Abstract
Cyclophilins constitute a family of proteins involved in many essential cellular functions. They have also been identified as a panallergen family able to elicit IgE-mediated hypersensitivity reactions. Moreover, it has been shown that human cyclophilins are recognized by serum IgE from patients sensitized to environmental cyclophilins. IgE-mediated autoreactivity to self-antigens that have similarity to environmental allergens is often observed in atopic disorders. Therefore comparison of the crystal structure of human proteins with similarity to allergens should allow the identification of structural similarities to rationally explain autoreactivity. A new cyclophilin from Aspergillus fumigatus (Asp f 27) has been cloned, expressed and showed to exhibit cross-reactivity in vitro and in vivo. The three-dimensional structure of cyclophilin from the yeast Malassezia sympodialis (Mala s 6) has been determined at 1.5 Å (1 Å=0.1 nm) by X-ray diffraction. Crystals belong to space group P41212 with unit cell dimensions of a=b=71.99 Å and c=106.18 Å. The structure was solved by molecular replacement using the structure of human cyclophilin A as the search model. The refined structure includes all 162 amino acids of Mala s 6, an active-site-bound Ala-Pro dipeptide and 173 water molecules, with a crystallographic R- and free R-factor of 14.3% and 14.9% respectively. The overall structure consists of an eight-stranded antiparallel β-barrel and two α-helices covering the top and bottom of the barrel, typical for cyclophilins. We identified conserved solvent-exposed residues in the fungal and human structures that are potentially involved in the IgE-mediated cross-reactivity.
Keywords: allergy, Aspergillus fumigatus, autoreactivity, Malassezia sympodialis, recombinant allergen, self antigen
Abbreviations: 3D, three-dimensional; ABPA, allergic bronchopulmonary aspergillosis; AE, atopic eczema; CsA, cyclosporin A; CyP, cyclophilin; DTT, dithiothreitol; EU, ELISA units; IDT, intradermal skin test; MnSOD, manganese-dependent superoxide dismutase; Ni-NTA, Ni2+-nitrilotriacetate; rmsd, root mean square deviation; SPT, skin prick test
INTRODUCTION
CyPs (cyclophilins) constitute a family of cytosolic proteins which play a pivotal role in protein folding through enzymatic catalysis of the peptidyl-prolyl cis–trans isomerization reaction [1]. The primary structure of this important enzyme is highly conserved among phylogenetically distant species, indicating involvement of CyP in basic cellular functions [2]. Belonging to the family of immunophilins, CyP binds the immunosuppressive drug CsA (cyclosporin A) [1]. The complex of CyP with CsA binds and inhibits the protein phosphatase calcineurin [3], thus suppressing the signal transduction in T-cells [4]. Therefore CsA is one of the most important immunosuppressant drugs used for prevention of graft rejection after transplant surgery [5]. CyPs from Aspergillus fumigatus {Asp f 11 [6] and Asp f 27 (the present study)}, the aetiological agent identified in the majority of Aspergillus-related human diseases, and from Malassezia sympodialis (Mala s 6 [7]), a skin-colonizing yeast involved in the pathophysiology of AE (atopic eczema) [formerly known as AD (atopic dermatitis)] [8], have been isolated as IgE-binding proteins from phage surface-displayed cDNA libraries. Later, CyP was isolated as an allergen also from birch (Betula verrucosa) pollen [9] and carrots (Daucus carota) [10], demonstrating that this molecular structure can elicit IgE-mediated responses in different types of allergic disorders. Inhalation of environmental allergens from pollens, dust mites, animal dander and fungi is the most common cause of IgE-mediated antibody responses in humans. The symptoms related to allergen exposure range from rhinoconjunctivitis to AE, asthma and fatal anaphylaxis, and the incidence of allergic disease has increased dramatically in recent years [11]. Although the mechanisms leading to allergy are quite well understood [12], our knowledge of the repertoire of molecular structures that are able to induce a switch to IgE production is still incomplete. A common hallmark of allergenic structures is their ability to induce cross-linking of FcεRI (high-affinity IgE receptor)-bound IgE on effector cells of sensitized individuals, thus causing immediate release of anaphylactogenic mediators [12]. Although the 3D (three-dimensional) structures of some allergens are known [13], the question of whether a protein exhibits special structural characteristics that are responsible for its allergenicity is poorly understood. However, elucidation of the 3D structuress of allergens has contributed greatly to our understanding of cross-reactivity among similar structures derived from phylogenetically distant allergenic sources [14]. Among these, MnSOD (manganese-dependent superoxide dismutase) of A. fumigatus [15] has been shown to cross-react with a wide variety of MnSODs [16], including human MnSOD [17]. Cross-reactivity between allergens and closely related similar human antigens is often observed in allergic individuals suffering from chronic atopic diseases [8,17,18], and the availability of the 3D structures of a given allergen and its comparable human antigen allows a detailed study of the residues involved in cross-reactivity [15]. These studies provide strong evidence for in vitro and in vivo humoral and cell-mediated IgE autoreactivity in patients suffering from long-lasting atopic diseases [17,18] potentially contributing to exacerbation and/or perpetuation of chronic allergic reactions [8,19]. Recently, up-regulation of human MnSOD in eczematous skin of patients suffering from AE has been shown, providing a direct link between IgE-sensitization, cross-reactive structures and pathogenesis of the disease [8]. In the presents study, we cloned, produced and characterized Asp f 27, a new CyP from A. fumigatus, and the 3D structure of CyP from M. sympodialis (Mala s 6), a lipophilic yeast involved in the pathogenesis of AE [7,8], compared the structures with the solved structure of human CyP B [20], and defined the solvent-accessible amino acid residues shared by the two crystal structures. These amino acid residues potentially involved in IgE-mediated cross-reactivity among CyPs offer an explanation for the IgE-mediated autoreactivity found in clinically distinct chronic atopic diseases.
