Abstract
The EGFR (epidermal growth factor receptor; ErbB1) is frequently the subject of genetic changes in human tumours which contribute to the malignant phenotype by altering EGFR signalling. Examples of such genetic changes include overexpression, extracellular domain deletions and point mutations, and small deletions in the tyrosine kinase domain. We hypothesized that a point mutation in one of the EGFR ligand-binding domains would increase the affinity of EGFR for NRG2β (neuregulin-2β), which is not a potent stimulus of signalling by EGFR-Wt (wild-type EGFR). This mutation would permit NRG2β stimulation of EGFR signalling in settings in which NRG2β does not normally do so. To test this hypothesis, we have generated and evaluated various EGFR alleles containing mutations at Val441 and Ser442. NRG2β is a much more potent stimulus of the EGFR-S442F mutant than of EGFR-Wt. Furthermore, the affinity of NRG2β for the EGFR-S442F mutant is greater than the affinity of NRG2β for EGFR-Wt. Finally, the EGFR-S442F mutant constitutively suppresses apoptosis via phosphoinositide 3-kinase and Akt signalling but is not highly tyrosine phosphorylated in the absence of ligand. These results suggest that mutations in the EGFR ligand-binding domain in tumours may permit potent stimulation of EGFR signalling by ligands that are not normally potent EGFR agonists, thereby providing for a novel mechanism by which EGFR signalling may be deregulated. These results also suggest that novel EGFR mutations and signalling activities may be responsible for deregulated EGFR signalling in tumour cells.
Keywords: affinity, apoptosis, constitutive signal transduction, epidermal growth factor receptor, neuregulin-2β, phosphoinositide 3-kinase
Abbreviations: ECL, enhanced chemiluminescence; EGF, epidermal growth factor; EGFR, EGF receptor; EGFR-Wt, wild-type EGFR; IL3, interleukin-3; NRG, neuregulin; PEOE, partial equalization of orbital electronegativities; PI3K, phosphoinositide 3-kinase; TGFα, transforming growth factor α
INTRODUCTION
The ErbB family of receptor tyrosine kinases consists of four members, including the EGFR [EGF (epidermal growth factor) receptor; HER1/ErbB1], ErbB2 (HER2/Neu), ErbB3 (HER3) and ErbB4 (HER4). The naturally occurring agonists for these receptors are members of the EGF family of peptide hormones. The signalling network composed of these ligands and receptors plays important roles in mammalian development [1]. Moreover, increased signalling by EGFR or ErbB2 contributes to many types of human tumours [1,2].
Deregulated EGFR signalling may arise via a number of mechanisms. EGFR overexpression correlates with a more aggressive phenotype and poorer patient prognosis in a number of tumour types [2]. The EGFR VIII mutant is frequently detected in glioblastomas. This mutant contains a deletion in the EGFR extracellular domain that causes constitutive EGFR dimerization, phosphorylation and coupling with downstream effectors [2–5]. Substitution of an arginine residue for Leu858 in the EGFR kinase domain or deletion of Leu747 to Pro753 in the EGFR kinase domain results in increased EGFR coupling with antiapoptotic signalling pathways and increased sensitivity to the EGFR tyrosine kinase inhibitor gefitinib [6,7]. Moreover, substitution of methionine for Thr790 in the EGFR kinase domain confers resistance to gefitinib, possibly through steric hindrance of inhibitor binding [8,9]. Indeed, cells harbouring the EGFR T790M mutation retain sensitivity to irreversible EGFR tyrosine kinase inhibitors, which can overcome low affinity for EGFR [10].
We have hypothesized that EGFR mutants that display increased affinity for a ligand that normally displays minimal affinity for EGFR could now signal in the presence of that ligand, thereby dysregulating ligand-induced EGFR signalling and cellular growth control. However, efforts to generate such EGFR mutants have been hindered by a paucity of appropriate information concerning the interactions of EGF family hormones with their receptors.
Recent findings have addressed these issues. The EGF family hormone NRG2β (neuregulin-2β) exhibits only low affinity for EGFR but is a high-affinity agonist for ErbB4 [11,12]. We have previously shown that a hydrophobic residue at position 45 of NRG2β is sufficient and necessary for high-affinity NRG2β binding to ErbB4 [13,14]. Modelling studies based on the structure of ligand–EGFR complexes [15,16] indicate that the hydrophobic side chain of Phe45 of NRG2β appears to interact with a hydrophobic pocket in ErbB4 defined in part by Leu437 and Lys438 [13,17]. Indeed, in the present study, we demonstrate that substitution of a phenylalanine residue for Ser442 in EGFR is sufficient to markedly increase the affinity of NRG2β for EGFR and to permit more potent stimulation of EGFR tyrosine phosphorylation by NRG2β. Surprisingly, the EGFR-S442F mutant displays ligand-independent coupling with suppression of apoptosis. We discuss these findings in the context of the roles that EGFR mutations may be playing in tumorigenesis.
EXPERIMENTAL
Cell lines and cell culture
The Ψ2 and PA317 cell lines are gifts from Daniel DiMaio (Yale University, New Haven, CT, U.S.A.). The 32D mouse myeloid cell line was purchased from the American Type Culture Collection (Manassas, VA, U.S.A.). Cell lines were maintained according to the vendor's instructions or published procedures [19,20].
Reagents
Recombinant human EGF was purchased from Sigma. Recombinant NRG2β was generated as previously described [12,13]. 125I-EGF (105.8 μCi/μg) was purchased from Amersham. The EGFR tyrosine kinase inhibitor PD153035 was purchased from Tocris and the PI3K (phosphoinositide 3-kinase) inhibitors LY294002 and wortmannin were purchased from Calbiochem.
Plasmids and plasmid construction
The recombinant retrovirus expression vector pLXSN/EGFR [18] was used as a template for site-directed mutagenesis (Stratagene, La Jolla, CA, U.S.A.). Primer sequences are available upon request.
Production of recombinant retroviruses and retroviral infections
The EGFR constructs and established procedures [19] were used to generate high-titre amphotropic recombinant retrovirus stocks. Briefly, the constructs were transfected into the Ψ2 ecotropic retrovirus packaging cell line to generate low-titre ecotropic retrovirus stocks. The PA317 amphotropic retrovirus packaging cell line was infected with the ecotropic retrovirus particles to generate high-titre amphotropic retrovirus stocks. We used these stocks and established procedures [20] to generate pooled 32D mouse myeloid cell lines that express EGFR-Wt (wild-type EGFR) or mutant EGFR.
Analyses of EGFR and Akt expression and phosphorylation
We adapted established procedures [21,22] to assay ligand stimulation of EGFR tyrosine phosphorylation. Briefly, 32D/EGFR cells were incubated for 24 h in a medium devoid of serum to reduce basal EGFR tyrosine phosphorylation. Chilled aliquots containing 107 cells were incubated with ligand on ice for 7 min and lysed in an iso-osmotic buffer containing Nonidet P40 (50 mM Tris, pH 7.4, 120 mM NaCl and 0.5% Nonidet P40). Concanavalin A–Sepharose beads were used to precipitate glycoproteins (which include EGFR) from cleared cell lysates. The precipitates were resolved by SDS/PAGE and electroblotted on to nitrocellulose. EGFR tyrosine phosphorylation was assayed using an anti-phosphotyrosine mouse monoclonal antibody (Upstate), a goat anti-mouse antibody conjugated with horseradish peroxidase (Pierce), and ECL® (enhanced chemiluminescence; Amersham). EGFR expression was evaluated using an anti-EGFR sheep polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), a sheep anti-rabbit antibody (Pierce), a goat anti-rabbit antibody conjugated with horseradish peroxidase (Pierce) and ECL®.
