Abstract
A galactose-specific C-type lectin has been purified from a pupal extract of Drosophila melanogaster. This lectin gene, named DL1 (Drosophila lectin 1), is part of a gene cluster with the other two galactose-specific C-type lectin genes, named DL2 (Drosophila lectin 2) and DL3 (Drosophila lectin 3). These three genes are expressed differentially in fruit fly, but show similar haemagglutinating activities. The present study characterized the biochemical and biological properties of the DL1 protein. The recombinant DL1 protein bound to Escherichia coli and Erwinia chrysanthemi, but not to other Gram-negative or any other kinds of microbial strains that have been investigated. In addition, DL1 agglutinated E. coli and markedly intensified the association of a Drosophila haemocytes-derived cell line with E. coli. For in vivo genetic analysis of the lectin genes, we also established a null-mutant Drosophila. The induction of inducible antibacterial peptide genes was not impaired in the DL1 mutant, suggesting that the galactose-specific C-type lectin does not participate in the induction of antibacterial peptides, but possibly participates in the immune response via the haemocyte-mediated mechanism.
Keywords: C-type lectin, Drosophila, galactose-specific lectin, immunity, phagocytosis
Abbreviations: CBB, Coomassie Brilliant Blue; FBS, foetal bovine serum; GFP, green fluorescent protein; IMD, immune deficiency; LPS, lipopolysaccharide; NF-κB, nuclear factor κB; PGRP, peptidoglycan-recognition protein; RACE, rapid amplification of cDNA ends; RT, reverse transcription; UTR, untranslated region
INTRODUCTION
Insects have an effective defence system against microbial infection that shares many characteristics with the innate immune system of vertebrates [1,2]. These systems discriminate among various classes of infecting micro-organisms and mount a proper immune response. In the case of Drosophila melanogaster, the infection of Gram-negative bacteria engenders the expression of antibacterial peptide genes via the IMD (immune deficiency) pathway; on the other hand, the infection of fungi engenders the expression of antifungal peptide genes via the Toll pathway [1–5]. These selective responses are based on molecules that specifically recognize pathogens. These proteins, called pattern-recognition receptors, bind to microbial components such as LPS (lipopolysaccharide) or peptidoglycans, which trigger the activation of corresponding immune responses.
In addition to such recent progress in the investigation of immune-regulation pathways, many lectins have been purified from various insect species such as flesh fly and cockroach [6,7]. Some of them putatively participate in self-defence by recognizing polysaccharide chains on the surface of pathogens. Some insect lectins are believed to bind to microbial components such as LPS, activate prophenoloxidase, and to enhance cellular immune responses by modulating haemocytes.
Insect immune-system-activation mechanisms have been uncovered using D. melanogaster as a model organism by genetic and molecular studies [1–5]. For that reason, further analysis of lectins in Drosophila will elucidate their biological role in immune systems. Drosophila is estimated to have more than 30 C-type lectin genes [8]. However, little is known about the participation of lectin in Drosophila immunity [9,10].
Previously, we purified a galactose-specific C-type lectin from a pupal extract of Drosophila, which agglutinated, trypsinized and glutaraldehyde-fixed bovine red blood cells [9]. This lectin is partly glycosylated and is believed to form a trimer. In addition, cDNA cloning of the lectin gene revealed that the 20 N-terminal residues out of 186 residues were a leader peptide and that most of the other residues corresponded to the carbohydrate-recognition domain of C-type lectin. Expression of the lectin gene in larvae was enhanced by body injury, suggesting that this lectin is a defence molecule.
Genomic analysis of this lectin gene suggested that the lectin gene consists of a gene cluster with the other C-type lectin genes [11]. This result implied that the original lectin gene was multiplicated, possibly to diversify or amplify its function. The biological meaning of the multiplication can be clarified through comparative analysis of these lectins and mutant analysis of this gene cluster.
MATERIALS AND METHODS
Drosophila stocks and mutagenesis
D. melanogaster stocks were kept on a standard medium at 25 °C.
The EP(2)1173 strain was obtained from Exelixis FlyStation. The P element was transposed by crossing with yw; CyO/Sp; TMS Sb P{ry+(Δ2-3)}(99B)/TM3 jumpstarter males; the F1 male flies with both the Sp and Sb markers were crossed with yw; CyO/Gla balancer females. F2 flies with different eye colour from that of their progenitors were collected. Flies with the P element on the second chromosome were selected by crossing with yw flies. Positive F3 males were balanced by CyO. The transposed lines (250 lines) were screened by genomic PCR using primers for the inverted repeat of the P element and the genomic sequences around the lectin gene locus, which were designed to detect amplification of the PCR products when the P element was inserted near the respective lectin genes. The PCR product derived from a P-transposed line #914 was sequenced on subcloned fragments using a DNA Sequencing kit (Applied Biosystems). The P insertion site was mapped through comparison with the Drosophila genomic sequence. The P element was excised from the chromosome of #914 by crossing with the jumpstarter males again to generate the null-mutant for the lectin gene. The F1 male flies with the Cy marker were then crossed with w1118; CyO/Gla balancer females. F2 flies with white eyes were collected and were used to initiate 79 independent P-excised lines. The P-excised lines were screened by genomic PCR using appropriate primers for genomic sequences. The PCR product derived from a P-excised line E136 was sequenced; the deletion was mapped through comparison with the genomic sequence.
We also established transgenic flies to rescue the expression of the DL1 gene. The genomic sequence from 0.98 kb 5′-upstream to 0.04 kb 3′-downstream of the DL1 gene was inserted into pP{CaSpeR-4}. Flies carrying this construct on the third chromosome were obtained by P element-mediated germline transformation into w1118 flies.
The GFP (green fluorescent protein)-tagged balancer chromosome CyO P{w+mC=GAL4-Kr.C}DC3 P{w+mC=UAS-GFP.S65T}DC7 [12] was used to discriminate between heterozygous and homozygous larvae for the mutation.
RT (reverse transcription)–PCR analysis
Total RNA from tissues of Canton-S flies was isolated using an RNeasy Mini Kit (Qiagen). RT–PCR was carried out according to standard procedures with gene-specific primers. First-strand cDNA was synthesized from 0.25 μg of total RNA using StrataScript reverse transcriptase (Stratagene).
cDNA and genome analysis
The cDNA for the lectin genes were obtained using PCR from a Drosophila adult cDNA library (Stratagene) using the primers for the lectin genes and the vector. The PCR products were sequenced on subcloned fragments as mentioned above. The 5′-region was isolated by a 5′-RACE (rapid amplification of cDNA ends) method using a Marathon cDNA amplification kit (BD Biosciences/Clontech). Genomic PCR was performed using isolated DNA from Canton-S adult flies and appropriate primers.
Northern blot analysis
Total RNA from Canton-S flies was isolated using TRIzol® reagent (Invitrogen). RNA from each sample was separated on 1.2% agarose/formaldehyde gel and was transferred to a GeneScreen Plus membrane (NEN Life Science Products/PerkinElmer). Hybridization with the probe was performed in ExpressHyb Hybridization Solution (BD Biosciences/Clontech) at 68 °C for 2 h. The following probes [γ-32P]dCTP-labelled with a random primer labelling kit (TaKaRa Holdings) were used for hybridization: DL1 and DL2, 470 bp of the translational region on the second exon; DL3, 327 bp of the translational region on the second exon. Probes for rp49 (ribosomal protein 49), Cecropin A and Defensin were prepared from the genomic clone. After washing with 0.1× SSC (15 mM NaCl and 5 mM sodium citrate) containing 0.1% (w/v) SDS, the membrane was exposed on X-ray film.
Plasmid construction and transfection experiment
Expression vectors were constructed by inserting the cDNA into pAC5.1/V5-His A (Invitrogen), which encoded the protein with V5 epitope and His6 tags at the C-terminus. A vector inserted with the cDNA including the stop codon was also constructed to obtain the constructs encoding the intact lectins for stable transfection. Drosophila Schneider-2 cells (Invitrogen) were cultured at 25 °C in DES® (Drosophila Expression System) expression medium (Invitrogen) supplemented with 10% FBS (foetal bovine serum), 50 units/ml penicillin and 50 μg/ml streptomycin. The cells were transiently transfected with the lectin expression vector or the β-galactosidase expression vector (pAC5.1/V5-His/lacZ; Invitrogen) by calcium phosphate transfection, followed by incubation until expression of the recombinant protein reached the maximum concentration. For stable transfections, the cells were co-transfected with the expression vector and the selection vector pCoHygro (Invitrogen). Hygromycin-B-resistant cells were selected by culture in the medium containing 300 μg/ml hygromycin B. After establishment of the stable cell lines, cells were cultured in Schneider's Drosophila medium (Invitrogen) supplemented with 10% FBS, 50 units/ml penicillin, 50 μg/ml streptomycin and 300 μg/ml hygromycin B.