MATERIALS AND METHODS
Cloning and production of CyPs
Asp f 11, a CyP of A. fumigatus, and human CyP B were cloned and produced as described [6]. A new CyP of A. fumigatus formally termed Asp f 27 was detected during a high-throughput screening program of cDNA libraries displayed on phage surface [21]. The gene encoding Asp f 27 was amplified by PCR using the primers 5′ BamHI, 5′-CGGCGGATCCATGGTTGTCAAGACTTTCTTC-3′, and 3′ HindIII, 5′-GCCCAAGCTTACAGCTGACCACAGTCGG-3′, subcloned as a BamHI/HindIII fragment into pQE 30 (Qiagen) and transformed into Escherichia coli M15 cells followed by DNA sequence verification. To produce recombinant proteins, E. coli M15 cells were grown at 37 °C in LB (Luria–Bertani) medium to a D600 of 0.6, induced with 1 mM IPTG (isopropyl β-D-thiogalactoside), incubated at 30 °C for 16 h, harvested by centrifugation at 6000 g for 10 min at 4 °C and stored at −20 °C. The cell pellet was resuspended in lysis buffer (50 mM sodium phosphate, 300 mM NaCl and 10 mM imidazole, pH 8.0) and lysed using a French press. Insoluble material was removed by centrifugation at 20000 g for 20 min at 4 °C.
His6-tagged recombinant proteins were purified by Ni-NTA (Ni2+-nitrilotriacetate)-affinity chromatography using a 5 ml HiTrap chelating HP column (Amersham Biosciences). Proteins were eluted in a linear buffer gradient (10–250 mM imidazole, 50 mM sodium phosphate and 300 mM NaCl, pH 8.0) and dialysed against distilled water. Molecular size, purity and enzymatic activity of the recombinant CyP proteins were assessed as described in [6].
For crystallization of Mala s 6, a thrombin-cleavable His6-tagged protein was designed. The original Mala s 6 cDNA [7] was amplified from the original clone by PCR using Pfu Turbo DNA polymerase (2.5 units/μl) (Stratagene) with the primers 5′ BamHI, 5′-CGGCGGATCCATGTCTAACGTTTTCTTCG-3′, and 3′ HindIII, 5′-GCCCAAGCTTTTAGCACACACCGCACTTG-3′. The PCR product was digested with BamHI and HindIII restriction endonucleases (NEB), cleaned with QIAquick PCR purification kit (Qiagen) and ligated into a modified pQE32 vector (Qiagen) containing an N-terminal His6 tag followed by a thrombin cleavage site and a BamHI restriction site (HHHHHHLVPRGS). The ligation mixture was transformed into E. coli strain M15, and the sequence of picked clones containing inserts of the correct size was verified by DNA sequencing. A correct clone was used to produce and purify His6-tagged protein processed as described for Asp f 11 [6]. The N-terminal His6 tag of Mala s 6 was cleaved off using thrombin (20 units/mg of protein) in 300 mM NaCl, 50 mM Tris/HCl, 2 mM CaCl2, pH 7.5, by incubation for 30 h at 22 °C. The cleaved protein was purified further by gelfiltration on a Superdex 75 column (FPLC; Amersham Biosciences) equilibrated with 100 mM NaCl and 50 mM Tris/HCl, pH 7.0. The eluted protein was diluted 1:10 with water and 2 mM DTT (dithiothreitol) and concentrated to 10 mg/ml. Correct cleavage and purity were assessed by SDS/PAGE.
Subjects, routine assessments and skin tests
Sera from 40 patients suffering from AE sensitized to M. sympodialis and from 40 patients sensitized to A. fumigatus selected according to clinical history and immediate SPT (skin prick test) reactivity to the respective fungal extract were analysed together with sera from 15 healthy controls. Allergen-specific IgE to extracts was quantitatively determined by ImmunoCAP® m3 (A. fumigatus) and m70 (M. sympodialis) using the CAP system (Pharmacia Diagnostics) according to the package inserts. SPTs were performed with A. fumigatus extract purchased from ALK as commercially available SPT solution and with an in-house M. sympodialis extract prepared as described in [8]. Tests were considered positive if a weal surface area >9 mm2 surrounded by an erythema was induced in the absence of reaction to the negative 0.9% saline control [22]. IDTs (intradermal skin tests) with human recombinant CyP A, CyP B and Asp f 11 were performed as described in [17,18]. The protocol was carried out according to a clinical protocol approved by the ethical committee and all participants gave written informed consent after a full explanation of the procedure given individually before testing.
Immunoassays for IgE antibody binding to CyPs
IgE binding to recombinant CyPs was determined by a standard direct solid-phase ELISA in polystyrene microtitre plates (Maxisorp™; Nunc) coated and processed as described in [6,22]. Results were expressed as EU (ELISA units)/ml calibrated against the absorbency of an in-house reference standard defined arbitrarily as 100 EU/ml for each allergen tested [22]. Inhibition ELISA was performed using 1:10 diluted patients' sera and BSA as a negative control as described in [6]. Percentage inhibition was calculated from the absorbency of the serial dilutions of serum containing BSA as negative control in fluid phase set as 0% inhibition.
IgE immunoblots
Recombinant CyPs were separated by SDS/PAGE on 12% gels (Novex) under denaturing reducing conditions, transferred to Hybond™ ECL® (enhanced chemiluminescence) nitrocellulose membranes (Amersham Biosciences) and processed as described in [6]. Membranes were incubated with sera of patients sensitized to either A. fumigatus or M. sympodialis and positive in IgE ELISA with Asp f 11 or Mala s 6 coated to the solid phase. After incubation with mouse anti-human IgE monoclonal antibody TN-142, specific IgE-binding was detected with horseradish-peroxidase-conjugated goat anti-mouse IgG (Dako) and bands were visualized with SuperSignal® West Pico Chemiluminescent Substrate (Pierce) on Hyperfilm™ ECL® (Amersham Biosciences) as described in [6].
Crystallization and data collection for Mala s 6
Crystallization was performed using the hanging drop vapour diffusion method at 16 °C. The protein solution (6 mg/ml, 80 mM Ala-Pro and 2 mM DTT) was mixed in a 2:1 ratio with reservoir solution [1.5 M ammonium sulphate, 12% (v/v) glycerin and 0.1 M Tris/HCl, pH 8.5]. The drop was equilibrated against 500 μl of reservoir solution. After several months, a crystal grew to a size of 300 μm×300 μm×250 μm, and the drop solution had turned cryoprotectant. Therefore the crystal was directly flash-cooled in a stream of gaseous nitrogen at 100 K. A dataset was collected to 1.5 Å (1 Å=0.1 nm) resolution on the synchrotron beamline X06SA at Swiss Light Source (Villigen, Switzerland) at 100 K. Data were processed and scaled using DENZO and SCALEPACK of the HKL program package [23]. The crystal belongs to the tetragonal space group P41212, with cell parameters a=b=71.99 Å and c=106.18 Å (Table 1).