Chemilumigrams were digitized using a UMAX Astra 2400S scanner at 600 dpi resolution. Images were manipulated using Adobe Photoshop and the bands were quantified using NIH Image. As a control, multiple samples of cells were stimulated with 10 nM EGF and concanavalin A precipitates were prepared and pooled. Increasing amounts of the precipitates (1–100%) were loaded on to a gel and immunoblotted using an anti-phosphotyrosine antibody to construct a loading response curve. A curve of best fit was generated using Microsoft Excel; the coefficients of correlation exceeded 0.96. The loading response curves were used to determine the band intensity that represents half-maximal receptor phosphorylation. This value was then used to calculate agonist EC50 values.
To assay ligand-induced EGFR coupling with Akt phosphorylation, 32D/EGFR cells were treated with ligand as described above. Cell lysates were resolved by SDS/PAGE and electroblotted on to nitrocellulose. Akt phosphorylation was detected using a rabbit polyclonal anti-phospho-Akt (phospho-Ser437) antibody (Cell Signaling Technology), a goat anti-rabbit antibody conjugated with horseradish peroxidase (Pierce) and ECL®. Akt expression was detected using a rabbit polyclonal anti-Akt antibody (Cell Signaling Technology), a goat anti-rabbit antibody conjugated with horseradish peroxidase (Pierce) and ECL®.
Analysis of ligand binding to EGFR
32D/EGFR cells or 32D/LXSN vector control cells were seeded in a 96-well plate at a density of 106 cells/well in 100 μl of PBS. Samples were incubated on ice for 2 h with 0.1–30 nM 125I-EGF (Amersham), and then transferred to a filter plate and washed using a cell harvester. The filter plate was dried and Microscint PS scintillation fluid (Packard) was added to each well. Radioscintography was performed using a TopCount scintillation counter. To determine the affinity of NRG2β for EGFR, we briefly incubated cells with 10 nM–10 μM NRG2β, and then incubated the cells on ice for 2 h with 1 nM 125I-EGF. Radioligand binding was assayed as described in [14]. Specific binding was determined by subtracting the amount of 125I-EGF bound to the 32D/LXSN cells. GraphPad Prism software was used to construct nonlinear regression two-site binding curves using the 125I-EGF binding data and to determine ligand dissociation constants (Kd). The software was also used to plot nonlinear regression curves for the competition binding data and to calculate inhibition constants (Ki).
Molecular modelling of a region of the extracellular domain of the EGFR-S442F mutant that surrounds Phe442
We depicted the area adjacent to Ser442 of the extracellular domain of the EGFR-Wt using the MOE (Molecular Operating Environment) version 2004.03 (Chemical Computing Group) program. The basis for this visualization is the crystal structure of the partially liganded EGFR-Wt (Protein Data Bank accession number 1NQL) [23]. (Modelling performed using the crystal structure of EGFR fully bound to EGF [16] yielded similar results.) We generated the S442F mutant by mutating the serine to phenylalanine. Hydrogens were added to the structure and partial charges were calculated using the Gasteiger [PEOE (partial equalization of orbital electronegativities)] force-field. An energy minimization of the mutated residue was performed using the MMFF94s force-field set to a 0.05 gradient with chiral constraints turned on. Molecular surfaces for the EGFR-Wt structure and for the model of the EGFR-S442F mutant were generated by adding hydrogens to the molecule and calculating the partial charge using the Gasteiger (PEOE) force-field.
Analysis of ligand-induced EGFR coupling with IL3 (interleukin-3)-independence
The 32D mouse myeloid cell line requires IL3 for survival and proliferation and 32D cells that lack IL3 undergo apoptosis. EGF stimulation of exogenously expressed EGFR in 32D cells causes IL3-independence [11]. Thus we used established procedures [12,13,18,21,22,24] to compare ligand-induced EGFR coupling with IL3-independence in 32D cells that express EGFR-Wt or the EGFR-S442F mutant. Briefly, quiescent 32D cells were seeded in a 24-well dish at a density of 105 cells/ml in medium lacking IL3, in medium supplemented with IL3, or in medium lacking IL3 and supplemented with EGF (0.1–30 nM) or NRG2β (10–300 nM). The cells were incubated for 4 days and counted using a haemocytometer. Three distinct responses were observed. Some cultures displayed cell viability of greater than 90% and a viable cell density of greater than 3×105 cells/ml; these cells were judged to be proliferating. Some cultures displayed cell viability of greater than 90% and a viable cell density of between 3×104 and 3×105 cells/ml; these cells were judged to be surviving but not proliferating. Some cultures displayed cell viability of less than 10% and a viable cell density of below 3×104 cells/ml; these cells were judged to be dying from apoptosis.
Inhibitor studies
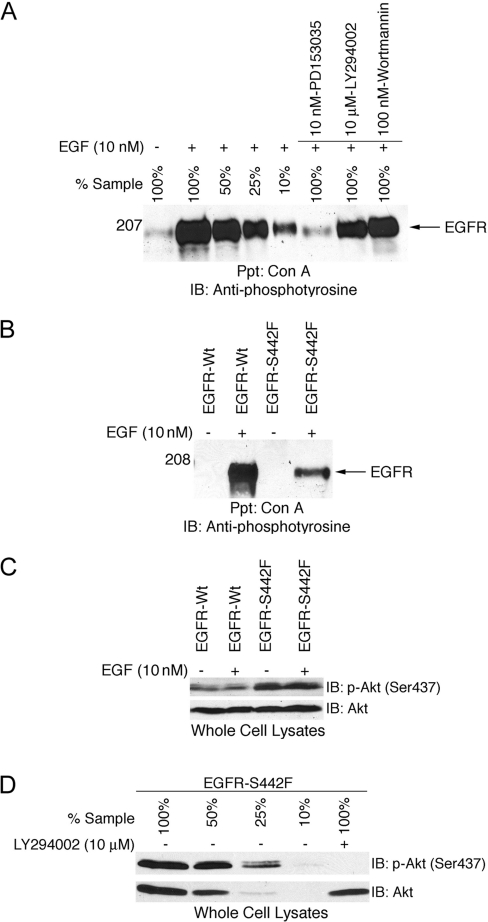
To assay inhibition of EGFR tyrosine phosphorylation, 32D/EGFR cells were incubated in a medium devoid of serum for 24 h. The cells were incubated with 10 nM PD153035, 10 mM LY294002 or 100 nM wortmannin for 2 h at 37 °C. Cells were stimulated with 10 nM EGF and EGFR tyrosine phosphorylation was assayed as described above. To assay inhibition of Akt phosphorylation, 32D/EGFR cells were incubated in a medium devoid of serum and IL3 for 8 h. Cells were treated with LY294002 (10 μM) and EGF as described above. Akt phosphorylation was assayed as described above.
