]
- 10.Koziak K., Kaczmarek E., Kittel A., Sevigny J., Blusztajn J. K., Schulte Am Esch J., II, Imai M., Guckelberger O., Goepfert C., Qawi I., et al. Palmitoylation targets CD39/endothelial ATP diphosphohydrolase to caveolae. J. Biol. Chem. 2000;275:2057–2062. doi: 10.1074/jbc.275.3.2057. [DOI] [PubMed] [Google Scholar]
- 11.Robson S. C., Daoud S., Begin M., Cote Y. P., Siegel J. B., Bach F. H., Beaudoin A. R. Modulation of vascular ATP diphosphohydrolase by fatty acids. Blood Coagul. Fibrinolysis. 1997;8:21–27. doi: 10.1097/00001721-199701000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Kittel A., Kaczmarek E., Sevigny J., Lengyel K., Csizmadia E., Robson S. C. CD39 as a caveolar-associated ecto-nucleotidase. Biochem. Biophys. Res. Commun. 1999;262:596–599. doi: 10.1006/bbrc.1999.1254. [DOI] [PubMed] [Google Scholar]
- 13.Shaul P. W., Anderson R. G. Role of plasmalemmal caveolae in signal transduction. Am. J. Physiol. 1998;275:L843–L851. doi: 10.1152/ajplung.1998.275.5.L843. [DOI] [PubMed] [Google Scholar]
- 14.Kittel A., Csapó Z., Jackson S., Csizmadia E., Robson S. C. Expression of nucleoside triphosphate diphosphohydrolase-1/CD39 and type-2 purinergic (P2) receptors in human placenta: co-localization of P2Y1 and NTPDase1/CD39 in caveolae. Eur. J. Histochem. 2004;48:253–260. [PubMed] [Google Scholar]
- 15.Nakamura M., Masuda H., Horii J., Kuma K., Yokoyama N., Ohba T., Nishitani H., Miyata T., Tanaka M., Nishimoto T. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to γ-tubulin. J. Cell Biol. 1998;143:1041–1052. doi: 10.1083/jcb.143.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishitani H., Hirose E., Uchimura Y., Nakamura M., Umeda M., Nishii K., Mori N., Nishimoto T. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene. 2001;272:25–33. doi: 10.1016/s0378-1119(01)00553-4. [DOI] [PubMed] [Google Scholar]
- 17.Wang D., Li Z., Messing E. M., Wu G. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J. Biol. Chem. 2002;277:36216–36222. doi: 10.1074/jbc.M205111200. [DOI] [PubMed] [Google Scholar]
- 18.Denti S., Sirri A., Cheli A., Rogge L., Innamorati G., Putignano S., Fabbri M., Pardi R., Bianchi E. RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. J. Biol. Chem. 2004;279:13027–13034. doi: 10.1074/jbc.M313515200. [DOI] [PubMed] [Google Scholar]
- 19.Ponting C., Schultz J., Bork P. SPRY domains in ryanodine receptors (Ca(2+)-release channels) Trends Biochem. Sci. 1997;22:193–194. doi: 10.1016/s0968-0004(97)01049-9. [DOI] [PubMed] [Google Scholar]
- 20.James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baykov A. A., Evtushenko O. A., Avaeva S. M. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 22.Emberley E. D., Gietz R. D., Campbell J. D., HayGlass K. T., Murphy L. C., Watson P. H. RanBPM interacts with psoriasin in vitro and their expression correlates with specific clinical features in vivo in breast cancer. BMC Cancer. 2002;2:1–7. doi: 10.1186/1471-2407-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y., Sun X., Enjyoji K., Robson S. C. CD39 interacts with RanBPM to directly modulate Ras activation and cellular proliferation in liver regeneration following partial hepatectomy. Hepatology. 2004;40:222A. [Google Scholar]
- 24.Robson S. C., Wu Y., Sun X., Knosalla C., Dwyer K., Enjyoji K. Ecto-nucleotidases of CD39 family modulate vascular inflammation and thrombosis in transplantation. Semin. Thromb. Hemost. 2005;31:217–233. doi: 10.1055/s-2005-869527. [DOI] [PubMed] [Google Scholar]
- 25.Deaglio S., Morra M., Mallone R., Ausiello C. M., Prager E., Garbarino G., Dianzani U., Stockinger H., Malavasi F. Human CD38 (ADP-ribosyl cyclase) is a counter-receptor of CD31, an Ig superfamily member. J. Immunol. 1998;160:395–402. [PubMed] [Google Scholar]
- 26.Deaglio S., Mehta K., Malavasi F. Human CD38: a (r)evolutionary story of enzymes and receptors. Leuk. Res. 2001;25:1–12. doi: 10.1016/s0145-2126(00)00093-x. [DOI] [PubMed] [Google Scholar]
- 27.Zhong X., Malhotra M., Guidotti G. Regulation of yeast ectoapyrase Ynd1p activity by activator subunit Vma13p of vacuolar H+-ATPase. J. Biol. Chem. 2000;275:35592–35599. doi: 10.1074/jbc.M006932200. [DOI] [PubMed] [Google Scholar]
- 28.Robson S. C., Enjyoji K., Goepfert C., Imai M., Kaczmarek E., Lin Y., Sevigny J., Warny M. Modulation of extracellular nucleotide-mediated signaling by CD39/nucleotide triphosphate diphosphohydrolase-1. Drug Dev. Res. 2001;53:193–207. [Google Scholar]
- 29.Mikolajczyk M., Shi J., Vaillancourt R. R., Sachs N. A., Nelson M. The cyclin-dependent kinase 11(p46) isoform interacts with RanBPM. Biochem. Biophys. Res. Commun. 2003;310:14–18. doi: 10.1016/j.bbrc.2003.08.116. [DOI] [PubMed] [Google Scholar]
- 30.Hughes P. E., Renshaw M. W., Pfaff M., Forsyth J., Keivens V. M., Schwartz M. A., Ginsberg M. H. Suppression of integrin activation: a novel function of a Ras/Raf-initiated MAP kinase pathway. Cell (Cambridge, Mass.) 1997;88:521–530. doi: 10.1016/s0092-8674(00)81892-9. [DOI] [PubMed] [Google Scholar]
- 31.Saras J., Heldin C. H. PDZ domains bind carboxy-terminal sequences of target proteins. Trends Biochem. Sci. 1996;21:455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- 32.Hung A. Y., Sheng M. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 2002;277:5699–5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- 33.Fan J. S., Zhang M. Signaling complex organization by PDZ domain proteins. Neurosignals. 2002;11:315–321. doi: 10.1159/000068256. [DOI] [PubMed] [Google Scholar]
- 34.Nourry C., Grant S. G., Borg J. P. PDZ domain proteins: plug and play! Science STKE 2003. 2003. p. RE7. [DOI] [PubMed]
- 35.Hall R. A., Ostedgaard L. S., Premont R. T., Blitzer J. T., Rahman N., Welsh M. J., Lefkowitz R. J. A C-terminal motif found in the β2-adrenergic receptor, P2Y1 receptor and cystic fibrosis transmembrane conductance regulator determines binding to the Na+/H+ exchanger regulatory factor family of PDZ proteins. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8496–8501. doi: 10.1073/pnas.95.15.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]