Table 1. Data collection and refinement statistics.
The data were collected at 100 K. Numbers in parentheses are for the highest resolution shell. Rfree was calculated using a test set of 5%.
| (a) Data collection | |
|---|---|
| Parameter | Value |
| X-ray source | X06SA |
| Wavelength (Å) | 1.0001 |
| Unit cell axes a=b, c (Å) | 71.99, 106.18 |
| Resolution (Å) | 42.7–1.5 (1.55–1.50) |
| Unique reflections | 45175 |
| Redundancy | 14.2 |
| Completeness (%) | 99.4 (100.0) |
| Rsym (%) | 6.1 (30.7) |
| Average I/σ | 26.9 (8.5) |
| Space group | P41212 |
| Protomers per ASU | 1 |
| (b) Refinement statistics | |
| Parameter | Value |
| Resolution (Å) | 36.7–1.5 |
| Number of reflections (working/test) | 42825/2272 |
| Number of atoms | 1409 |
| Rcryst (%) | 14.3 |
| Rfree (%) | 14.9 |
| Mean B factor (Å2) | |
| All atoms | 22.7 |
| Main chain atoms | 19.6 |
| Side chain atoms | 21.9 |
| Dipeptide atoms | 23.2 |
| Solvent atoms | 37.0 |
| Rmsd bond lengths (Å) | 0.015 |
| Rmsd bond angles (°) | 1.63 |
Structure determination and refinement
The structure was solved by molecular replacement using MOLREP [24]. A polyalanine model of human CyP A (PDB code 2CPL) served as a search model. One molecule was located in the asymmetric unit, which yields a Matthews coefficient (Vm) of 4.0 Å3/Da and a corresponding solvent content of 69.0%. Rigid body refinement using REFMAC [25] as implemented in the CCP4 program suite [26] resulted in R and Rfree values of 46.2 and 45.5% respectively. Further refinement was performed with REFMAC, manual rebuilding and correction with XtalView [27]. A total of 173 water molecules were introduced using ARP [28]. Final rounds of refinement were carried out with individual anisotropic B factors. The side chain of Gln87 was modelled as double conformation with occupancies of 0.5 each. Statistics from data collection and refinement are provided in Table 1. The stereochemical quality of the final model was assessed using PROCHECK [29] and WHATCHECK [30]. Anisotropic validation was performed with PARVATI [31]. Secondary-structure elements were assigned automatically with DSSP [32].
Calculation of the solvent-accessible area
Solvent-accessible surface areas were calculated with the program NACCESS (http://wolf.bms.umist.ac.uk/naccess), an implementation of the Lee and Richards solvent accessibility algorithm [33], using a probe radius of 1.4 Å and a slice width of 0.01 Å. Ligands and water molecules were omitted from the calculation. The relative residue accessibility is the ratio of the accessible area of a residue in the model to the accessible area of that residue in an extended Ala-Xaa-Ala tripeptide.
RESULTS
Molecular cloning of Asp f 27 and production of recombinant CyPs
Screening of a phage surface-displayed cDNA library of A. fumigatus yielded a vast variety of clones carrying inserts encoding putative IgE-binding proteins [21]. Among these, 173 clones encoded Asp f 11, an already known allergen of the mould identified as a member of the CyP family [6], and clone E6n01 coded for a variant of CyP sharing 61% sequence identity with Asp f 11 at the primary structure level. The cDNA contained a 492 bp open reading frame, coding for a 163-amino-acid protein with the conserved active-site residues of CyP (Figure 1) and a calculated molecular mass of 17.74 kDa. The new allergen cannot be considered as an isoform of Asp f 11 according to the recommendations of the allergen nomenclature committee (http://www.allergen.org) and was therefore termed Asp f 27. PCR-amplified DNA encoding the full-length Asp f 27 was ligated into pQE 30, expressed as an N-terminally His6-tagged protein in E. coli, and purified by Ni-NTA-affinity chromatography. The purified protein migrated as a single band of the expected size on SDS/PAGE (12% gradient gels), as was the case for Mala s 6, Asp f 11 and human CyP B produced and purified in the same way (Figure 2A). All recombinant CyPs were able to catalyse the cis–trans isomerizaion of N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (results not shown), and enzymatic activity was considered to be proof for native-like folding [6].
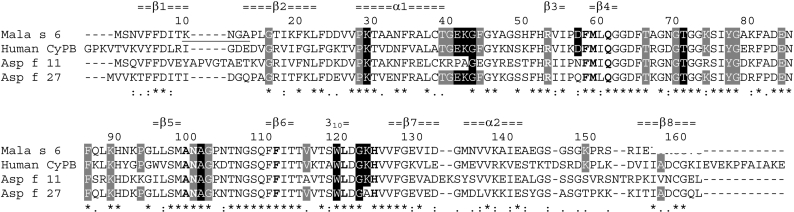
Figure 1. Structural alignment of Mala s 6 and human CyP B combined with a sequence alignment of Asp f 11 and Asp f 27.
The sequence of human CyP B corresponds to the mature form, but is missing six N-terminal amino acids. The top line shows the secondary structure assigned by DSSP [32], and the second line shows the sequence numbering of Mala s 6. Identical (*), strongly similar (:) and weakly similar (.) amino acids in all sequences are indicated. Active-site residues are in bold. The two loops, which adopt a different conformation in Mala s 6 compared with human CyP B, are underlined. Identical residues that are at least 50% or between 30 and 50% solvent-exposed in Mala s 6 and human CyP B are shown on grey and black backgrounds respectively. Residues which are also conserved in Asp f 11 or Asp f 27 are coloured correspondingly.
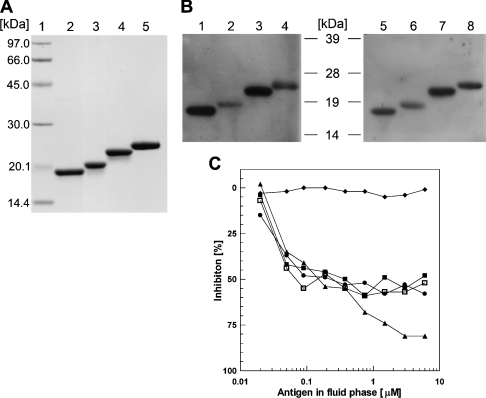
Figure 2. Protein purification, Western blot analysis and inhibition ELISA.