RESULTS
The potential NRG2β binding site of ErbB4 includes Leu437 and Lys438, which correspond to Val441 and Ser442 in EGFR
NRG2β exhibits only low affinity for EGFR. However, NRG2β is a high-affinity agonist for ErbB4 and a hydrophobic phenylalanine residue at residue 45 of NRG2β is critical for high-affinity binding to ErbB4 [11–14]. Modelling studies suggest that the hydrophobic side chain of Phe45 of NRG2β interacts with a hydrophobic pocket in ErbB4 defined in part by Leu437 and Lys438 [13,14,17], which correspond to EGFR Val441 and Ser442 (Figure 1). Thus we hypothesized that changing EGFR Val441 or Ser442 to the corresponding ErbB4 leucine or lysine residues would increase the affinity of NRG2β for EGFR. We also hypothesized that substituting a hydrophobic leucine or phenylalanine residue for Ser442 of EGFR would increase the affinity of NRG2β for EGFR. Consequently, we generated four EGFR point mutants (V441L, S442K, S442L and S442F) and stably expressed them in the 32D mouse myeloid cell line, which is devoid of endogenous ErbB family receptor expression [11].
Figure 1. Alignment of the ErbB4 potential ligand-binding domain with the corresponding region in EGFR, ErbB2 and ErbB3.
Indicated in boldface, underlined type are the amino acids in ErbB4 that are being implicated in NRG2β binding. The corresponding amino acids of EGFR, ErbB2 and ErbB3 are indicated by boldface, underlined type. Also shown is the relative affinity of NRG2β for each receptor.
NRG2β stimulates the tyrosine phosphorylation of the EGFR-S442F mutant
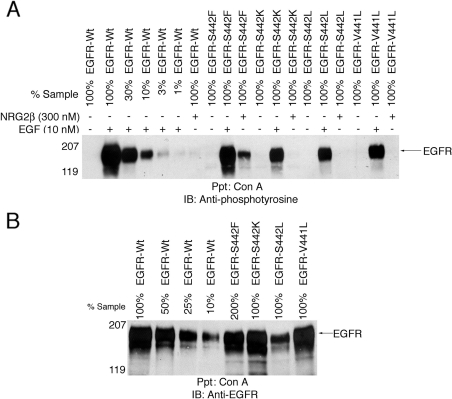
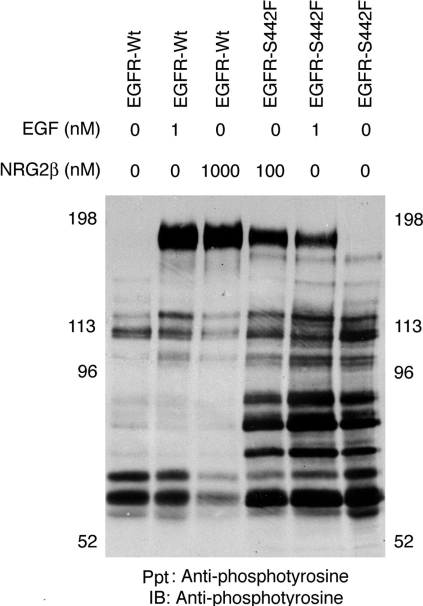
We stimulated 32D cells that express the EGFR-Wt or the EGFR mutants with EGF (10 nM) or NRG2β (300 nM). EGF markedly stimulated the tyrosine phosphorylation of EGFR-Wt and of the mutants. NRG2β stimulated the tyrosine phosphorylation of the EGFR-S442F mutant to a much greater extent than EGFR-Wt or the other mutants (Figure 2A). The minimal amount of NRG2β-induced tyrosine phosphorylation displayed by EGFR-Wt or the S442K and V441L mutants was not due to reduced expression relative to S442F (Figure 2B).
Figure 2. NRG2β stimulates the tyrosine phosphorylation of the EGFR-S442F mutant.
Following ligand stimulation, EGFR tyrosine phosphorylation and expression were analysed by immunoblotting as described above. A loading response curve constructed using positive control lysates was used for analysis. The position of the molecular mass standards is indicated. EGFR is represented by a band of approx. 180 kDa. The results shown are representative of three independent experiments. (A) 32D cells that express EGFR-Wt or the S442F, S442K, S442L or V441L mutant were stimulated with 10 nM EGF or 300 nM NRG2β. (B) The expression of EGFR-Wt or the EGFR mutants was analysed in the absence of ligand stimulation. Anti-EGFR, a sheep polyclonal anti-EGFR antibody; Anti-phosphotyrosine, a mouse monoclonal anti-phosphotyrosine antibody; Con A, concanavalin A–Sepharose beads; IB, the immunoblotting reagent; Ppt, the precipitating reagent.
NRG2β stimulates the tyrosine phosphorylation of the EGFR-S442F mutant more potently than it stimulates the tyrosine phosphorylation of EGFR-Wt
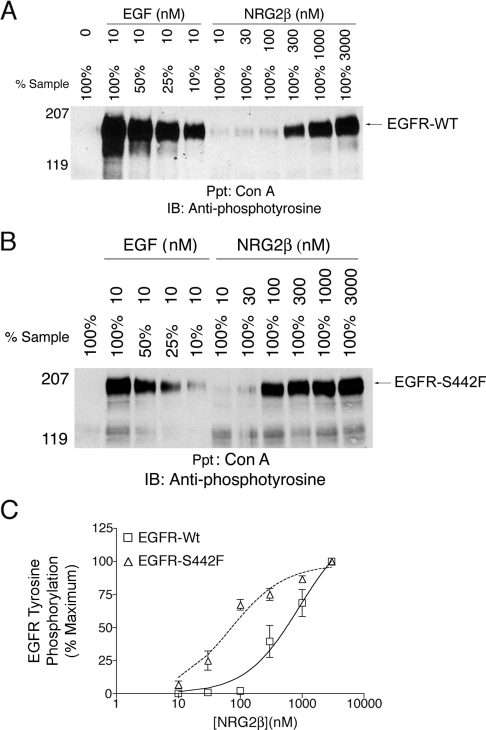
At high concentrations, NRG2β stimulated abundant tyrosine phosphorylation of both EGFR-Wt (Figure 3A) and of the S442F mutant (Figure 3B). However, the EC50 for NRG2β stimulation of EGFR-Wt is approx. 890 nM, whereas the EC50 for NRG2β stimulation of the S442F mutant is approx. 70 nM (Figure 3C). This difference suggests that NRG2β has a higher affinity for the S442F mutant than it has for EGFR-Wt. The S442F mutant is expressed at a slightly lower level in the 32D cells than is EGFR-Wt (Figure 2B). This suggests that the observed increase in NRG2β potency is not due to greater expression of S442F.
Figure 3. NRG2β stimulates the tyrosine phosphorylation of the EGFR-S442F mutant more potently than it stimulates the tyrosine phosphorylation of EGFR-Wt.
Following ligand stimulation, EGFR tyrosine phosphorylation was analysed as described above. (A) 32D cells that express EGFR-Wt were stimulated with increasing concentrations of NRG2β. 32D cells that express EGFR-Wt were stimulated with 10 nM EGF for comparative purposes. Results shown are representative of three independent experiments. The position of each of the molecular mass standards is indicated. Tyrosine-phosphorylated EGFR is represented by a band of approx. 180 kDa. (B) In parallel, 32D cells that express the EGFR–S442F mutant were stimulated with increasing concentration of NRG2β. 32D cells that express the EGFR–S442F mutant were stimulated with 10 nM EGF for comparative purposes. Results shown are representative of three independent experiments. The position of the molecular mass standards is indicated. Tyrosine-phosphorylated EGFR is represented by a band of approx. 180 kDa. (C) The Figure shows a representation of the means calculated from three independent experiments. Error bars represent the standard error of the means.