Detection of the recombinant lectins with anti-(His6 tag) antibody
The transfected cells were lysed in 50 μl of 50 mM Tris/HCl, pH 7.8, containing 150 mM NaCl, 1% Nonidet P-40, 1 mM PMSF, 1 μg/ml pepstatin A and 1 μg/ml leupeptin. Protein concentration in the lysate was measured using a protein assay kit (Bio-Rad Laboratories). Then, 10 μl of the culture medium or 36 μg of the cell lysate was separated by SDS/PAGE (15% gels) and blotted on to an Immobilon transfer membrane (Millipore) that was subsequently incubated at 4 °C for 16 h in blocking solution [1% (w/v) casein]. The blot was probed with His·Tag monoclonal antibody (Novagen) at room temperature (25 °C) for 1 h. A goat anti-mouse horseradish-peroxidase-conjugated antibody (Promega) was used as the secondary antibody; target protein detection was performed using ImmunoStar reagents (Wako Pure Chemical Industries).
Assay of haemagglutinating activity
The stable cell line was cultured in Schneider's Drosophila medium supplemented with 50 units/ml penicillin and 50 μg/ml streptomycin for 4 days. Haemagglutinating activity (titre) was expressed as in [9]. The effect of sugars on the activity was measured as the minimum concentration required for inhibition of two titres.
Detection of the recombinant intact DL1
The medium containing 20 μg of protein was separated by SDS/PAGE (15% gels), and proteins were detected with CBB (Coomassie Brilliant Blue) stain. For detection of recombinant lectin by Western blot analysis, 10 μl of the culture medium was separated by SDS/PAGE (15% gels), and proteins were blotted on to an Immobilon transfer membrane. It was then incubated at 4 °C for 16 h in blocking solution [5% (w/v) dried skimmed milk]. The blot was probed with an affinity-purified rabbit anti-peptide antibody against CVKAEPFTKINDGYYFFGT-NH2 at room temperature for 1 h. A goat anti-rabbit horseradish-peroxidase-conjugated antibody (Promega) was used as the secondary antibody, and peroxidase activity was detected as mentioned above.
Purification of the recombinant DL1
Harvested stable cells were suspended in Schneider's Drosophila medium supplemented with 50 units/ml penicillin and 50 μg/ml streptomycin to adjust the cell density to 6×106 cells/ml. The cells were then cultured for 1 week with stirring. The culture medium was filtered through a 0.22 μm pore-size filter, concentrated by ultrafiltration, then dialysed against buffered insect saline (10 mM Tris/HCl, pH 7.9, containing 130 mM NaCl, 5 mM KCl and 1 mM CaCl2). The medium was applied to an immobilized D-galactose column (Pierce Biotechnology), and the column was then washed with the saline. Thereafter, the galactose-binding proteins were eluted with saline containing 0.2 M galactose. The elution was concentrated using Centricon-10 (Amicon), then gel-filtered with Superose 12HR10/30 using an FPLC system (Amersham Biosciences). The fractions containing the purified recombinant protein were dialysed against the saline. Proteins from each step were separated by SDS/PAGE (15% gels) and detected by silver stain.
Determination of N-terminal amino acid sequence of the recombinant DL1
The recombinant DL1 (10 μg) was subjected to SDS/PAGE (15% gels) and then blotted on to Immobilon-PSQ transfer membrane (Millipore). After staining with CBB, the recombinant protein band was excised and analysed using a protein sequencer (PPSQ-21; Shimadzu).
Digestion with N-glycosidase
The recombinant DL1 (2 μg) was treated with N-glycosidase F (Roche Diagnostics) in 250 mM Tris/HCl, pH 7.9, containing 0.5% (w/v) SDS, 50 mM 2-mercaptoethanol, 50 mM PMSF, 7 mg/ml leupeptin and 1 mM EDTA at 37 °C for 18 h. Then the protein was separated by SDS/PAGE (15% gels) and stained with CBB.
Microbial strains
Escherichia coli K-12 W3110, Enterobacter cloacae (A.T.C.C. 13047), Erwinia carotovora [CFBP (Collection Française de Bacteries Pathogenes, 194, 1401, 1488, 2140 and 2141], Erwinia chrysanthemi (CFBP 1446, 2811 and 3477), Pseudomonas aeruginosa (A.T.C.C. 10145), Salmonella serotype Minnesota (A.T.C.C. 9700), Salmonella serotype Typhimurium [IFO (Institute for Fermentation), Osaka, Japan, 14193), Serratia marcescens (A.T.C.C. 13880), Sphingomonas paucimobilis (A.T.C.C. 29837), Bacillus megaterium (A.T.C.C. 14581), Bacillus subtilis (A.T.C.C. 6051), Enterococcus faecalis (A.T.C.C. 19433), Micrococcus luteus (A.T.C.C. 4698), Staphylococcus aureus (Cowan 1 strain), Beauveria bassiana (A.T.C.C. 9453) and Metarhizium anisopliae (A.T.C.C. 22099) were used. All bacterial strains except Sphingomonas paucimobilis were cultured at 30 °C in a medium comprising 1% (w/v) peptone, 0.3% (w/v) meat extract and 0.5% (w/v) NaCl, with the pH adjusted to 7.2. Sphingomonas paucimobilis was cultured in brain heart infusion broth (Difco Laboratories). Fungal strains were cultured on potato dextrose agar plates.
Binding assay of DL1 to micro-organisms
Each bacterial strain or spores from fungal strain (300 μg) was washed with buffered insect saline and centrifuged at 20000 g at 4 °C for 2 min. The harvested micro-organisms were then suspended in 30 μl of the saline containing 400 ng of recombinant DL1 and incubated at 4 °C for 1 h. After washing with 100 μl of saline three times, the micro-organism-binding proteins were released by suspension in 15 μl of saline containing 50 mM galactose or mannose. Eluted proteins were subjected to Western blot analysis with the anti-peptide antibody, as mentioned above.
Agglutination of E. coli
E. coli was transformed with the GFP expression vector pEGFP (BD Biosciences/Clontech). These transformants were cultured at 30 °C in a medium containing 50 μg/ml ampicillin and 0.4 mM IPTG (isopropyl β-D-thiogalactoside). E. coli (2×107 cells) was mixed with the recombinant DL1 and incubated at 4 °C for 1 h. The bacteria were harvested, then washed with buffered insect saline. The bacteria were fixed with 4% (w/v) paraformaldehyde, washed with the saline, and then applied to a flow cytometer (EPICS Elite ESP; Beckman Coulter). The mean GFP fluorescence intensity of the 10000 counts was calculated.
Flow-cytometric analysis of the association of E. coli with mbn-2 cells
Drosophila mbn-2 cells were cultured at 25 °C in Schneider's Drosophila medium supplemented with 12% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. The cell suspension was diluted with fresh medium to adjust the cell density to 8×105 cells/ml. Then the culture was continued for 3–4 days. For experiments, 2×106 mbn-2 cells were harvested and resuspended in 1.8 ml of Schneider's Drosophila medium supplemented with 170 μg/ml chloramphenicol instead of penicillin and streptomycin. The GFP-labelled E. coli (2×108 cells) were washed with Schneider's Drosophila medium, then harvested by centrifugation at 800 g for 3 min and resuspended in 0.2 ml of medium containing the recombinant DL1 and 50 mM galactose or xylose. Xylose was used instead of mannose as a non-hapten sugar because mannose had inhibited the DL1-independent association of mbn-2 cells with E. coli (results not shown). The suspension was incubated at 4 °C for 1 h, mixed with a suspension of mbn-2 cells and then incubated at 27 °C for 10 min with rotating. The harvested mbn-2 cells were washed with buffered insect saline, fixed with 4% (w/v) paraformaldehyde, washed with the saline and then resuspended in 1 ml of the saline containing 10 μg/ml propidium iodide. Then the suspension was applied to a flow cytometer. The mean GFP fluorescence intensity of the 10000 propidium-iodide-positive counts was calculated. The E. coli cells that were agglutinated, but not associated with mbn-2 cells, were readily discernible from mbn-2 cells by the propidium iodide fluorescence intensity.