(A) Recombinant proteins (5 μg) were separated by SDS/PAGE and were stained with Coomassie Blue. Molecular-mass standards (in kDa) are indicated in lane 1. Lane 2, Mala s 6; lane 3, Asp f 27; lane 4, Asp f 11; lane 5, human CyP B. (B) Specific IgE binding of recombinant CyPs analysed by Western blotting using serum pools from individuals sensitized to M. sympodialis (left) and A. fumigatus (right). Lanes 1 and 5, Mala s 6; lanes 2 and 6, Asp f 27; lanes 3 and 7, Asp f 11; lanes 4 and 8, human CyP B. Molecular-mass sizes are indicated in kDa. (C) Competitive inhibition of IgE binding to recombinant human CyP B coated on a solid phase. Pooled serum from A. fumigatus-sensitized patients was pre-incubated with increasing amounts of recombinant human CyP B (▲), Mala s 6 (●), Asp f 27 (□), Asp f 11 (■) or BSA as negative control (◆). Pre-incubated serum samples were transferred to wells coated with human CyP B, and IgE bound was analysed by ELISA.
Demonstration of IgE antibody responses to recombinant CyPs
The IgE-binding capacity of the recombinant CyPs was investigated by Western blotting using serum pools from individuals sensitized to M. sympodialis and A. fumigatus (Figure 2B). These results show that the CyPs of different origin are cross-reactive, and the specificity of the antigen–antibody interaction was demonstrated by inhibition ELISAs (Figure 2C). The prevalence of IgE binding to the different CyPs was assessed using a standard solid-phase ELISA in groups of patients with AE sensitized to M. sympodialis or patients suffering from ABPA (allergic bronchopulmonary aspergillosis) and therefore sensitized to A. fumigatus. Both conditions represent chronic relapsing atopic diseases with a strong inflammatory background. Results were expressed as arbitrary EU/ml and were considered positive when determinations exceeded three times the mean EU/ml value of a control group comprising 15 healthy individuals regarded as background value for the ELISA determination of allergen-specific IgE.
The results reported in Figure 3 suggest a prevalence of approx. 50 and 75% of Asp f 11 and Asp f 27 among patients suffering from ABPA and AE respectively. The incidence of sensitization against Mala s 6 was approx. 30% among both patients suffering from AE and those suffering from ABPA. These results, together with the results of the cross-inhibition ELISAs reported in Figure 2(C), suggest an extended cross-reactivity between these similar molecular structures. In accordance with results reported previously [6], patients sensitized to either of the environmental allergens also show cross-reactivity both in Western blot and in ELISA analyses with the human self-antigen CyP B (Figure 2B), now traceable back to structural features shown by comparison of the 3D structure of Mala s 6 solved during this work and the known 3D structure of human CyP B (see below).
Figure 3. IgE binding to the allergens determined with an antigen-specific ELISA.
Antigen-specific ELISA is compared with the binding of a reference serum arbitrarily assigned to 100 EU/ml [22]. (A) Sensitization to Mala s 6 (●), Asp f 11 (■), Asp f 27 (□) and human CyP B (▲) in a group of 40 AE patients sensitized to M. sympodialis. (B) Sensitization to the same allergens in a group of 40 A. fumigatus-sensitized patients suffering from ABPA. The lower level of assay sensitivity (hatched area) is indicated and corresponds to three times the mean EU/ml values of a healthy control group of 15 individuals. Mean values are indicated by horizontal lines.
The biological activity of human CyP A and CyP B was assessed by quantitative IDT of four ABPA patients showing IgE antibodies against all CyPs, three healthy controls, and three ABPA patients with CyP-specific IgE antibodies below background (Table 2). These results show that positive IDTs were obtained using 0.1–100 ng/ml recombinant Asp f 11, human CyP A or CyP B, and that all patients showing skin test reactivity also had detectable CyP-specific IgE levels in serum. In contrast, non-allergic controls and ABPA patients without detectable serum IgE antibodies to CyP were negative in IDTs using up to 10 μg/ml, demonstrating highly specific hypersensitivity responses in CyP-sensitized individuals (Table 2).
Table 2. Induction of immediate skin reactions with recombinant CyPs.
Values are in mm2 calculated according to the formula [(D1+D2)/2]2 [22]. hCyP, human CyP.
| Weal surface area (mm2) | IgE (EU/ml)* | |||||||
|---|---|---|---|---|---|---|---|---|
| Individual | Histamine | NaCl | Asp f 11 | hCyP A | hCyP B | Asp f 11 | Mala s 6 | hCyp B |
| ABPA patients sensitized to CyP | ||||||||
| Patient 1 | 361 | 0 | 625 | 1215 | 1089 | 319 | 146 | 137 |
| Patient 2 | 324 | 0 | 400 | 169 | 225 | 216 | 76 | 129 |
| Patient 3 | 484 | 0 | 572 | 246 | 225 | 262 | 95 | 111 |
| Patient 4 | 245 | 0 | 256 | 151 | 121 | 156 | 111 | 98 |
| ABPA patients not sensitized to CyP | ||||||||
| Patient 5 | 484 | 0 | 0 | 0 | 0 | 19 | 23 | 28 |
| Patient 6 | 289 | 0 | 0 | 0 | 0 | 16 | 6 | 8 |
| Patient 7 | 529 | 0 | 0 | 0 | 0 | 18 | 26 | 9 |
| Healthy controls | ||||||||
| Patient 8 | 436 | 0 | 0 | 0 | 0 | 10 | 15 | 21 |
| Patient 9 | 387 | 0 | 0 | 0 | 0 | 14 | 12 | 16 |
| Patient 10 | 414 | 0 | 0 | 0 | 0 | 11 | 13 | 15 |
*Relative level of allergen-specific IgE; cut-off value 45 EU/ml (see Figure 3).
Overall structure of Mala s 6 CyP
The X-ray structure of Mala s 6 was solved by molecular replacement and was refined to a resolution of 1.5 Å, with R and Rfree values of 14.3 and 14.9% respectively. Of the non-glycine and non-proline residues, 87.0% have main-chain dihedral angels in the most favoured regions of the Ramachandran plot, with the remaining ones located in the additional allowed regions. Data collection and refinement statistics are shown in Table 1.