EGF stimulates the tyrosine phosphorylation of EGFR-Wt and of the EGFR-S442F mutant with similar potency
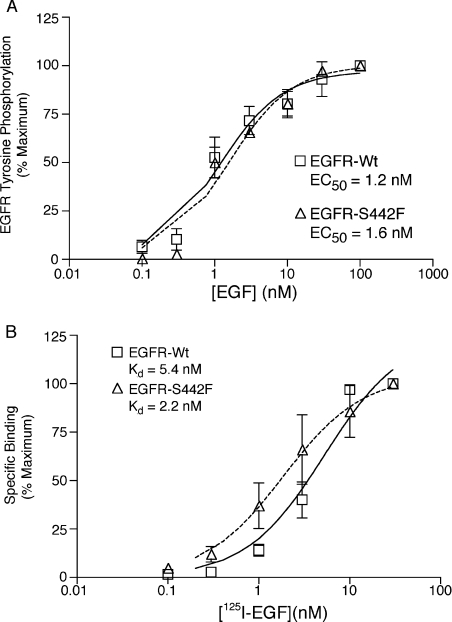
Next we considered the possibility that the apparent increased affinity of the S442F mutant for NRG2β may not be specific for NRG2β. We assayed tyrosine phosphorylation of EGFR-Wt or of S442F following stimulation with increasing concentrations of EGF. The EC50 for EGF stimulation of tyrosine phosphorylation of EGFR-Wt is approx. 1.2 nM, whereas the EC50 for EGF stimulation of tyrosine phosphorylation of S442F is approx. 1.6 nM (Figure 4A). This slight difference in EGF potency may be due to the slightly lower level of S442F expression (Figure 2B), suggesting that the increased potency of S442F stimulation by NRG2β is specific to this ligand and that S442F displays increased affinity for NRG2β.
Figure 4. EGF stimulates the tyrosine-phosphorylation of EGFR-Wt and of the EGFR-S442F mutant with similar potency and binds the receptors with similar affinity.
(A) Following stimulation with EGF, the phosphorylation of EGFR-Wt or of the EGFR-S442F mutant was analysed as described above. The Figure shows a representation of the means for three independent experiments. Error bars represent the standard error of the means. EGF EC50 values (with respect to stimulation of EGFR-Wt or of the EGFR-S442F mutant) are indicated. (B) The specific binding of 125I-EGF to EGFR-Wt or to the EGFR-S442F mutant was analysed as described above. The Figure shows a representation of the means for three independent experiments. Error bars represent the standard error of the means. EGF dissociation constants (Kd) (with respect to binding to EGFR-Wt or to the EGFR-S442F mutant) are indicated.
EGF binds EGFR-Wt and the EGFR–S442F mutant with similar affinity
We formally tested the latter prediction by determining the affinity of EGF for 32D cells that express EGFR-Wt or the S442F mutant. The dissociation constant (Kd) of radiolabelled EGF for EGFR-Wt is 5.4 nM and is 2.2 nM for S442F (Figure 4B). Thus, in concordance with the potency data, EGF appears to have similar affinity for EGFR-Wt and for S442F. We believe that the apparent minimal difference in the affinity of EGF for EGFR-Wt and for S442F is the result of experimental variability.
NRG2β inhibits EGF binding to the EGFR-S442F mutant more potently than it inhibits EGF binding to the EGFR-Wt
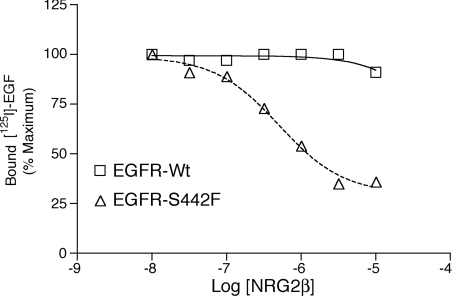
Our inability to generate radiolabelled NRG2β of sufficient specific radioactivity prevents us from directly measuring the affinity of NRG2β for EGFR-Wt and for the EGFR-S442F mutant. Instead, we adapted the EGF binding assay to assess whether the difference in potency of NRG2β with respect to EGFR-Wt and to S442F reflects a difference in the affinity of NRG2β for EGFR-Wt and for S442F. We performed competition binding experiments using 125I-EGF and increasing concentrations of unlabelled NRG2β. At 10 μM, NRG2β only partially abrogates the binding of 125I-EGF to EGFR-Wt. In contrast, the binding of 125I-EGF to S442F is markedly reduced by 10 μM NRG2β, and NRG2β exhibits an inhibitory constant (Ki) of approx. 218 nM (Figure 5). This apparent increased affinity of the S442F mutant for NRG2β is consistent with the fact that NRG2β is a more potent agonist for the S442F mutant than it is for EGFR-Wt.
Figure 5. NRG2β inhibits EGF binding to the EGFR-S442F mutant more potently than it inhibits EGF binding to the EGFR-Wt.
32D cells that express the EGFR-Wt or the EGFR-S442F mutant were incubated with increasing concentrations of NRG2β and 1 nM 125I-EGF. The net binding of 125I-EGF to EGFR was determined as described above. The graph shown is representative of results obtained from three independent experiments.
The ability of an unlabelled ligand to abrogate binding of a radiolabelled ligand to a receptor is a function of the concentration of the radiolabelled ligand as well as the relative affinities of the ligands for the receptor. Thus, given the relatively high concentration of radiolabelled EGF (1 nM) and the high affinity of EGF for EGFR-Wt, it is not surprising that 10 μM NRG2β only minimally abrogates labelled EGF binding to EGFR-Wt, despite the fact that 3 μM NRG2β stimulates abundant tyrosine phosphorylation of EGFR-Wt.
The EGFR-S442F mutation appears to create a hydrophobic bulge on the surface of the EGFR
The lone electron pair present in the side chain hydroxy group of the Ser442 residue of EGFR-Wt is present on the surface of the molecule [16,23] and is depicted in purple in Figure 6(A). We used this structure to develop a model for the structure of the extracellular domain of the S442F mutant. The bulky aromatic side chain of the Phe442 residue of S442F is predicted to be present on the cell surface and to create a hydrophobic bulge (depicted in green in Figure 6B) that should enable hydrophobic–hydrophobic interactions with Phe45 of NRG2β.
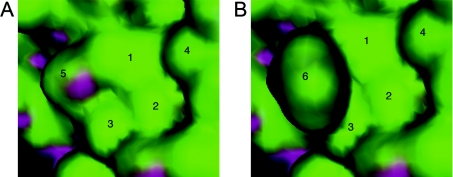
Figure 6. The S442F mutation in EGFR appears to create a hydrophobic bulge in the EGFR ligand-binding domain.
(A) The structure of a portion of the extracellular domain of EGFR-Wt is depicted. (B) A computer-generated model of a portion of the extracellular domain of the EGFR-S442F mutant is depicted. For both images, the area surrounding Ser442 or Phe442 is depicted. Green indicates a hydrophobic surface and purple indicates areas of electron lone pairs capable of hydrogen bonding. Residues are numbered as follows: (1) Val441; (2) Ser464; (3) Gly465; (4) Ile462; (5) Ser442; and (6) Phe442.