Southern blot analysis
Genomic DNA (5 μg) prepared from third instar larvae was digested with 30 units of PstI (TaKaRa Holdings) for 18 h; it was then separated by agarose gel electrophoresis. The DNA was transferred to a GeneScreen Plus membrane. After UV cross-linking, hybridization with the probe was performed in ExpressHyb hybridization solution at 60 °C for 2 h. The same probes as those for Northern blot analysis were used for hybridization. After washing with 0.1×SSC containing 0.1% (w/v) SDS, the membrane was exposed on X-ray film.
Infection experiment
Natural infection of third instar larvae with E. coli was performed following the method of Basset et al. [13] for phytopathogenic bacteria Erwinia carotovora.
RESULTS
A gene cluster of three C-type lectin genes at 37D6 on the genome of D. melanogaster
The Berkeley Drosophila Genome Project revealed that the Drosophila genome has more than 30 C-type lectin-like genes [8]. Among these genes, the CG33532 and CG33533 genes, known together as the CG9978 gene, are localized next to the galactose-specific C-type lectin gene locus examined in [9]. To find out whether the predicted genes were expressed, we performed RT–PCR analysis using Canton-S third instar larvae, pupae and adults. Gene-specific amplifications of both carbohydrate-recognition domains were detected during those developmental stages (results not shown).
To elucidate the complete structure of these genes, we obtained their corresponding cDNAs from an adult cDNA library of Canton-S, and the genome fragments from Canton-S adults. We also performed 5′-RACE to get the 5′-terminal region. The intron splice sites and the plural polyadenylation sites were identified through comparison of the cDNA and genome sequences. As shown in Figure 1(A), we named these three lectins on 37D6 as DL1 (identical with the lectin studied in [9]), DL2 (CG33532) and DL3 (CG33533), according to their order on the genome. Each cDNA consists of the short first exon and the second exon containing the carbohydrate-recognition domain. Each of the three genes has a promoter-like sequence with a TATA box, a cap site at the upstream of the coding region [14], and a polyadenylation signal sequence at the downstream. The gaps between these lectin genes were very short; the putative TATA sequence for the DL2 genes overlaps the 3′-UTR (untranslated region) of the DL1 gene, and only 25 bp of nucleotides exist between the polyadenylation site for the DL2 genes and the putative TATA sequence for the DL3 gene.
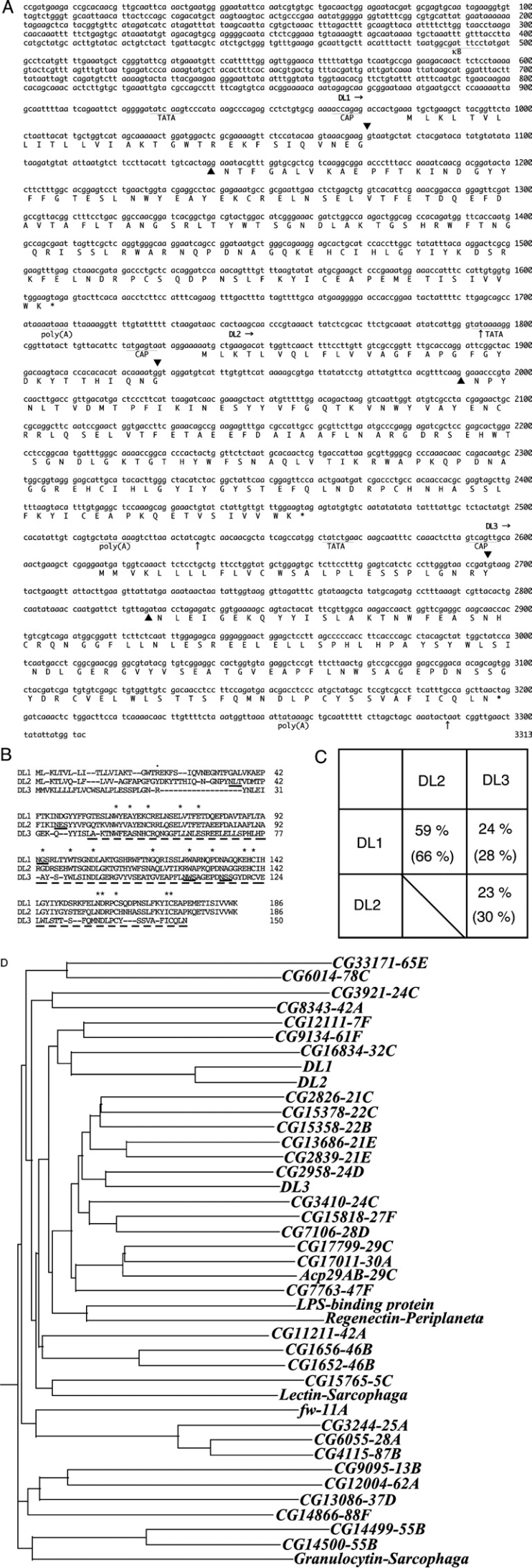
Figure 1. Gene structure of Drosophila lectins on 37D.
(A) Genomic sequence of the lectin genes locus. Intron splice sites are indicated by ▲ for donor and acceptor sites. Possible NF-κB site (κB), and TATA, cap and polyadenylation [poly(A)] signal sequences are dotted underlined. DL1, DL2 and DL3 are translated using one-letter symbols below their respective exons. The polyadenylation sites are indicated with arrows. (B and C) Comparison of the amino acid sequences of DL1, DL2 and DL3. (B) Gaps are introduced to obtain maximum matching. The N-terminal residue of DL1 is indicated by a dot. Residues corresponding to the carbohydrate-recognition domain are broken-underlined. Asterisks indicate residues conserved among various C-type lectins. Possible N-glycosylation sites are indicated by solid underlining [34]. (C) Identities between the lectins at the amino acid level. Identities of the carbohydrate-recognition domains are shown in parentheses. (D) Phylogenetic analysis of Drosophila C-type lectins. Carbohydrate-recognition domains of these proteins were aligned using the ClustalW program. Based on alignments, a phylogenetic tree was constructed using the dendrogram method. Gene names are shown with cytological locations. For comparison, C-type lectins identified from Sarcophaga peregrina [35,36] and Periplaneta americana [37,38] were analysed together.
The amino acid sequences of three lectins are compared in Figures 1(B) and 1(C). The number of amino acid residues of DL2 is equal to that of DL1; their identity is 59% in the full-length and 66% in the carbohydrate-recognition domain. Their homology is extremely high, as shown by phylogenetic analysis of Drosophila C-type lectins (Figure 1D). In contrast, DL3 has fewer amino acid residues, and DL3 has relatively low homology with DL1 and DL2. The hydrophobic N-terminal regions of DL2 and DL3, like DL1 [9], suggest that they are also secreted proteins.
Expression analysis of the lectin genes
We analysed expression during development using Northern blotting to compare expression of the DL1, DL2 and DL3 genes. As reported previously [9], expression of the DL1 gene was detected from the larval to adult stage (Figure 2A). When probed for the DL2 gene, 0.8 and 1.6 kb RNA bands were found; both bands were detected from the late pupal stage. Expression of the DL3 gene was detected only at the adult stage. For all three lectin genes, 0.8 kb bands were consistent with their gene structures, although the structure for the additional minor 1.6 kb bands of the DL2 gene was unclear. Three lectin genes all showed the highest expression at the adult stage, but the expression at the pupal stage was different between the DL1 and DL2 genes: the expression of the DL1 gene was higher at the early pupal stage than at the late pupal stage, but the expression of the DL2 gene was higher at the late pupal stage. These results indicate that the expression of these lectin genes was differentially regulated. As DL1 gene expression has been known to be enhanced on injury at the larval stage [9], we also examined whether DL2 and DL3 gene expression was changed on injury or septic injury with E. coli at larval and adult stages; no change was detected (results not shown).