There is one monomer per asymmetric unit. This results in a Vm of 4.0 Å3/Da and a corresponding solvent content of 69.0%. For such a high solvent content, the excellent resolution is remarkable. The monomeric state was also observed in solution in gel-filtration experiments on a Superdex 75 column (results not shown).
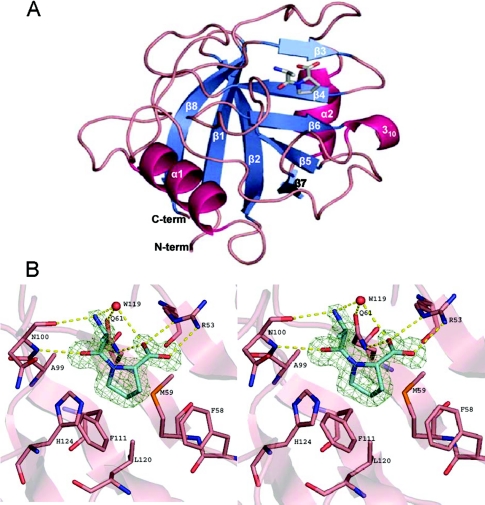
The final model consists of all the amino acids of Mala s 6 (amino acids 1–162), a dipeptide (Ala-Pro) bound to the active site, a glycerin molecule bound on the surface and 173 water molecules. As in all known CyP structures, the fold consists of an eight-stranded antiparallel β-barrel, and two α-helices covering the top and the bottom of the barrel, with an additional small 310-helix formed by Ser118, Trp119 and Leu120 (Figure 4A).
Figure 4. Overall structure and active site of Mala s 6.
(A) Cartoon representation of Mala s 6. The fold consists of an eight-stranded antiparallel β-barrel and two α-helices covering the top and the bottom of the barrel. There is an additional small 310-helix formed by Ser118 Trp119 and Leu120. The Ala-Pro ligand (shown as capped stick model) is situated in a hydrophobic pocket made up of β-sheets 3–6 and adjacent loops. (B) Stereo view of the active site with the bound dipeptide Ala-Pro. The cis form of the dipeptide is well defined in the electron density as shown by an Fo−Fc omit map contoured at 4.5 σ. The side chain of the proline residue makes hydrophobic interactions with the side chains of Phe58, Met59, Phe111, Leu120 and His124 of Mala s 6, while the N- and C-termini of Ala-Pro form hydrogen bonds with Arg53, Gln61, Asn100 and a water molecule (W119).
The two conserved cysteine residues (Cys38 and Cys159) in Mala s 6 could theoretically form a disulphide bond; however, in the structure, they are present in the reduced form. The sulphur atoms are well defined in the electron density, are separated by 5.22 Å and are solvent-inaccessible. Simple rotation of the sulphur atom about the Cα–Cβ cysteine side chain bonds towards each other would allow disulphide bond formation with a bond length of approx. 2 Å and χ torsion angles around 180°.
Active site of Mala s 6 CyP
The dipeptide Ala-Pro binds into the active site, which is responsible for the cis–trans isomerization reaction. It is well defined in the electron density (Figure 4B). The active site lies in a hydrophobic pocket, which is formed by residues of β-strands 3–6 and adjacent loops. The peptide bond of the ligand Ala-Pro is in the cis conformation, with an ω torsion angle of −0.2°. The binding of the dipeptide involves hydrogen bonds and Van der Waals interactions with the active-site residues Arg53, Phe58, Met59, Gln61, Ala99, Asn100, Phe111, Leu120 and His124 (Figure 4B). The proline side chain sits in the hydrophobic pocket, which is made up by the side chains of Phe58, Met59, Phe111, Leu120 and His124 of Mala s 6. The alanine residue of the dipeptide forms two hydrogen bonds with Asn100 and one with a water molecule: O of alanine with N of Asn100 (distance: 2.96 Å), N of alanine with O of Asn100 (2.85 Å), and N of alanine with water119 (2.82 Å). The C-terminal carboxy group of the proline residue forms four H-bonds: O of proline with NH2 of Arg53 (2.88 Å), O of proline with NE of Arg53 (2.80 Å), O of proline with NE2 of Gln61 (2.80 Å), and O of proline with water119 (3.08 Å). Water119 forms an additional H-bond with OE1 of Gln61 (2.81 Å).
The active-site residues are highly conserved among all CyPs. The binding mode of Ala-Pro to Mala s 6 is identical with that in the human CyP A–Ala-Pro complex (PDB code 1CYH). It involves the same residues and the same hydrogen bonding. For human CyP A, it has been questioned whether Ala-Pro acts as a competitive inhibitor of the cis–trans isomerization function or as a substrate which is subject to cis–trans isomerization [34].
Superposition of Mala s 6 CyP on human CyP B reveals putative IgE-binding residues
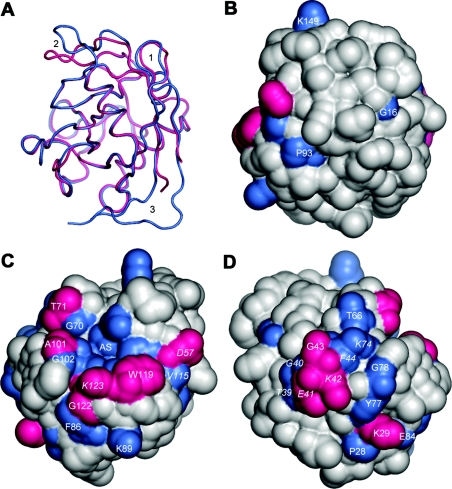
Mala s 6 shares 60% identity with human CyP B. The structure of the mature form, lacking the signal sequence and the subsequent six N-terminal amino acids, was solved by X-ray crystallography. On the structural level, Mala s 6 and human CyP B superpose well with an rmsd (root mean square deviation) of 1.05 Å for all Cα atoms (Figure 5A). There are structural differences in two loops and in the extensions of the N- and C-termini of human CyP B. The loop connecting the first two β-strands in Mala s 6 (Lys10–Ala13) and the ten-amino-acid loop connecting the second α-helix to the next β-strand in Mala s 6 (Glu143–Ser152) adopt a different conformation than the corresponding loops in human CyP B. There is very little sequence identity in these loops (Figure 1). The extension of the C-terminus (11 residues) of human CyP B prolongs the last β-strand, makes a turn and forms, together with the extension of the N-terminus (four residues), an additional β-sheet.