EGFR-Wt and the EGFR-S442F mutant display differential ligand-induced and ligand-independent coupling with IL3-independence
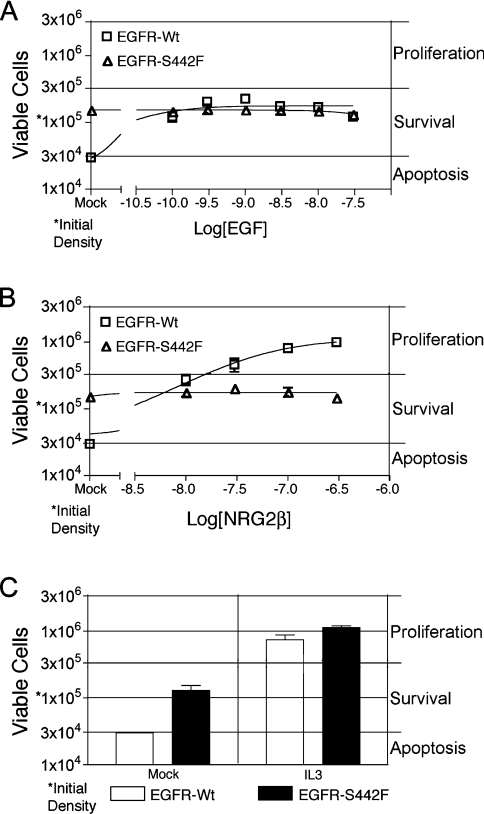
Next we assessed whether ligand-induced EGFR coupling with biological responses is altered in the S442F mutant. 32D cells are dependent on IL3 for survival and proliferation and IL3 with-drawal causes apoptosis. In 32D/EGFR cells, EGF stimulates EGFR coupling with antiapoptotic signalling pathways and IL3-independence [11,25]. Consequently, we tested whether EGF or NRG2β stimulates receptor coupling with IL3-independence in the 32D cell lines (Figure 7). In the absence of IL3, 32D cells that express EGFR-Wt remained at the seeding density and a high level of viability following stimulation with EGF (Figure 7A). These results indicate that EGF stimulates EGFR coupling with IL3-independent survival. In contrast, NRG2β stimulated an increase in the viable cell density of 32D cells that express EGFR, indicating that it stimulates EGFR coupling with IL3-independent proliferation (Figure 7B). Naturally, both ligands failed to stimulate any IL3-independence in 32D cells devoid of ectopic EGFR expression (results not shown). These results suggest that EGF and NRG2β stimulate EGFR coupling with distinct signalling pathways.
Figure 7. EGFR-Wt and the EGFR-S442F mutant display differential ligand-induced and ligand-independent coupling with IL3-independence.
IL3-independence of 32D cells that express the EGFR-Wt or the EGFR-S442F mutant was assessed following ligand stimulation as described above. Briefly, cells were seeded in IL3-free medium at a density of 1×105 cells/ml and were incubated for 4 days with (A) increasing concentrations of EGF or (B) increasing concentrations of NRG2β. (C) As a control, cells were incubated with 10% medium conditioned for 5 days by the WeHi-3B mouse myelomonocytic leukaemia cell line. This conditioned medium contains IL3. Following incubation for 4 days, viable cell density was calculated using a haemacytometer. The results shown are averages of four independent experiments. Error bars represent the standard error of the means. In many cases, the error bars do not extend beyond the boundaries of the symbol used to depict the mean data point.
We were surprised to note that, unlike EGFR-Wt, S442F couples with IL3-independent survival in the absence of ligand stimulation (Figures 7A–7C). The results shown in Figures 2(A) and 3(B) failed to suggest that S442F displays substantial ligand-independent tyrosine phosphorylation. Moreover, neither EGF nor NRG2β stimulates coupling of S442F with IL3-independent proliferation (Figures 7A–7C). Thus the S442F mutation alters ligand-dependent and -independent EGFR signalling.
Next, we addressed the possibility that the 32D/S442F cells had acquired a somatic mutation that rendered them IL3-independent. We also addressed the possibility that S442F is coupled with IL3-independence independently of EGFR tyrosine kinase activity. We assayed ligand-dependent and -independent coupling of EGFR-Wt and of S442F with IL3-independence in the presence of the EGFR tyrosine kinase inhibitor PD153035. PD153035 potently inhibits EGF-induced EGFR coupling with IL3-independence in 32D cells that express EGFR-Wt (IC50=0.9±0.1 nM; n=3). This value is very similar to the IC50 value (2.6 nM) for PD153035 (with respect to inhibition of EGFR tyrosine phosphorylation) in BaF3 mouse lymphoid cells and is much lower than the IC50 value for PD153035 with respect to inhibition of ErbB2 (143 nM) and ErbB4 (49 nM) tyrosine phosphorylation [26]. Moreover, in the presence of IL3, 1000 nM PD15035 fails to inhibit proliferation of 32D cells that express EGFR-Wt or S442F. Thus these results indicate that PD153035 is indeed specific for EGFR.
PD153035 potently inhibits ligand-independent coupling of S442F with IL3-independence (IC50=0.4±0.2 nM; n=3). Moreover, PD153035 potently inhibits coupling of S442F with IL3-independence in the presence of EGF (IC50=2.4±0.2 nM; n=3). These results suggest that EGFR tyrosine kinase activity is required to couple S442F with IL3-independence in 32D cells. Moreover, the fact that PD153035 inhibits the coupling of EGFR-Wt and S442F with IL3-independence with very similar potencies suggests that the 32D cells that express S442F have not acquired a somatic mutation that renders them IL3-independent.
EGFR-Wt and the EGFR-S442F mutant couple with distinct signalling pathways
Next, we sought to compare signalling events downstream of EGFR-Wt and S442F. Lysates prepared from mock stimulated cells or from cells stimulated with EGF or NRG2β were analysed by anti-phosphotyrosine immunoprecipitation and immunoblotting (Figure 8). Ligand stimulation of EGFR-Wt and of S442F resulted in the tyrosine phosphorylation of a protein of approx. 180 kDa; presumably this is EGFR. Three bands of approx. 60–80 kDa represent tyrosine-phosphorylated proteins that appear to be specifically and constitutively coupled with S442F. These may be specific substrates for the kinase domain of S442F or they may be ligands for phosphorylated tyrosine residues specific to S442F.
Figure 8. EGFR-Wt and the EGFR-S442 mutant couple with distinct signalling pathways.
32D cells that express the EGFR-Wt or the EGFR-S442F mutant were treated with PBS, 1 nM EGF or 1000 nM NRG2β. Protein tyrosine phosphorylation was analysed by immunoprecipitation and immunoblotting with an anti-phosphotyrosine antibody (4G10). The position of the molecular mass standards is indicated. The band of approx. 180 kDa is presumed to be tyrosine-phosphorylated EGFR.