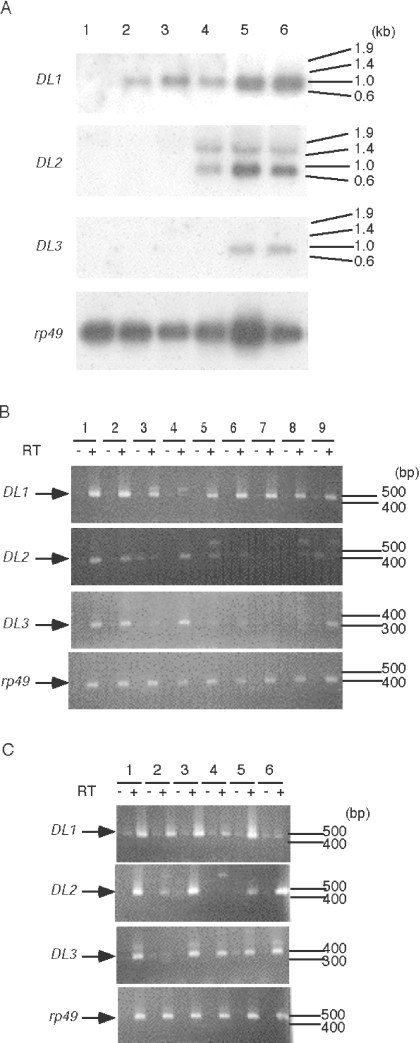
Figure 2. Comparisons of expression profiles of the lectin genes.
(A) Northern blot analysis of the expression of the lectin genes during development. Total RNA was extracted from Drosophila Canton-S at different developmental stages and then subjected to Northern blot hybridization. RNA was extracted from embryos (lane 1), third instar larvae (lane 2), early pupae (lane 3), late pupae (lane 4), female adults (lane 5) and male adults (lane 6). The Drosophila rp49 gene probe was used as a loading control. Sizes are indicated in kb. (B and C) RT–PCR analysis of the expression of lectin genes in Drosophila tissues. Representative RT–PCR for the lectin genes and the rp49 gene is shown. Sizes are indicated in bp. (B) RT–PCR for larval tissues. Total RNA from the whole body (lane 1), cuticle and muscles (lane 2), salivary glands (lane 3), midgut and Malpighian tubules (lane 4), fat body (lane 5), tracheae (lane 6), brain and eye-antennal discs (lane 7), leg discs and wing discs (lane 8), and haemocytes (lane 9) were analysed. (C) RT–PCR for adult tissues. Total RNA from female whole body (lane 1), ovaries (lane 2), male whole body (lane 3), testes (lane 4), cuticle and muscles (lane 5), and midgut and Malpighian tubules (lane 6) were analysed. Arrows indicate positions of gene-specific amplification.
We also investigated the expression of these lectin genes in various tissues. During the larval stage, the expression of all three genes was detectable by RT–PCR. Figure 2(B) shows that the expression of the DL1 gene was detected in all examined tissues except the midgut and Malpighian tubules. On the other hand, the expression of the DL2 and DL3 genes was relatively tissue-specific. The expression of these genes was detected in cuticle and muscles, midgut and Malpighian tubules, and fat bodies. Additional expression of the DL3 gene was also detected in haemocytes. These results indicate that the expression pattern of the DL1 gene differs greatly from that of the DL2 and DL3 genes at the larval stage.
At the adult stage, expression of the DL1 gene was detected in cuticle and muscles, ovaries, and testes, but not in the midgut or Malpighian tubules (Figure 2C). However, expression of the DL2 and DL3 genes was detected in midgut or Malpighian tubules. Differences in DL2 and DL3 gene expression were found, especially in reproductive organs; the DL2 gene was expressed in ovaries, but the DL3 gene was expressed in testes. These results indicate that the expression patterns of the DL1, DL2 and DL3 genes are also different in the adult stage.
Localization of the recombinant lectins at the transfected cells
The lectins expressed in fat body and haemocytes are expected to be secreted proteins to haemolymph [7]. To determine the secretion of DL1, DL2 and DL3, we transfected Drosophila Schneider-2 cells with the expression vector for each lectin and localization of the recombinant protein with the His6 tag was analysed. Figure 3(A) shows that the recombinant DL1, DL2 and DL3 were all detected mainly in the culture medium on the condition that β-galactosidase, transfected as a control, was detected in the cell lysate as a 120 kDa band. This result indicates that these lectins are all secreted proteins, as suggested by hydropathy profiles of these proteins.
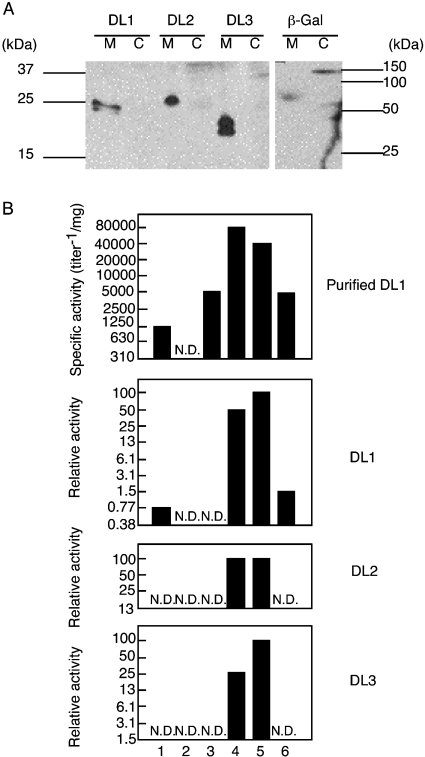
Figure 3. Comparisons of localization and haemagglutinating activity of lectins.
(A) Expression of recombinant proteins from the transfected Schneider-2 cells. From the culture medium (M) or cell lysate (C) of the transiently transfected cells with the expression vector for each lectin or the β-galactosidase expression vector (β-Gal), recombinant proteins were detected using Western blot analysis using anti-(His6 tag) antibody. (B) Agglutinating activity of the recombinant intact lectins against red blood cells from various species. Culture media from the stably transfected Schneider-2 cells were serially diluted and mixed with bovine (lane 1), chicken (lane 2), horse (lane 3), porcine (lane 4), rabbit (lane 5) or sheep (lane 6) red blood cells. Results of the purified DL1 are also shown (see Figure 4). The highest level of activity of the culture medium in a series was normalized as 100; the representative results are given as relative activity (percentages). N.D. means not detected (<630, <0.77, <25 and <3.1 titre−1·mg−1 for the purified DL1 and the culture medium from the Schneider-2 cells transfected with expression vectors for DL1, DL2 and DL3 respectively). No haemagglutinating activities were detected when the culture medium from Schneider-2 cells transfected with the mock expression vector was analysed.
Recombinant proteins of DL1 and DL2 were detected as having a larger molecular size than that of DL3. That result is consistent with the estimated values of the deduced amino acid sequences from their cDNAs. Two bands of DL3 suggest the existence of the N-glycosylated form, which is supported by the existence of possible N-glycosylation sites, as indicated in Figure 1(B).
Activity of the recombinant lectins
We stably transfected Schneider-2 cells to make them secrete each recombinant intact lectin constitutively. We thereby determined the activity of these lectins. We investigated haemagglutinating activity of the culture medium using bovine, chicken, horse, porcine, rabbit and sheep red blood cells. We clarified that the recombinant DL1 had agglutinating activity against porcine and rabbit red blood cells (Figure 3B). Weaker agglutinating activity, or none at all, was detected when the other red blood cells were used. Activity against bovine red blood cells was not detected because the cells were not treated with trypsin and glutaraldehyde, as in [9]. Similar results were obtained when a culture medium containing recombinant DL2 or DL3 was examined, suggesting that these lectins had binding activity with similar specificity to polysaccharide chains. Furthermore, haemagglutinating activities of these lectins were all inhibited by addition of EDTA, indicating that they were C-type lectins.
Next, we examined the effect of sugars on the haemagglutinating activity of these lectins. Table 1 shows that the activity of the recombinant DL1 was inhibited by galactose or oligosaccharides containing galactose (raffinose and lactose). Similar results were obtained when the culture medium containing the recombinant DL2 or DL3 was examined, suggesting that all of these lectins are galactose-specific.
Table 1. Competing effects of sugars on agglutinating activity of the recombinant lectins against rabbit red blood cells.