Figure 5. Superposition of Mala s 6 on human CyP B.
(A) Superposition of Mala s 6 (pink) on human CyP B (blue) reveals large structural similarities with minor deviations: the loop after the first β-strand (1) and the loop after the second α-helix (2) adopt different conformations. The N- and C-terminal extensions of human CyP B form an additional β-sheet (3). (B)–(D) Solvent-accessible surfaces of Mala s 6 showing putative IgE-binding amino acids. Identical residues that are at least 50% solvent-exposed in Mala s 6 and human CyP B are shown in pink. Identical residues that are at least 30% solvent-exposed and the conserved active-site residues are in blue. Residues conserved between Mala s 6 and human CyP B, but not in Asp f 11 and/or Asp f 27, are in italic. (B) The back side, the top and the bottom of the molecule are devoid of conserved surface patches, owing to lack of similarity, to conformational differences or to the additional β-sheet in human CyP B. The view and scale are the same as in (A). (C) This side of the molecule shows a conserved patch, made up by the active-site residues (AS) and flanking residues. The solvent-accessible surface area is quite large (1017 Å2) and might represent two or more overlapping B-cell epitopes. The view is as in (B), but is rotated by 100° about the y-axis. (D) The other side shows two conserved surface patches: a lower patch (Pro28-Lys29, Tyr77-Gly78, Glu84) and an upper patch (Thr39–Phe44, Thr66, Lys74). Both patches could account for cross-reactive IgE-binding epitopes in Mala s 6 and human CyP B. View is as in (B), but is rotated by 220° about the y-axis.
Since Mala s 6 and human CyP B show cross-reactivity, they must share common IgE-binding epitopes. Only those residues that are at least partly exposed to solvent can contribute to antigen–antibody interactions in native proteins. Thus solvent-accessible residues conserved in both proteins are potentially involved in the IgE-mediated cross-reactivity. A sequence alignment of Mala s 6 and human CyP B shows that a total of 88 of the 162 aligned residues are identical (Figure 1). Based on the solved structures, the solvent-accessible surface areas of Mala s 6 and human CyP B were calculated with the program NACCESS, using a probe radius of 1.4 Å. Of the 88 identical amino acids, 30 are at least 30% solvent-exposed, and, of those, ten are at least 50% solvent-exposed, in both structures and are likely to form cross-reactive IgE-binding regions. The residues which are identical and at least 30% or 50% solvent-exposed in Mala s 6 and human CyP B, as well as the conserved active-site residues, were mapped on the solvent-accessible surface of Mala s 6 (Figures 5B–5D). The Figures reveal contiguous patches conserved at sequence and structural level which might represent conformational cross-reactive IgE-binding epitopes. Interestingly, there are no conserved patches on the top, the bottom and the back of the molecule (Figure 5B). Lys149 on the top lies in the conformationally different loop after the second α-helix and thus cannot act as a cross-reactive IgE-binding residue, despite its conservation in the sequence of both molecules (Figure 1). Furthermore, this amino acid is not conserved in Asp f 11 and Asp f 27 and thus could not account for cross-reactivity. The residues at the back are poorly conserved, and the residues on the bottom are masked by the additional N- and C-terminal β-strand of human CyP B. Figure 5(C) shows a conserved patch which is made up by the active site and flanking residues (Arg53, Asp57–Met59, Gln61, Gly70-Thr71, Ala99–Gly102, Phe111, Val115, Trp119-Leu120, Gly122-Lys123). The rmsd values for the Cα and all heavy atoms of these amino acids resulting from the superposition of Mala s 6 and the human Cyp B are 0.23 and 0.71 Å respectively. The solvent-accessible surface area is quite large at 1017 Å2, and might thus represent two or more overlapping B-cell epitopes.
The other side of the molecule reveals two conserved patches which might be responsible for two different IgE-binding epitopes (Figure 5D). The lower patch consists of amino acids Pro28-Lys29, Tyr77-Gly78 and Glu84, covering a surface area of 386 Å2. The structural comparison for the Cα and all heavy atoms of the patch amino acids yielded rmsd values of 0.23 and 1.04 Å respectively. The upper patch is made up by the six-amino-acid loop (Thr39–Phe44) after the first α-helix and by residues Thr66 and Lys74. A high structural conservation of this patch is revealed by comparison of the amino acids involved, expressed by rmsd values of 0.21 and 0.82 Å for the C and all heavy atoms respectively. The patch covers a solvent-accessible surface area of 597 Å2, fulfilling the criteria for a putative antigen–antibody interaction quite well.
Mala s 6 is also cross-reactive with the two isoforms Asp f 11 and Asp f 27. The structures of these two proteins are not known, but they can be compared on the primary-sequence level with Mala s 6 and human CyP B (Figure 1). Eighteen identical residues that are at least 30% solvent-exposed are shared among all four proteins. Of these, six residues are at least 50% solvent-exposed. In the conserved patch around the active site (Figure 5C), all but three (Asp57, Val115, Lys123) amino acids are conserved among the four proteins. These three residues lie on the edge of the patch and therefore they probably do not have a large effect on IgE binding. The lower patch in Figure 5(D) is conserved completely in all of the proteins and could thus accounts for their overall cross-reactivity. Interestingly, the upper patch is not conserved in Asp f 11 and cannot be involved in the cross-reactivity of Asp f 11 with Mala s 6.
DISCUSSION
It has been demonstrated that sera from patients suffering from chronic allergic diseases with an inflammatory background contain IgE antibodies against a vast variety of self antigens [16–19]. The structures were derived from either screening of human cDNA expression libraries with patients' sera [19] or from direct cloning of human (phylogenetically highly conserved) proteins sharing sequence similarity with environmental allergens [6,17,18] and were postulated to be implicated in many clinical syndromes, including AE [8,19]. However, not many crystal structures of allergens in general, and of cross-reactive allergens in particular, are available which would allow the study of antibody-mediated cross-reactivity in detail [13].