The EGFR-S442F mutant couples with IL3-independence via PI3K and Akt
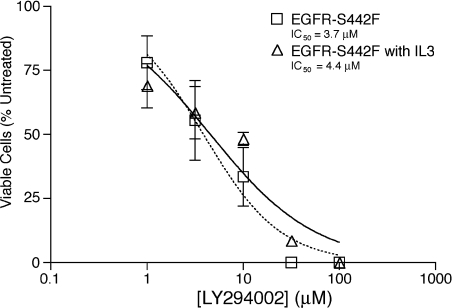
The IL3 receptor is coupled with survival and proliferation through PI3K [27]. Thus we hypothesized that S442F is constitutively coupled with IL3-independent survival via PI3K. We treated 32D cells that express S442F with the PI3K antagonist LY294002 in the presence and absence of IL3 and calculated the potency of LY294002 inhibition of cell survival or proliferation (Figure 9). The IC50 of LY294002 was 3.7 μM in the absence of IL3 and 4.4 μM in the presence of IL3. The similarity of these IC50 values suggests that S442F, like the IL3 receptor, suppresses apoptosis via PI3K. Indeed, wortmannin, another PI3K inhibitor, inhibits coupling of S442F with IL3-independence (results not shown).
Figure 9. The EGFR-S442F mutant couples with IL3-independence via PI3K.
32D cells that express the EGFR-S442F mutant were seeded at a density of 1×105 cells/ml in a medium containing IL3 or devoid of IL3. The effects of incubation for 4 days in increasing concentrations of the PI3K inhibitor LY294002 were determined. The results shown are the means for four independent experiments. Error bars represent the standard error of the means.
Nonetheless, we wished to rule out the possibility that wortmannin and LY294002 were inhibiting coupling of the EGFR–S442F mutant with IL3-independence by directly inhibiting EGFR tyrosine kinase activity. The EGFR tyrosine kinase inhibitor PD153035 markedly inhibited EGF-induced tyrosine phosphorylation of S442F. In contrast, a physiologically relevant concentration of wortmannin (100 nM) and of LY294002 (10 μM) only slightly inhibited EGF-induced tyrosine phosphorylation of S442F (Figure 10A). Ligand dose–response experiments indicate that the level of EGFR tyrosine phosphorylation displayed following treatment with 100 nM wortmannin or 10 μM LY294002 is sufficient to couple with IL3-independence (results not shown). Thus the concentrations of wortmannin and LY294002 sufficient to disrupt coupling of the S442F mutant with IL3-independence are not sufficient to cause a physiologically relevant reduction in EGFR tyrosine phosphorylation. Thus it appears that wortmannin and LY294002 disrupt S442F coupling with IL3-independence by inhibiting PI3K activity.
Figure 10. PI3K antagonists do not markedly inhibit EGFR tyrosine-phosphorylation but do block the constitutive coupling of the EGFR-S442F mutant with Akt phosphorylation.
(A) 32D cells that express the EGFR-S442F mutant were incubated with 10 nM PD153035, 10 μM LY294002 or 100 nM wortmannin. Following stimulation with 10 nM EGF, EGFR tyrosine phosphorylation was analysed as described above. (B) 32D cells that express EGFR-Wt or the EGFR-S442F mutant were incubated in a medium devoid of serum and IL3. The cells were then stimulated with 10 nM EGF. EGFR tyrosine phosphorylation and (C) Akt phosphorylation (upper panel) and expression (lower panel) were analysed as described above. (D) 32D cells that express the EGFR-S442F mutant were incubated in a medium devoid of serum and IL3 and treated with 10 μM LY294002 or DMSO. Akt phosphorylation and expression were analysed as described above.
Since suppression of apoptosis by S442F is sensitive to PI3K antagonists, we hypothesized that S442F is coupled with Akt, a known PI3K effector. EGF stimulated a dramatic increase in the phosphorylation of EGFR-Wt and a moderate increase in the phosphorylation of S442F (Figure 10B). In studies using the same lysates, EGF did not stimulate coupling of EGFR-Wt with Akt phosphorylation (Figure 10C). Moreover, S442F is constitutively coupled with Akt phosphorylation (Figure 10C). These differences in Akt phosphorylation do not appear to be due to differences in Akt expression (Figure 10C). Thus these results provide additional evidence that S442F suppresses apoptosis via PI3K and Akt.
Finally, we evaluated whether the PI3K antagonist LY294002 inhibits the phosphorylation of Akt in cells that express S442F. We treated 32D cells that express S442F with a physiologically relevant concentration of LY294002 (10 μM) and assayed Akt phosphorylation and expression (Figure 10D). LY294002 markedly inhibited Akt phosphorylation but did not affect Akt expression. These results suggest that S442F suppresses apoptosis via PI3K coupling with Akt and not via some other PI3K effector.
DISCUSSION
The EGFR-S442F mutant displays increased affinity for NRG2β
In the present paper, we demonstrate that NRG2β is a more potent agonist for the EGFR-S442F mutant than for EGFR-Wt. Moreover, NRG2β only partially abrogates EGF binding to EGFR-Wt but completely abrogates EGF binding to S442F. Collectively, these results indicate that mutating a single amino acid in the ligand-binding pocket of EGFR-Wt to a hydrophobic residue increases the affinity of NRG2β for the EGFR.
It is unclear whether the increased affinity of S442F for NRG2β will result in increased stimulation of this mutant by NRG2β in vivo. Nonetheless, the results presented in this study suggest that relatively minor mutations in the EGFR extracellular domain may expand the repertoire of potent EGFR agonists and permit ligand-induced EGFR signalling in tissues that normally do not exhibit such signalling. As yet there are no data supporting the existence of such gain-of-function EGFR mutants in tumours. However, the germline EGFR-R497K polymorphism results in decreased TGFα (transforming growth factor α) affinity for EGFR and decreased EGF and TGFα potency with respect to mitogenesis [28]. Moreover, colorectal tumours that are homozygous for the EGFR-R497K polymorphism appear to be less aggressive [29]. Thus the proof-of-principle data presented here justify screening tumour samples for gain-of-function EGFR mutant alleles analogous to the S442F mutant described here.
The EGFR-S442F mutant does not display detectable ligand-independent tyrosine phosphorylation but constitutively suppresses apoptosis via PI3K and Akt
Here we demonstrate that the EGFR-S442F mutant constitutively suppresses apoptosis via PI3K and Akt. S442F does not appear to display increased basal (ligand-independent) tyrosine phosphorylation, yet suppression of apoptosis by S442F requires EGFR kinase activity. These results suggest that phosphorylation of a substrate other than the EGFR by S442F may be required for suppression of apoptosis by S442F. Indeed, three tyrosine phosphoproteins appear to be specifically and constitutively coupled with S442F. Identification of these phosphoproteins and investigation of the roles that they play in signalling by S442F are under way.
Another potential explanation for these results is that S442F is coupled with suppression of apoptosis via phosphorylation on only a very small set of EGFR tyrosine residues and that this low level of phosphorylation is undetectable using non-specific anti-phosphotyrosine antibodies. This model is consistent with our observations that ligand concentrations sufficient to stimulate ErbB receptor coupling with downstream signalling events may not be sufficient to induce detectable ErbB receptor tyrosine phosphorylation. This model is also consistent with the observation that EGFR kinase domain mutants that are coupled with suppression of apoptosis in NSCLC (non-small-cell lung carcinoma) cell lines sensitive to gefitinib are preferentially phosphorylated on Tyr845, Tyr992 and Tyr1068 [7]. However, our unpublished results fail to indicate that S442F is constitutively phosphorylated on any of these three residues (J. L. Gilmore and D. J. Riese, II, unpublished work). Thus genetic and biochemical analyses of the sites of tyrosine phosphorylation of S442F may be warranted to address this issue.