The assay was performed with sugars at concentrations of 1, 3, 10, 30 and 100 mM.
| Minimum concentration (mM) | ||||
|---|---|---|---|---|
| Sugar | Purified DL1 | DL1 | DL2 | DL3 |
| D-Galactose | 3 | 3 | 3 | 3 |
| Raffinose | 3 | 3 | 3 | 3 |
| Lactose | 10 | 10 | 10 | 3 |
| D-Glucose | 100 | 30 | 100 | 30 |
| D-Xylose | 100 | 30 | 100 | 30 |
| D-Mannose | >100 | >100 | >100 | >100 |
| Fructose | >100 | >100 | >100 | >100 |
| Sucrose | >100 | >100 | >100 | >100 |
| Trehalose | >100 | >100 | >100 | >100 |
| N-Acetylgalactosamine | >100 | >100 | >100 | >100 |
| N-Acetylglucosamine | >100 | >100 | >100 | >100 |
Preparation of recombinant DL1
DL1, DL2 and DL3 have similar haemagglutinating activity. For that reason, we focused our attention on one lectin, DL1, for further biochemical characterization. We expressed the recombinant protein of DL1 in the stably transfected Schneider-2 cells to obtain a large amount of DL1. From the culture medium, additional 19 kDa and 20 kDa proteins were detected, as shown in Figure 4(A) (lane 2). Both 19 and 20 kDa proteins were detected using Western blot analysis with the anti-peptide antibody for DL1 (Figure 4A, lane 4). As reported previously [9], the native DL1 is partly N-glycosylated, therefore the 20 kDa protein seemed to be the N-glycosylated form of the 19 kDa protein. We purified the recombinant protein by affinity chromatography on a galactose column, followed by gel-filtration; thereby, both 19 and 20 kDa proteins were purified (Figure 4B).
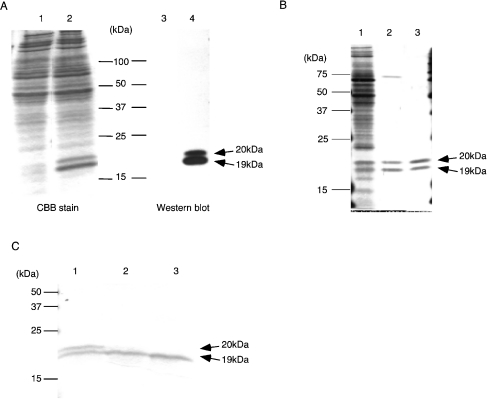
Figure 4. Preparation of recombinant DL1.
(A) Constitutive expression of the recombinant DL1 from the culture medium of the stable transfectants of Schneider-2 cells. Culture medium of the stable transfectant with mock (lanes 1 and 3) or the expression vector for the intact DL1 (lanes 2 and 4) was separated by SDS/PAGE (15% gels), followed by CBB staining (lanes 1 and 2) or Western blot analysis using anti-peptide antibody for DL1 (lanes 3 and 4). (B) Purification of the recombinant DL1. The culture medium of the stable transfectant of Schneider-2 cells with the expression vector for DL1 (lane 1), proteins purified from the culture medium with galactose-affinity column (lane 2) and proteins subsequently purified using gel-filtration (lane 3) were separated by SDS/PAGE (15% gels), followed by silver staining. (C) Digestion of the purified recombinant DL1 with N-glycosidase. Purified lectin (10 μg) was digested with increasing amounts of N-glycosidase F for 18 h at 37 °C. Then the mixtures were subjected to electrophoresis under denaturing conditions. Amounts of N-glycosidase used were: lane 1, 0 units; lane 2, 0.3 unit; lane 3, 1 unit. Arrows indicate positions of the 19 and 20 kDa proteins. Molecular-mass sizes are indicated in kDa.
As is evident from Figure 4(C), because of N-glycosidase treatment, the 20 kDa band disappeared and the 19 kDa band was intensified, indicating that the 20 kDa protein is a glycosylated form of the 19 kDa protein. The N-terminal residue of the purified lectin was determined as arginine at position 21 from the first methionine, which was the same as the native DL1 [9].
This recombinant protein had agglutinating activity against porcine and rabbit red blood cells, which was similar to the unpurified culture medium of the transfectant (Figure 3B). Table 1 shows that effects of various sugars on the haemagglutinating activity of the purified recombinant DL1 were identical with that of the native protein purified from Drosophila. Therefore this purified recombinant DL1 is available for in vitro investigation of the biochemical function of the lectin.
Binding of DL1 to bacteria
We examined the binding activity of DL1 to micro-organisms. Each bacterial strain was mixed with the purified recombinant DL1. Then, the binding lectin was eluted with galactose (a hapten sugar) or mannose, followed by Western blot analysis to detect the lectin eluted from bacteria. Of the nine Gram-negative and five Gram-positive bacterial species that we examined, the binding lectin was detected when E. coli (K-12 W3110) was used (Table 2 and Figure 5A). Binding with Erwinia chrysanthemi strains, which are phytopathogenic bacteria that can infect Drosophila naturally [13], was also detected. Nevertheless, the binding was weaker than that with E. coli. Furthermore, the binding lectin to E. coli was not eluted with mannose, indicating that binding was dependent on the activity as a lectin (Figure 5A). We also tested spores from two fungal strains, but the binding lectin was not detected (Table 2).
Table 2. Detection of the binding of DL1 to microbial strains.
+, Binding detected; −, binding not detected.
| Strain | Binding |
|---|---|
| Gram-negative bacteria | |
| Escherichia coli | + |
| Enterobacter cloacae | − |
| Erwinia carotovora | |
| CFBP194 | − |
| CFBP1401 | − |
| CFBP1488 | − |
| CFBP2140 | − |
| CFBP2141 | − |
| Erwinia chrysanthemi | |
| CFBP1446 | + |
| CFBP2811 | + |
| CFBP3477 | + |
| Pseudomonas aeruginosa | − |
| Salmonella typhimurium | − |
| Salmonella minnesota | − |
| Serratia marcescens | − |
| Sphingomonas paucimobilis | − |
| Gram-positive bacteria | |
| Bacillus megaterium | − |
| Bacillus subtilis | − |
| Enterococcus faecalis | − |
| Micrococcus luteus | − |
| Staphylococcus aureus | − |
| Fungi | |
| Beauveria bassiana | − |
| Metarhizium anisopliae | − |
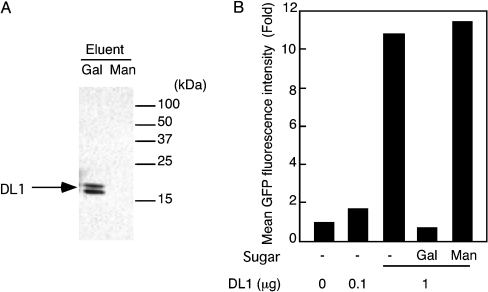
Figure 5. Binding of DL1 to E. coli.
(A) Detection of the bound DL1 to E. coli. The purified recombinant DL1 (400 ng) was mixed with 300 μg of bacteria. Binding lectin was then eluted with 50 mM sugars. The eluted lectin was detected using Western blot analysis. An arrow indicates the position of DL1. Gal, eluted with galactose; Man, eluted with mannose. Sizes are indicated in kDa. (B) Agglutination of E. coli with DL1. Increasing amounts of the purified recombinant DL1 were mixed with 2×107 cells of the GFP-labelled E. coli; the mean GFP fluorescence intensity from the representative experiment was then measured using a flow cytometer. Inhibitory effects by 50 mM sugars are also shown. Gal, galactose; Man, mannose.
The native DL1 forms a homotrimer [9]. Therefore it is likely that the DL1 protein binds bacteria and then agglutinates it. For that reason, we examined whether or not the lectin agglutinated E. coli. The GFP-labelled E. coli was mixed with the purified recombinant DL1, before measuring the fluorescence intensity using a flow cytometer. Figure 5(B) shows that the mean fluorescence intensity increased by the addition of the lectin in a dose-dependent manner. The total fluorescence intensity of the bacterial suspension was unaffected by the addition of the lectin (results not shown). Consequently, increased fluorescence intensity means an increase in the number of E. coli cells per count: the E. coli cells were agglutinated. Agglutination of E. coli was inhibited completely by galactose, but not by mannose, suggesting that the effect was dependent on the activity of DL1 as a lectin.