To date, the atomic details of the interaction between antibody and antigen are known for more than 30 antibody–antigen complexes [13]. The B-cell epitope, which is buried upon antibody binding, is, in all cases, conformational. The epitope is made up of residues which lie on different surface loops forming discontinuous B-cell epitopes. They occupy a buried surface with an area in the range 560–860 Å2, and consist of ten to twenty amino acids that are in contact with antibody residues [35]. The whole surface of a protein is potentially antigenic. Therefore a typical globular 20 kDa allergen can accommodate at most five to ten antibodies at the same time [13]. The only method to determine the complete structure of a B-cell epitope is to co-crystallize the allergen with a monoclonal antibody Fab fragment and solve the X-ray structure of the complex. Because monoclonal human IgE is difficult to obtain, antibodies from other sources are currently used. The first structure of an allergen–Fab complex that was solved was the structure of a complex between the major birch pollen allergen Bet v 1 and the Fab fragment of the murine monoclonal IgG1 antibody BV16 [36].
We used an alternative approach to identify putative B-cell epitopes of CyPs that are involved in IgE-mediated cross-reactivity by determining shared features at the levels of primary and tertiary structure. Notably, cross-reactivity between human and fungal CyPs can be demonstrated in vitro (Figure 2) as well as in vivo (Table 2). The crystal structure of M. sympodialis CyP was determined at 1.50 Å resolution and compared with the structure of human CyP B, which had been determined at 1.85 Å resolution [20]. As expected from the primary structure, which shows 88 identical amino acids among 162 aligned (Figure 1), the comparison revealed a high similarity between the two structures (Figure 5A). Although the majority of the amino acids are identical, only 30 are >30% and only ten are >50% solvent-exposed in both structures and are therefore likely to be accessible for interactions with cross-reactive antibodies (Figures 5B–5D). Thus a large number of the conserved amino acids are located in the core of the protein and are not accessible for antigen–antibody interactions. The conserved surface-exposed residues are scattered over the whole sequence (Figure 1) and are thus likely to define discontinuous structures found in B-cell epitopes [35]. These amino acids are clustered over the surface, forming three patches covering solvent-accessible surface areas of 386, 597 and 1017 Å2 respectively (Figures 5C and 5D). Based on the extent of the solvent-accessible surface area, it can be postulated that these patches can accommodate three or more B-cell epitopes which involve 15–22 amino acid residues on different surface loops and occupy a buried surface in the range 540–890 Å2 [35].
Although the crystal structures of Asp f 11 and Asp f 27 are not yet solved, modelling experiments based on the human CyP B structure as template show comparable results with those obtained by comparison of the CyP B and Mala s 6 crystal structures (results not shown), offering a plausible explanation for the IgE-mediated cross-reactivity between CyPs demonstrated by Western blot analysis and inhibition ELISA (Figure 2) and in other studies [6]. However, the observed cross-reactivity between fungal and human CyPs, as deduced from ELISA inhibition experiments (Figure 2C), was always approx. 50% if a single patient's serum was also analysed (results not shown), which is not compatible with a complete cross-reactivity. This indicates that primary sensitization to a fungal CyP induces polyclonal responses eliciting additional IgE antibody responses raised to surface areas of the CyP not conserved between the similar molecules. Based on these results, the actual contribution of a conserved solvent-exposed residue to the binding of IgE and to the cross-reactivity among different CyPs can be investigated by site-directed mutagenesis, and this might lead to the production of an engineered molecule lacking IgE-binding capacity and thus be useful as a safe candidate vaccine [37].
However, the most exciting issue will be to use the sequence information and the recombinant proteins to investigate in more detail the role played by these cross-reactive structures in the pathogenesis of chronic inflammatory allergic diseases. The first promising results have been obtained with MnSOD, a pan-allergen involved in autoreactivity [17], which has opened the way for a more detailed understanding of the mechanisms involved in eliciting and maintaining clinically severe atopic disorders.
Acknowledgments
We are grateful to M. G. Grütter and P. Mittl for their support and the opportunity to collect test data on their home source, and to G. Menz for performing IDTs. Data collection was performed at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland, and we thank T. Tomizaki for his great technical support. This work was supported by the Swiss National Science Foundation grant no. 3100-063381/2 by the OPO foundation, Zürich, and by the Swedish Research Council grant nos. 74F-15193 and 74X-7924.
References
- 1.Takahashi N., Hayano T., Suzuki M. Peptidyl-prolyl cis–trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature (London) 1989;337:473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- 2.Trandinh C. C., Pao G. M., Saier M. H., Jr Structural and evolutionary relationships among the immunophilins: two ubiquitous families of peptidyl-prolyl cis–trans isomerases. FASEB J. 1992;6:3410–3420. doi: 10.1096/fasebj.6.15.1464374. [DOI] [PubMed] [Google Scholar]
- 3.Liu J., Farmer J. D., Jr, Lane W. S., Friedman J., Weissman I., Schreiber S. L. Calcineurin is a common target of cyclophilin–cyclosporin A and KFBP–FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 4.Jain J., McCaffrey P. G., Miner Z., Kerppola T. K., Lambert J. N., Verdine G. L., Curran T., Rao A. The T-cell transcription factor NAFTp is a substrate for calcineurin and interacts with Fos and Jun. Nature (London) 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 5.Borel J. F. Pharmacology of cyclosporine (sandimmune). IV. Pharmacological properties in vivo. Pharmacol. Rev. 1990;41:259–371. [PubMed] [Google Scholar]