The implications of these results on attempts to define the roles that EGFR plays in tumorigenesis may be profound. These results indicate that screening tumour cells for high levels of EGFR tyrosine phosphorylation may not identify all tumours that exhibit constitutive EGFR coupling with biological responses. Furthermore, the use of focus formation assays or colony formation assays in semisolid medium to identify constitutively active EGFR alleles may miss mutants such as S442F that are not constitutively coupled with mitogenesis but are instead constitutively coupled with the suppression of apoptosis.
NRG2β stimulates EGFR coupling with suppression of apoptosis and mitogenesis, whereas EGF stimulates EGFR coupling with suppression of apoptosis but not mitogenesis
A plausible explanation for these results is that different ErbB receptor agonists may induce different conformational changes in the same cognate receptor. These differential conformational changes may then be translated into differential sites of receptor tyrosine phosphorylation and differential coupling with biological responses. Indeed, differential juxtapositioning of the receptor monomers within constitutively active homodimeric ErbB2 mutants can result in differential coupling of these mutants with biological responses [30]. Similar findings are observed with a set of constitutively active homodimeric ErbB4 mutants [19].
Different ErbB4 agonists stimulate distinct patterns of ErbB4 tyrosine phosphorylation and differential coupling with downstream signalling pathways and biological responses [12,13,31–33]. However, the individual sites of differential ligand-induced ErbB4 tyrosine phosphorylation have yet to be identified and the physiological relevance of these sites of differential phosphorylation has yet to be determined. Likewise, betacellulin and EGF stimulate EGFR tyrosine phosphorylation on different tyrosine residues with differential kinetics. Again, the relevance of these ligand-specific differences in sites of EGFR tyrosine phosphorylation has not been conclusively established [34]. Finally, the differential effects of the EGFR agonists TGFα and amphiregulin on the morphology of MDCK (Madin–Darby canine kidney) cells do not reflect differences in the affinity of these ligands for EGFR but instead appear to reflect differential ligand-induced EGFR coupling with mitogen-activated protein kinase and Src. However, the mechanism for ligand-specific EGFR signalling has yet to be elucidated [35].
Preliminary immunoblotting data (J. L. Gilmore and D. J. Riese, II, unpublished work) from our laboratory obtained using site-specific anti-phospho-EGFR antibodies do not indicate that EGF and NRG2β stimulate a qualitative difference in EGFR phosphorylation at Tyr845, Tyr992, Tyr1045 and Tyr1068. This is not entirely unexpected given that we predict that NRG2β stimulates EGFR phosphorylation on a residue that is not phosphorylated following EGF stimulation. Phosphopeptide mapping of the sites of EGFR tyrosine phosphorylation following stimulation with EGF and NRG2β appears to be necessary to address this issue definitively.
Finally, it is possible that differential coupling of the EGFR-Wt with Akt phosphorylation following EGF and NRG2β stimulation may account for the difference in the physiological effects of stimulation with these two ligands. NRG2β stimulation may induce PI3K coupling with Akt, whereas EGF stimulation may induce PI3K coupling with a different PI3K effector. However, a mechanism by which this differential PI3K coupling may occur or may be regulated is not apparent.
Nonetheless, the results presented here clearly indicate that the efficacy of an EGFR agonist is not a function of its potency or affinity for EGFR. This is consistent with results from our laboratory indicating that the efficacy of NRG2 isoforms (with respect to stimulation of ErbB4 coupling with biological responses) is regulated independently of their potency and affinity for ErbB4 [12,13]. Moreover, these results suggest that it may be possible to create mutants of EGFR agonists that retain high-affinity EGFR binding yet fail to stimulate EGFR coupling with mitogenesis or suppression of apoptosis. This hypothesis is supported by preliminary results from our laboratory indicating that the NRG2β–Q43L mutant fails to stimulate ErbB4 coupling with downstream signalling events and biological responses and competitively antagonizes stimulation of ErbB4 signalling by wild-type NRG2β. Obviously, analogous mutants of EGFR agonists would hold significant potential as cancer chemotherapeutical agents.
Acknowledgments
We thank E. Williams for her assistance in generating the EGFR point mutants. We acknowledge support from the National Institutes of Health (R21CA080770 and R21CA089274 to D.J.R.), the U.S. Army Medical Research and Material Command (DAMD17-00-1-0415, DAMD17-00-1-0416 and DAMD17-02-0130 to D.J.R.), the Indiana Elks Foundation (to D.J.R.) and the American Cancer Society (IRG-58-006 to the Purdue Cancer Research Center).
References
- 1.Yarden Y., Sliwkowski M. X. Untangling the ErbB signaling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Hynes N. E., Lane H. A. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 3.Ekstrand A. J., Sugawa N., James C. D., Collins V. P. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc. Natl. Acad. Sci. U.S.A. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wikstrand C. J., Hale L. P., Batra S. K., Hill M. L., Humphrey P. A., Kurpad S. N., McLendon R. E., Moscatello D., Pegram C. N., Reist C. J. Monoclonal antibodies against EGFRVIII are tumor specific and react with breast and lung carcinomas and malignant gliomas. Cancer Res. 1995;55:3140–3148. [PubMed] [Google Scholar]
- 5.Sugawa N., Ekstrand A. J., James C. D., Collins V. P. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc. Natl. Acad. Sci. U.S.A. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch T. J., Bell D. W., Sordella R., Gurubhagavatula S., Okimoto R. A., Brannigan B. W., Harris P. L., Haserlat S. M., Supko J. G., Haluska F. G., et al. Activating mutations in the epidermal growth factor receptor underling responsiveness of non-small-cell lung cancer to Gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 7.Sordella R., Bell D. W., Haber D. A., Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi S., Boggon T. J., Dayaram T., Janne P. A., Kocher O., Meyerson M., Johnson B. E., Eck M. J., Tenen D. G., Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 9.Pao W., Miller V. A., Politi K. A., Riely G. J., Somwar R., Zakowski M. F., Kris M. G., Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLos Med. 2005;2:225–235. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak E. L., Sordella R., Bell D. W., Godin-Heymann N., Okimoto R. A., Brannigan B. W., Harris P. L., Driscoll D. R., Fidias P., Lynch T. J., et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinkas-Kramarski R., Shelly M., Guarino B. C., Wang L. M., Lyass L., Alroy I., Alimandi M., Kuo A., Moyer J. D., Lavi S., et al. ErbB tyrosine kinases and the two neuregulin families constitute a ligand–receptor network. Mol. Cell. Biol. 1998;18:6090–6101. doi: 10.1128/mcb.18.10.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hobbs S. S., Coffing S. L., Le A. T. D., Cameron E. M., Williams E. E., Andrew M., Blommel E. N., Hammer R. P., Chang H., Riese D. J., II Neuregulin isoforms exhibit distinct patterns of ErbB family receptor activation. Oncogene. 2002;21:8442–8452. doi: 10.1038/sj.onc.1205960. [DOI] [PubMed] [Google Scholar]