Effect of DL1 on the association of mbn-2 cells with E. coli
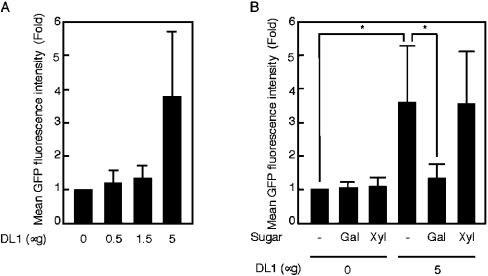
In an innate immune system, bacterial aggregation by lectin often engenders bacterial clearance by scavenger cells [7]. For that reason, we examined whether the binding of DL1 to E. coli affected the association of haemocytes, Drosophila blood cells, with E. coli or not. The GFP-labelled E. coli was mixed with and without the purified recombinant DL1, then added to cultured mbn-2 cells, a Drosophila haemocyte-derived cell line. The GFP fluorescence associated with mbn-2 cells was measured using a flow cytometer, and the mean fluorescence intensity of 10000 mbn-2 cells was calculated. Figure 6(A) shows that the mean fluorescence intensity associated with mbn-2 cells was increased by the addition of the lectin in a dose-dependent manner. That effect was inhibited markedly by galactose, but not by xylose, suggesting that the effect was dependent on the activity as a lectin, which was the same as the binding of the lectin to the bacteria (Figure 6B). That result strongly suggests that the binding of DL1 to E. coli enhanced the association of haemocytes with the bacteria.
Figure 6. Effect of DL1 on the association of mbn-2 cells with E. coli.
(A) Increasing amounts of the purified recombinant DL1 was mixed with 2×108 cells of the GFP-labelled E. coli; then the mixture was mixed with 2×106 mbn-2 cells. The mean GFP fluorescence intensity associated with mbn-2 cells was measured using a flow cytometer. Results are means±S.D. (n=3). (B) Inhibition of the effects of 50 mM sugars on the association of DL1 with E. coli. Gal, galactose; Xyl, xylose. Results are means±S.D. (n=4); *P<0.05.
Generation of the lectin locus deletion mutant
The lectin genes DL1, DL2 and DL3 localize at 37D on the second chromosome. From a database, we found the P-insertion line EP(2)1173 in which the P element was inserted 27 kb 3′-downstream of the lectin genes (Figure 7A). The P-insertion site was on the 5′-UTR of the Sd gene. We used the P{ry+(Δ2-3)}(99B) jumpstarter element to mobilize the inserted P element and screened transposed lines for novel insertion sites by PCR analysis to generate the mutant [15]. From 250 transposed lines on the second chromosome, one line, named #914, carried the P element 6.4 kb 3′-downstream of the DL1 gene (Figure 7A).
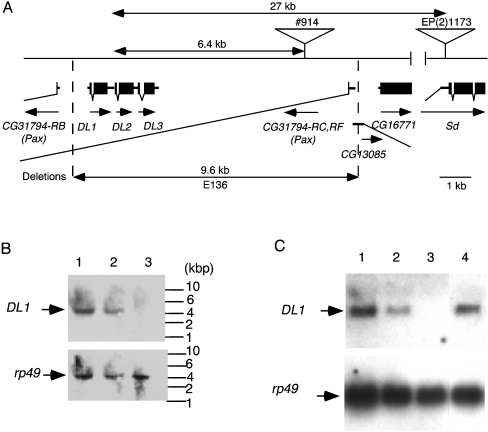
Figure 7. Generation of the deletion mutant of the lectin gene locus.
(A) Genomic map of 37D, the mutant and the progenitors. The #914 line was generated by local transposition of the P element from EP(2)1173, followed by generation of E136 by imprecise excision. Transcripts of the DL1 gene and the neighbouring genes are shown. Coding parts of the transcripts are shown as closed boxes; UTRs are shown as bars. Southern (B) and Northern (C) blot analysis of the DL1 gene. In the upper panel, the filter was probed for DL1 expression. In the lower panel, the same filter was reprobed with the rp49 probe as a loading control. Lane 1, w1118; lane 2, w1118; DL1E136/CyO; lane 3, w1118; DL1E136/DL1E136; lane 4, w1118; DL1E136/DL1E136; P{DL1}/P{DL1}. (B) Southern blot analysis of the DL1 gene. Sizes are indicated in kb. (C) Northern blot analysis of the DL1 gene. Total RNA isolated from the third instar larvae was analysed.
Homozygous flies for the #914 chromosome reached adulthood, but died soon after emerging from pupae. This phenotype reverted to wild-type after the precise excision of the transposed P element. This phenotype was mapped genetically to the cytological area containing the P-insertion site by deficiency mapping. These results indicate that the P-insertion at the site caused the phenotype.
We mobilized the inserted P element from the #914 line and used PCR analysis to screen the excision lines for deletions, thereby generating the null-mutant for the lectin genes. From 79 excised lines, one line, named E136, carried approx. 9.6 kb of deletions that covered the DL1, DL2 and DL3 genes (Figure 7A). The deletion site was determined precisely by sequencing the PCR product obtained from the analysis. This line also lacked the first exon of the CG31974-RC and -RF genes (Paxillin) and the part of the first exon of the CG13085 gene, a predicted gene whose expression and function are unknown.
Deletion of the DL1 gene from the E136 fly genome was confirmed using Southern blot analysis (Figure 7B). Figure 7(C) shows that the expression of the lectin gene in the mutant larvae was not detected by Northern blot analysis. These results indicate that the E136 line is the null-mutant for the DL1 gene.
Homozygous flies for the E136 chromosome, just as with the #914 line, reached adulthood, but the adults died soon after emerging from pupae. This phenotype was mapped to the cytological area containing the P-excised site by deficiency mapping, suggesting that the P-insertion and the following excision caused the phenotype. In addition, the phenotype was not rescued by the transgene of the DL1 gene. For that reason, the lethality was not likely to be the result of DL1 mutation; rather, it was caused by the other neighbouring gene's mutation.
Persistence of E. coli in the E136 larvae
We examined whether the E136 homozygous larvae had some defect in their defence against bacteria. As shown in Figure 5(A), DL1 recombinant protein strongly bound to an E. coli strain, so we considered the possibility that the lectin participated in eliminating the infecting bacteria. The E136 homozygous adult flies die soon after emerging from pupae, so we focused our attention on the immune response during the larval stage. Drosophila larvae are known to induce expression of various antibacterial peptide genes in response to infection with Gram-negative bacteria [1–5]. Expression was induced by IMD pathway activation, which is indispensable for defence against infection by Gram-negative bacteria [16]. We examined the expression of antibacterial peptide genes after infection with E. coli.
Merely pricking the body wall of Drosophila larvae induces the immune response [17]; we could not detect a difference in the induction of antimicrobial peptide genes by septic injury between the E136 larvae and wild-type (results not shown). Therefore we examined whether we could infect Drosophila larvae with the E. coli strain naturally, as do phytopathogenic bacteria Erwinia carotovora, to avoid pricking [13]. Expression of the gene for an antibacterial peptide Cecropin A1 was detected in fat body by the coexistence of larvae with the bacteria, indicating the natural infection of the Drosophila larvae with the E. coli strain and the consequent systemic immune response (results not shown).
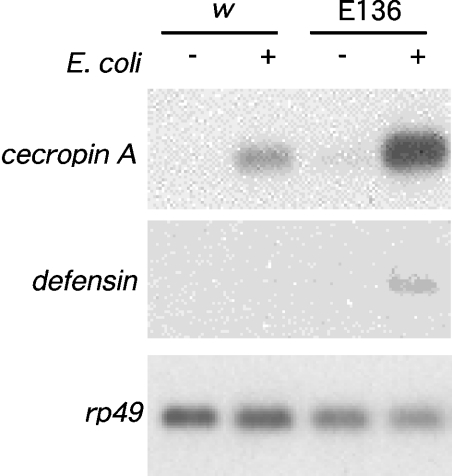
At that time, we infected the E136 larvae naturally with E. coli, and examined the expression of antibacterial peptide genes. Figure 8 shows that expression of the Cecropin A gene was detected in wild-type larvae 3 h after infection. The Cecropin A gene expression in the E136 larvae was more intensified than in the wild-type larvae. Such enhanced expression in the E136 larvae was also detected in another antibacterial peptide gene: Defensin. These results indicate that DL1 does not participate in the immune response by induction of the antibacterial peptides.
Figure 8. Expression of the antibacterial peptide genes after natural infection with E. coli in the lectin deletion mutant larvae.