- 6.Flückiger S., Fijten H., Whitley P., Blaser K., Crameri R. Cyclophilins, a new family of cross-reactive allergens. Eur. J. Immunol. 2002;32:10–17. doi: 10.1002/1521-4141(200201)32:1<10::AID-IMMU10>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 7.Lindborg M., Magnusson C. G., Zargari A., Schmidt M., Scheynius A., Crameri R., Whitley P. Selective cloning of allergens from the skin colonizing yeast Malassezia furfur by phage surface display technology. J. Invest. Dermatol. 1999;113:156–161. doi: 10.1046/j.1523-1747.1999.00661.x. [DOI] [PubMed] [Google Scholar]
- 8.Schmid-Grendelmeier P., Flückiger S., Disch R., Trautmann A., Wüthrich B., Blaser K., Scheynius A., Crameri R. IgE-mediated and T cell-mediated autoimmunity against manganese superoxide dismutase in atopic dermatitis. J. Allergy Clin. Immunol. 2005;115:1068–1075. doi: 10.1016/j.jaci.2005.01.065. [DOI] [PubMed] [Google Scholar]
- 9.Cadot P., Diaz J. F., Proost P., Van Damme J., Engelborghs Y., Stevens E. A., Ceuppens J. L. Purification and characterization of an 18-kd allergen of birch (Betula verrucosa) pollen: identification as a cyclophilin. J. Allergy Clin. Immunol. 2000;105:286–291. doi: 10.1016/s0091-6749(00)90078-2. [DOI] [PubMed] [Google Scholar]
- 10.Fujita C., Moriyama T., Ogawa T. Identification of cyclophilin as an IgE-binding protein from carrots. Int. Arch. Allergy Immunol. 2001;125:44–50. doi: 10.1159/000053795. [DOI] [PubMed] [Google Scholar]
- 11.Hopkin J. M. Mechanisms of enhanced prevalence of asthma and atopy in developed countries. Curr. Opin. Immunol. 1997;9:788–792. doi: 10.1016/s0952-7915(97)80179-3. [DOI] [PubMed] [Google Scholar]
- 12.Gould H. J., Sutton B. J., Beavil A. J., Beavil R. L., McCloskey N., Coker H. A., Fear D., Smurthwaite L. The biology of IgE and the basis of allergic disease. Annu. Rev. Immunol. 2003;21:579–628. doi: 10.1146/annurev.immunol.21.120601.141103. [DOI] [PubMed] [Google Scholar]
- 13.Aalberse R. C. Structural biology of allergens. J. Allergy Clin. Immunol. 2000;106:228–238. doi: 10.1067/mai.2000.108434. [DOI] [PubMed] [Google Scholar]
- 14.Holm J., Baerentzen G., Gajhede M., Ipsen H., Larsen J. N., Lowenstein H., Wissenbach M., Spangfort M. D. Molecular basis of allergic cross-reactivity between group 1 major allergens from birch and apple. J. Chromatogr. B Biomed. Sci. Appl. 2001;756:307–313. doi: 10.1016/s0378-4347(01)00089-5. [DOI] [PubMed] [Google Scholar]
- 15.Flückiger S., Mittl P. R., Scapozza L., Fijten H., Folkers G., Grutter M. G., Blaser K., Crameri R. Comparison of the crystal structures of the human manganese superoxide dismutase and the homologous Aspergillus fumigatus allergen at 2-Å resolution. J. Immunol. 2002;168:1267–1272. doi: 10.4049/jimmunol.168.3.1267. [DOI] [PubMed] [Google Scholar]
- 16.Mayer C., Hemmann S., Faith A., Blaser K., Crameri R. Cloning, production, characterization and IgE cross-reactivity of different manganese superoxide dismutases in individuals sensitized to Aspergillus fumigatus. Int. Arch. Allergy Immunol. 1997;113:213–215. doi: 10.1159/000237550. [DOI] [PubMed] [Google Scholar]
- 17.Crameri R., Faith A., Hemmann S., Jaussi R., Ismail C., Menz G., Blaser K. Humoral and cell-mediated autoimmunity in allergy to Aspergillus fumigatus. J. Exp. Med. 1996;184:265–270. doi: 10.1084/jem.184.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer C., Appenzeller U., Seelbach H., Achatz G., Oberkofler H., Breitenbach M., Blaser K., Crameri R. Humoral and cell-mediated autoimmune reactions to human acidic ribosomal P2 protein in individuals sensitized to Aspergillus fumigatus P2 protein. J. Exp. Med. 1999;189:1507–1512. doi: 10.1084/jem.189.9.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valenta R., Seiberler S., Natter S., Mahler V., Mossabeb R., Ring J., Stingl G. Autoallergy: a pathogenic factor in atopic dermatitis? J. Allergy Clin. Immunol. 2000;105:432–437. doi: 10.1067/mai.2000.104783. [DOI] [PubMed] [Google Scholar]
- 20.Mikol V., Kallen J., Walkinshaw M. D. X-ray structure of a cyclophilin B/cyclosporin complex: comparison with cyclophilin A and delineation of its calcineurin-binding domain. Proc. Natl. Acad. Sci. U.S.A. 1994;91:5183–5186. doi: 10.1073/pnas.91.11.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kodzius R., Rhyner C., Konthur Z., Buczek D., Lehrach H., Walter G., Crameri R. Rapid identification of allergen-encoding cDNA clones by phage display and high-density arrays. Comb. Chem. High Throughput Screen. 2003;6:147–154. doi: 10.2174/1386207033329751. [DOI] [PubMed] [Google Scholar]
- 22.Moser M., Crameri R., Brust E., Suter M., Menz G. Diagnostic value of recombinant Aspergillus fumigatus allergen I/a for skin testing and serology. J. Allergy Clin. Immunol. 1994;93:1–11. doi: 10.1016/0091-6749(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 25.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 27.McRee D. E. XtalView/Xfit: a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 28.Lamzin V. S., Wilson K. S. Automated refinement of protein models. Acta Crystallogr. D Biol. Crystallogr. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 30.Hooft R. W., Vriend G., Sander C., Abola E. E. Errors in protein structures. Nature (London) 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 31.Merritt E. A. Expanding the model: anisotropic displacement parameters in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1109–1117. doi: 10.1107/s0907444999003789. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 33.Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–380. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y., Ke H. Mechanisitc implication of crystal structures of the cyclophilin–dipeptide complexes. Biochemistry. 1996;35:7362–7368. doi: 10.1021/bi960278x. [DOI] [PubMed] [Google Scholar]
- 35.Laver W. G., Air G. M., Webster R. G., Smith-Gill S. J. Epitopes of protein antigens. Misconceptions and realities. Cell. 1990;61:553–556. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- 36.Mirza O., Henriksen A., Ipsen H., Larsen J. N., Wissenbach M., Spangfort M. D., Gajhede M. Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. J. Immunol. 2000;165:331–338. doi: 10.4049/jimmunol.165.1.331. [DOI] [PubMed] [Google Scholar]
- 37.Neudecker P., Lehmann K., Nerkamp J., Haase T., Wangorsch A., Fotisch K., Hoffmann S., Rosch P., Vieths S., Scheurer S. Mutational epitope analysis of Pur av1 (Prunus avium) and Api g 1 (Apium graveolens), the major allergens of cherry and celery: correlating IgE reactivity to three-dimensional structure. Biochem. J. 2003;376:97–107. doi: 10.1042/BJ20031057. [DOI] [PMC free article] [PubMed] [Google Scholar]