- 13.Hobbs S. S., Cameron E. M., Hammer R. P., Le A. T., Gallo R. M., Blommel E. N., Coffing S. L., Chang H., Riese D. J., II Five carboxyl-terminal residues of neuregulin2 are critical for stimulation of signaling by the ErbB4 receptor tyrosine kinase. Oncogene. 2004;23:883–893. doi: 10.1038/sj.onc.1207250. [DOI] [PubMed] [Google Scholar]
- 14.Hobbs S. S., Gallo R. M., Riese D. J., II Phe45 of NRG2β is critical for the affinity of NRG2β for ErbB4 and for potent stimulation of ErbB4 signaling by NRG2β. Growth Factors. 2005;23:273–283. doi: 10.1080/08977190500199345. [DOI] [PubMed] [Google Scholar]
- 15.Garrett T. P. J., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovercz G. O., Zhu H.-J., Walker F., Frenkel M. J., Hoyne P. A., et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 16.Ogiso H., Ishitani R., Nureki O., Fukal S., Yamanaka M., Kim J.-H., Saito K., Sakamoto A., Inoue M., Shirouzu M., et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domain. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 17.Luo C., Xu L., Zheng S., Luo X., Shen J., Jiang H., Liu X., Zhou M. Computational analysis of molecular basis of 1:1 interactions of NRG1β wild-type and variants with ErbB3 and ErbB4. Proteins Struct. Funct. Bioinform. 2005;59:749–756. doi: 10.1002/prot.20443. [DOI] [PubMed] [Google Scholar]
- 18.Riese D. J., II, van Raaij T. M., Plowman G. D., Andrews G. C., Stern D. F. The cellular response to neuregulins is governed by complex interactions of the erbB receptor family. Mol. Cell. Biol. 1995;15:5770–5776. doi: 10.1128/mcb.15.10.5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams E. E., Trout L. J., Gallo R. M., Pitfield S. E., Bryant I., Penington D. J., Riese D. J., II A constitutively active ErbB4 mutant inhibits drug-resistant colony formation by the DU-145 and PC-3 human prostate tumor cell lines. Cancer Lett. 2003;192:67–74. doi: 10.1016/s0304-3835(02)00690-0. [DOI] [PubMed] [Google Scholar]
- 20.Feroz K., Williams E., Riese D. J., II ErbB2 and ErbB3 do not quantitatively modulate ligand-induced ErbB4 tyrosine phosphorylation. Cell. Signalling. 2002;14:793–798. doi: 10.1016/s0898-6568(02)00019-0. [DOI] [PubMed] [Google Scholar]
- 21.Riese D. J., II, Kim E. D., Elenius K., Buckley S., Klagsbrun M., Plowman G. D., Stern D. F. The epidermal growth factor receptor couples the transforming growth factor-α, heparin-binding epidermal growth factor-like factor, and amphiregulin to Neu, ErbB-3, and ErbB-4. J. Biol. Chem. 1996;271:20047–20052. doi: 10.1074/jbc.271.33.20047. [DOI] [PubMed] [Google Scholar]
- 22.Riese D. J., II, Bermingham Y., van Raaij T. M., Buckley S., Plowman G. D., Stern D. F. Betacellulin activates the epidermal growth factor receptor and erbB-4 and induces cellular response patterns distinct from those stimulated by epidermal growth factor or neuregulin-β. Oncogene. 1996;12:345–353. [PubMed] [Google Scholar]
- 23.Ferguson K. M., Berger M. B., Mendrola J. M., Cho H.-S., Leahy D. J., Lemmon M. A. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 24.Riese D. J., II, Komurasaki T., Plowman G. D., Stern D. F. Activation of ErbB4 by the bifunctional epidermal growth factor family hormone epiregulin is regulated by ErbB2. J. Biol. Chem. 1998;273:11288–11294. doi: 10.1074/jbc.273.18.11288. [DOI] [PubMed] [Google Scholar]
- 25.Ettenberg S. A., Keane M. M., Nau M. M., Frankel M., Wang L. M., Pierce J. H., Lipkowitz S. Cbl-b inhibits epidermal growth factor receptor signaling. Oncogene. 1999;18:1855–1866. doi: 10.1038/sj.onc.1202499. [DOI] [PubMed] [Google Scholar]
- 26.VanBrocklin H. F., Lim J. K., Coffing S. L., Hom D. L., Negash K., Ono M. Y., Gilmore J. L., Bryant I., Riese D. J., II Anilinodialkoxyquinazolines: screening epidermal growth factor receptor tyrosine kinase inhibitors for potential tumor imaging probes. J. Med. Chem. 2005;48:7445–7456. doi: 10.1021/jm050607w. [DOI] [PubMed] [Google Scholar]
- 27.Songyang Z., Baltimore D., Cantley L. C., Kaplan D. R., Franke T. F. Interleukin 3-dependent survival by the Akt protein kinase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11345–11350. doi: 10.1073/pnas.94.21.11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moriai T., Kobrin M. S., Hope C., Speck L., Korc M. A variant epidermal growth factor receptor exhibits altered type α transforming growth factor α binding and transmembrane signaling. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10217–10221. doi: 10.1073/pnas.91.21.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W., Park D. J., Lu B., Yang D. Y., Gordon M., Groshen S., Yun J., Press O. A., Vallbohmer D., Rhodes K., et al. Epidermal growth factor receptor gene polymorphisms predict pelvic recurrence in patients with rectal cancer treated with chemoradiation. Clin. Cancer Res. 2005;11:600–605. [PubMed] [Google Scholar]
- 30.Burke C. L., Stern D. F. Activation of Neu (ErbB2) mediated by disulfide bond-induced dimerization reveals a receptor tyrosine kinase dimer interface. Mol. Cell. Biol. 1998;18:5371–5379. doi: 10.1128/mcb.18.9.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sweeney C., Lai C., Riese D. J., II, Diamonti A. J., Cantley L. C., Carraway K. L., III Ligand discrimination in signaling through an ErbB4 receptor homodimer. J. Biol. Chem. 2000;275:19803–19807. doi: 10.1074/jbc.C901015199. [DOI] [PubMed] [Google Scholar]
- 32.Sweeney C., Carraway K. L., III Ligand discrimination by ErbB receptors: differential signaling through differential phosphorylation site usage. Oncogene. 2000;19:5568–5573. doi: 10.1038/sj.onc.1203913. [DOI] [PubMed] [Google Scholar]
- 33.Amin D. N., Perkins A. S., Stern D. F. Gene expression profiling of ErbB receptor and ligand-dependent transcription. Oncogene. 2004;23:1428–1438. doi: 10.1038/sj.onc.1207257. [DOI] [PubMed] [Google Scholar]
- 34.Saito T., Okada S., Ohshima K., Yamada E., Sato M., Uehara Y., Shimizu H., Pessin J., Mori M. Differential activation of epidermal growth factor (EGF) receptor downstream signaling pathways by betacellulin and EGF. Endocrinology. 2004;145:4232–4243. doi: 10.1210/en.2004-0401. [DOI] [PubMed] [Google Scholar]
- 35.Chung E., Graves-Deal R., Franklin J. L., Coffey R. J. Differential effects of amphiregulin and TGF-α on the morphology of MDCK cells. Exp. Cell Res. 2005;309:149–160. doi: 10.1016/j.yexcr.2005.05.012. [DOI] [PubMed] [Google Scholar]