Total RNA was isolated from the third instar larvae of w1118 (w) or the DL1 mutant E136 3 h after infection with (+) or without (−) bacteria. Probes: Cecropin A (top panel), Defensin (middle panel) and rp49 as a loading control (bottom panel).
DISCUSSION
We analysed the expression and activity of three lectins arranged as a gene cluster at 37D on the genome of D. melanogaster. The genome structure and similarity of the sequences of these three genes implies that an original ancestral gene might be duplicated to create the DL3 gene and another gene, and that the latter gene was duplicated again to produce the DL1 and DL2 genes later on.
We showed that these three lectins are secreted galactose-specific C-type lectins and have similar binding specificities to the carbohydrate chains on red blood cells. The result for DL2 is exactly that expected from the high homology with DL1. However, the result for DL3 is not expected because DL3 has, at most, 30% identity in the carbohydrate-recognition domain with DL1 and DL2. Regarding Sarcophaga lectin, its carbohydrate-recognition domain had 30% identity with that of DL1, but Sarcophaga lectin showed high agglutinating activity against sheep red blood cells [18], which is different from that of DL1. Apparently, the multiplication of three genes at the locus does not produce variation in activity as haemagglutinin.
In spite of their similar activity, the expression patterns of these lectin genes differed markedly. First, expression of the DL1 gene was higher in the early pupal stage than in the late pupal stage, whereas the expression of the DL2 gene was higher at the late pupal stage. Secondly, in the midgut and Malpighian tubules at the larval stage, the DL1 gene was not expressed at detectable levels, but the DL2 and DL3 genes were. In contrast, in some other tissues where the DL1 gene was highly expressed, the DL2 and DL3 genes were not expressed. Finally, the expression of these genes in the reproductive organs differed from each other. These results are surprising because these lectin genes are adjacent to each other on the genome, and the specific regulation of these genes using the sequences on the gaps was thought to be improbable. Each gene's expression mechanism is yet to be elucidated, but the upstream and downstream sequences of the gene cluster are thought to act as cis-elements. Generation of lectins with different expression patterns presents the possibility of extending the original lectin function. It is an attractive question how the variation of the expression pattern was obtained following lectin gene multiplication.
Regulation of insect C-type lectin genes is not yet well characterized. It is noteworthy that a typical NF-κB (nuclear factor κB)-binding site exists upstream of the DL1 gene up-regulated by body injury; it is a common regulatory element among various insect immune genes [1–5,19,20]. Previously, we reported that the Sarcophaga lectin promoter was activated in various digestive tissues and fat body of transgenic Drosophila [21]. Common features of the regulation of insect lectin genes may be revealed in further investigations.
The DL1, DL2 and DL3 genes were expressed in various larval tissues including fat body and haemocytes, which are known as tissues that secrete various proteins to haemolymph. These lectins are secreted proteins, suggesting the localization of these lectins in haemolymph, which is similar to those of many lectins found in insects [6,7]. In haemocoel, the systemic immune system functions along with both humoral and cellular immune responses against invading pathogens. These results suggest that these lectins participate in immune responses.
By virtue of the D. melanogaster genome project, the Drosophila genome is predicted to encode more than 30 C-type lectins [8]. The predicted lectin genes are distributed to all chromosomes except the fourth and Y chromosomes, and many members of this family also arrange as gene clusters [11]. The possibility exists that they have a common ancestor until a recent stage in evolution. Comparative analysis of these Drosophila C-type lectin genes will shed light on the elucidation of the evolution of C-type lectins.
We demonstrated that DL1 bound to an E. coli and some Erwinia chrysanthemi strains. The most abundant polysaccharide chains on the surface of Gram-negative bacteria were those of LPS. However, haemagglutinating activity of DL1 against rabbit red blood cells was not inhibited by addition of commercially available LPS (results not shown), suggesting that DL1 did not bind to them via LPS, but via other polysaccharide chains on their surfaces.
In some other insect species, multiple lectins have been identified. In American cockroach, Periplaneta americana, a C-type lectin named LPS-binding protein binds to LPS from E. coli, but not to LPS from Salmonella serotype Minnesota [22]. On the other hand, another C-type lectin named Periplaneta lectin binds to both of them when their 2-oxo-3-deoxyoctonate residues are exposed [23]. Both lectins have been suggested to function as opsonins upon infection [23,24]. In another cockroach, Blaberus discoidalis, three C-type lectins with different sugar specificities and a β-1,3-glucan-specific lectin have been identified [25,26]. They promote the activation of the prophenoloxidase system and phagocytosis against different spectra of micro-organisms [27,28]. Therefore, although DL1 binds to the restricted strains of bacteria, other C-type lectins could recognize other microbial components in the same way. The C-type lectin family in Drosophila may recognize many strains of pathogens collectively.
Furthermore, DL1 increased the association of the Drosophila haemocyte-derived cell line with E. coli. Probably by agglutinating E. coli in haemolymph of Drosophila, a larger number of E. coli can be trapped by haemocytes via the receptor for E. coli on the cell surface, such as dSR-C1 [29]. The present paper makes the first biochemical suggestion of lectin participation in immunity in D. melanogaster.
We also established the null-mutant for the DL1/DL2/DL3 gene locus. However, expression of antibacterial peptide genes was not impaired in the E136 larvae, suggesting that DL1 does not participate in the elimination of bacteria by the induction of anti-bacterial peptide genes. Enhancement of the expression of the antibacterial peptide genes in the E136 larvae is likely to result from the increased number of bacteria that persist in the larvae, rather than the loss of the inhibitory effect of the DL1 gene on expression, since more infected bacteria were detected in the mutant larvae at the early stage of infection (results not shown). Although we could not have shown in the present study the function of DL1 in humoral immune response in vivo, cellular immune responses such as phagocytosis might be affected by the DL1 gene mutation, possibly through the decreased number of associated bacteria with haemocytes as shown in Figure 6. Further analyses using the mutant that we established might lead to the elucidation of the function of lectins in the Drosophila cellular immune response.
Recently in Drosophila, proteins of the PGRP (peptidoglycan-recognition protein) family have been found to participate in the recognition of infecting bacteria [30,31]. They discriminate the different structures of the peptidoglycans between Gram-positive and Gram-negative bacteria, and activate appropriate immune responses, i.e. the IMD pathway or the Toll pathway. However, in Gram-negative bacteria, peptidoglycans are enveloped in thick cell-wall layers. As a result, the existence of the other kind of recognition proteins that recognize the cell wall components of Gram-negative bacteria, preceding recognition by PGRPs, will facilitate the efficient initiation of the immune response. Some C-type lectins will be involved in Drosophila immunity as such pattern-recognition receptors. In addition, although we could not detect the change in DL2 and DL3 gene expression by septic injury with E. coli, a recent study using microarray analysis has revealed that expression of at least one of the DL2 and DL3 genes was up-regulated by infection with a protozoan parasite, Octosporea muscaedomesticae [32]. Little has been elucidated about the recognition mechanism of non-bacterial pathogens in Drosophila, although there are some reports of the interaction of lectins with them in other insect species [26,33]. These results support the possibility that some C-type lectins also participate in immune response against non-bacterial pathogens.
Acknowledgments
We thank Dr T. Kubo and Dr M. Morioka for the use of a peptide sequencer. This work was supported by CREST (Core Research for Evolutional Science and Technology) of the Japan Science and Technology Corporation (JST). We thank Dr Y. Tony Ip from the University of Massachusetts Medical School at whose laboratory several experiments were performed.
References
- 1.Hultmark D. Drosophila immunity: paths and patterns. Curr. Opin. Immunol. 2003;15:12–19. doi: 10.1016/s0952-7915(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann J. A. The immune response of Drosophila. Nature (London) 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 3.Brennan C. A., Anderson K. V. Drosophila: the genetics of innate immune recognition and response. Annu. Rev. Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- 4.Leclerc V., Reichhart J. M. The immune response of Drosophila melanogaster. Immunol. Rev. 2004;198:59–71. doi: 10.1111/j.0105-2896.2004.0130.x. [DOI] [PubMed] [Google Scholar]
- 5.Naitza S., Ligoxygakis P. Antimicrobial defences in Drosophila: the story so far. Mol. Immunol. 2004;40:887–896. doi: 10.1016/j.molimm.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Natori S. Insect lectins and innate immunity. Adv. Exp. Med. Biol. 2001;484:223–228. doi: 10.1007/978-1-4615-1291-2_21. [DOI] [PubMed] [Google Scholar]
- 7.Franc N. C., White K. Innate recognition systems in insect immunity and development: new approaches in Drosophila. Microbes Infect. 2000;2:243–250. doi: 10.1016/s1286-4579(00)00304-x. [DOI] [PubMed] [Google Scholar]
- 8.Dodd R. B., Drickamer K. Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology. 2001;11:71R–79R. doi: 10.1093/glycob/11.5.71r. [DOI] [PubMed] [Google Scholar]
- 9.Haq S., Kubo T., Kurata S., Kobayashi A., Natori S. Purification, characterization, and cDNA cloning of a galactose-specific C-type lectin from Drosophila melanogaster. J. Biol. Chem. 1996;271:20213–20218. doi: 10.1074/jbc.271.33.20213. [DOI] [PubMed] [Google Scholar]
- 10.Pace K. E., Lebestky T., Hummel T., Arnoux P., Kwan K., Baum L. G. Characterization of a novel Drosophila melanogaster galectin: expression in developing immune, neural, and muscle tissues. J. Biol. Chem. 2002;277:13091–13098. doi: 10.1074/jbc.M112105200. [DOI] [PubMed] [Google Scholar]
- 11.Theopold U., Rissler M., Fabbri M., Schmidt O., Natori S. Insect glycobiology: a lectin multigene family in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 1999;261:923–927. doi: 10.1006/bbrc.1999.1121. [DOI] [PubMed] [Google Scholar]
- 12.Casso D., Ramírez-Weber F. A., Kornberg T. B. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 2000;91:451–454. doi: 10.1016/s0925-4773(00)00248-3. [DOI] [PubMed] [Google Scholar]
- 13.Basset A., Khush R. S., Braun A., Gardan L., Boccard F., Hoffmann J. A., Lemaitre B. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3376–3381. doi: 10.1073/pnas.070357597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hultmark D., Klemenz R., Gehring W. J. Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell. 1986;44:429–438. doi: 10.1016/0092-8674(86)90464-2. [DOI] [PubMed] [Google Scholar]
- 15.Tower J., Karpen G. H., Craig N., Spradling A. C. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzou P., Reichhart J. M., Lemaitre B. Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2152–2157. doi: 10.1073/pnas.042411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bettencourt R., Tanji T., Yagi Y., Ip Y. T. Toll and Toll-9 in Drosophila innate immune response. J. Endotoxin Res. 2004;10:261–268. doi: 10.1179/096805104225004897. [DOI] [PubMed] [Google Scholar]
- 18.Komano H., Mizuno D., Natori S. Purification of lectin induced in the hemolymph of Sarcophaga peregrina larvae on injury. J. Biol. Chem. 1980;255:2919–2924. [PubMed] [Google Scholar]
- 19.Sun S. C., Faye I. Cecropia immunoresponsive factor, an insect immunoresponsive factor with DNA-binding properties similar to nuclear-factor κB. Eur. J. Biochem. 1992;204:885–892. doi: 10.1111/j.1432-1033.1992.tb16708.x. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi A., Matsui M., Kubo T., Natori S. Purification and characterization of a 59 kilodalton protein that specifically binds to NF-κB-binding motifs of the defense protein genes of Sarcophaga peregrina (the flesh fly) Mol. Cell. Biol. 1993;13:4049–4056. doi: 10.1128/mcb.13.7.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanji T., Kobayashi A., Natori S. Activation of the Sarcophaga lectin gene promoter in transgenic Drosophila. Arch. Insect Biochem. Physiol. 2002;50:131–138. doi: 10.1002/arch.10039. [DOI] [PubMed] [Google Scholar]
- 22.Jomori T., Kubo T., Natori S. Purification and characterization of lipopolysaccharide-binding protein from hemolymph of American cockroach Periplaneta americana. Eur. J. Biochem. 1990;190:201–206. doi: 10.1111/j.1432-1033.1990.tb15565.x. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki K., Kubo T., Natori S. A novel role of Periplaneta lectin as an opsonin to recognize 2-keto-3-deoxy octonate residues of bacterial lipopolysaccharides. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1993;106:675–680. doi: 10.1016/0305-0491(93)90148-x. [DOI] [PubMed] [Google Scholar]
- 24.Jomori T., Natori S. Function of the lipopolysaccharide-binding protein of Periplaneta americana as an opsonin. FEBS Lett. 1992;296:283–286. doi: 10.1016/0014-5793(92)80305-z. [DOI] [PubMed] [Google Scholar]
- 25.Chen C., Ratcliffe N. A., Rowley A. F. Detection, isolation and characterization of multiple lectins from the haemolymph of the cockroach Blaberus discoidalis. Biochem. J. 1993;294:181–190. doi: 10.1042/bj2940181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C., Rowley A. F., Newton R. P., Ratcliffe N. A. Identification, purification and properties of a β-1,3-glucan-specific lectin from the serum of the cockroach, Blaberus discoidalis which is implicated in immune defence reactions. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1999;122:309–319. doi: 10.1016/s0305-0491(99)00020-6. [DOI] [PubMed] [Google Scholar]
- 27.Chen C., Durrant H. J., Newton R. P., Ratcliffe N. A. A study of novel lectins and their involvement in the activation of the prophenoloxidase system in Blaberus discoidalis. Biochem. J. 1995;310:23–31. doi: 10.1042/bj3100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson R., Chen C., Ratcliffe N. A. Innate immunity in insects: the role of multiple, endogenous serum lectins in the recognition of foreign invaders in the cockroach, Blaberus discoidalis. J. Immunol. 1999;162:1590–1596. [PubMed] [Google Scholar]
- 29.Rämet M., Pearson A., Manfruelli P., Li X., Koziel H., Göbel V., Chung E., Krieger M., Ezekowitz R. A. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity. 2001;15:1027–1038. doi: 10.1016/s1074-7613(01)00249-7. [DOI] [PubMed] [Google Scholar]
- 30.Dziarski R. Peptidoglycan recognition proteins (PGRPs) Mol. Immunol. 2004;40:877–886. doi: 10.1016/j.molimm.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 31.Steiner H. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol. Rev. 2004;198:83–96. doi: 10.1111/j.0105-2896.2004.0120.x. [DOI] [PubMed] [Google Scholar]
- 32.Roxström-Lindquist K., Terenius O., Faye I. Parasite-specific immune response in adult Drosophila melanogaster: a genomic study. EMBO Rep. 2004;5:207–212. doi: 10.1038/sj.embor.7400073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mello C. B., Nigam Y., Garcia E. S., Azambuja P., Newton R. P., Ratcliffe N. A. Studies on a haemolymph lectin isolated from Rhodnius prolixus and its interaction with Trypanosoma rangeli. Exp. Parasitol. 1999;91:289–296. doi: 10.1006/expr.1998.4385. [DOI] [PubMed] [Google Scholar]
- 34.Marshall R. D. The nature and metabolism of the carbohydrate-peptide linkages of glycoproteins. Biochem. Soc. Symp. 1974;40:17–26. [PubMed] [Google Scholar]
- 35.Takahashi H., Komano H., Kawaguchi N., Kitamura N., Nakanishi S., Natori S. Cloning and sequencing of cDNA of Sarcophaga peregrina humoral lectin induced on injury of the body wall. J. Biol. Chem. 1985;260:12228–12233. [PubMed] [Google Scholar]
- 36.Fujita Y., Kurata S., Homma K., Natori S. A novel lectin from Sarcophaga: its purification, characterization, and cDNA cloning. J. Biol. Chem. 1998;273:9667–9672. doi: 10.1074/jbc.273.16.9667. [DOI] [PubMed] [Google Scholar]
- 37.Jomori T., Natori S. Molecular cloning of cDNA for lipopolysaccharide-binding protein from the hemolymph of the American cockroach, Periplaneta americana: similarity of the protein with animal lectins and its acute phase expression. J. Biol. Chem. 1991;266:13318–13323. [PubMed] [Google Scholar]
- 38.Arai T., Kawasaki K., Kubo T., Natori S. Cloning of cDNA for regenectin, a humoral C type lectin of Periplaneta americana, and expression of the regenectin gene during leg regeneration. Insect Biochem. Mol. Biol. 1998;28:987–994. doi: 10.1016/s0965-1748(98)00087-3. [DOI] [PubMed] [Google Scholar]