Abstract
The retinoblastoma protein pRb is required for cell-cycle exit of embryonic mammalian hair cells but not for their early differentiation. However, its role in postnatal hair cells is unknown. To study the function of pRb in mature animals, we created a new conditional mouse model, with the Rb gene deleted primarily in the inner ear. Progeny survive up to 6 months. During early postnatal development, pRb−/− hair cells continue to divide and can transduce mechanical stimuli. However, adult pRb−/− mice exhibit profound hearing loss due to progressive degeneration of the organ of Corti. We show that pRb is required for the full maturation of cochlear outer hair cells, likely in a gene-specific manner, and is also essential for their survival. In addition, lack of pRb results in cell division in postnatal auditory supporting cells. In contrast, many pRb−/− vestibular hair cells survive and continue to divide in adult mice. Significantly, adult pRb−/− vestibular hair cells are functional, and pRb−/− mice maintain partial vestibular function. Therefore, the functional adult vestibular pRb−/− hair cells, derived from proliferation of postnatal hair cells, are largely integrated into vestibular pathways. This study reveals essential yet distinct roles of pRb in cochlear and vestibular hair cell maturation, function, and survival and suggests that transient block of pRb function in mature hair cells may lead to propagation of functional hair cells.
Keywords: differentiation, proliferation, regeneration, maturation, conditional knockout
Hair cells of the inner ear perform the essential conversion of mechanical stimuli to neural signals for the senses of hearing and balance. Their development involves permanent exit from the cell cycle, fate determination, and differentiation into a functional hair cell. A basic helix–loop–helix transcription factor, Atoh1(Math1), has been identified as a key gene determining hair cell fate (1, 2). Other pathways, most notably the retinoic acid and Notch pathways, are involved in the induction of hair cells either from the progenitor cell pool or from the supporting cell pool at early stages of development (3–5). Hair cells of mammalian inner ears, unlike those in lower vertebrates, do not undergo spontaneous regeneration, even though vestibular supporting cells retain a limited capacity to divide (6, 7). As a consequence, damage to hair cells often results in irreversible deficits in hearing and balance. A key question in hair cell development is the identity of genes that regulate permanent cell-cycle exit of sensory progenitors and genes that maintain the quiescent postmitotic state of hair cells and supporting cells.
Some negative regulators of cell proliferation have been identified, notably p27kip1 (8, 9), p19ink4d (10), and pRb (11, 12), which control cell-cycle exit in the inner ear. p27kip1 is primarily involved in cell-cycle arrest of inner ear sensory progenitor cells and cochlear supporting cells, whereas p19ink4d plays a role in hair cell survival. Rb plays an essential role in cell-cycle exit of sensory progenitor cells and in maintenance of the quiescent state of early hair cells and supporting cells (11). In embryos, absence of pRb at embryonic day 10 (E10) causes an overproduction of sensory progenitor cells, which subsequently differentiate into hair cells and supporting cells. Remarkably, pRb−/− hair cells and supporting cells also continue to differentiate and express cellular markers appropriate for their embryonic stages. Furthermore, pRb−/− hair cells are able to transduce mechanical stimuli and appear capable of forming synapses with ganglion neurons. In mouse embryos, therefore, early differentiation and functional maturation of hair cells are pRb-independent.
The multiple roles of pRb have been well studied (13, 14). Underphosphorylated pRb interacts with its cellular targets (notably the E2F family of transcription factors) and suppresses their transcriptional activities, whereas phosphorylation of pRb by cyclin-dependent kinases releases pRb from its binding to E2F family members and promotes the transition from G1 to S phases of the cell cycle (15).
Homozygous Rb knockout animals die between E12.5 and E14.5 and exhibit severe defects in multiple systems, including neurogenesis defects (13). pRb becomes essential immediately after neuronal fate determination, and lack of pRb causes virtually all neuronal populations to undergo apoptosis (16, 17). However, in mice with Rb specifically deleted in the CNS, some neuronal populations increase because of aberrant S-phase entry, and they appear to differentiate relatively normally without apoptosis (18–20). Therefore, the role of pRb in differentiation of CNS cells, in addition to cell-cycle arrest, is likely cell type-dependent.
To understand the long-term roles of pRb in hair cell development and its effects on hearing and balance we created an inner ear Rb conditional knockout mouse model. Characterization of the inner ear of postnatal pRb−/− mice revealed essential and specific roles for pRb in the maturation and survival of auditory hair cells and somewhat different roles in the vestibular system. The studies support the concept that transient inactivation of pRb could aid in hair cell regeneration.
Results
Inner-Ear-Rb-Conditional Knockout Mice.
We created conditional Rb knockout mice to delete the Rb gene primarily in the inner ear. pRbflox/flox mice were bred to transgenic mice with cre recombinase under the control of the promoter of Pou4f3 (Pou4f3-cre), a hair cell-specific transcription factor (21). Pou4f3-pRb−/− mice survived postnatally for up to 6 months (the latest stage studied).
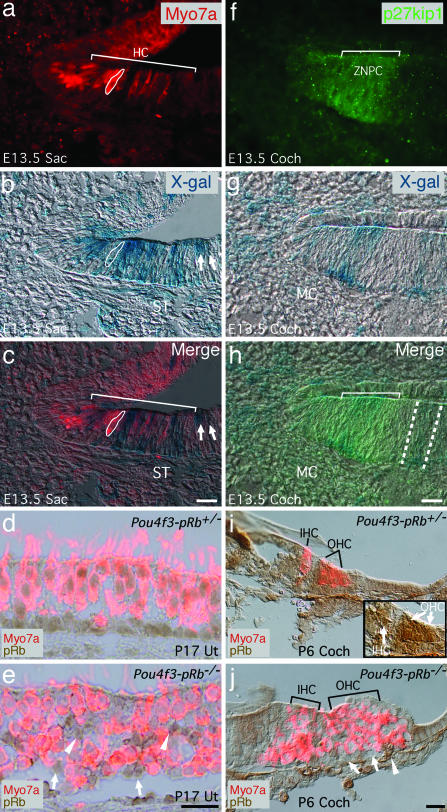
To identify the temporal–spatial expression pattern of Pou4f3-cre in the inner ear, Pou4f3-cre mice were bred to ROSA26 mice (22), and cre-driven β-galactosidase expression in the Pou4f3-Rosa26 inner ear was identified by X-gal histochemistry. Pou4f3 is first expressed in vestibular hair cells at E12.5 and in cochlear basal hair cells at E14.5, after Atoh1 expression, and is subsequently maintained in postnatal hair cells (23). In the immature utricles of E12.5 Pou4f3-Rosa26 mice, X-gal labeling appeared primarily in a region expressing Isl1, which represented the sensory epithelium with young hair cells and supporting cells (24) and which lacked Pax2 expression (Fig. 8, which is published as supporting information on the PNAS web site). At E13.5, X-gal staining was detected in vestibular hair cells identified by myosin VIIa (Myo7a) expression (Fig. 1a–c) and in the p27kip1-positive zone of nonproliferating cells (ZNPC) of the primordial cochlea that harbors sensory progenitor cells (Fig. 1 f–h) (25). In the E13.5 saccule, although most X-gal-positive cells were also Myo7a-positive, some were not (Fig. 1c, arrows). We believe that these are committed but undifferentiated hair cells, because Pou4f3 expression precedes that of Myo7a (23), because later in development X-gal staining was restricted to the vestibular hair cells (Fig. 9b, which is published as supporting information on the PNAS web site), and because the pRb−/− phenotype was associated with vestibular hair cells but not supporting cells (Fig. 1e). Pou4f3 expression in E13.5 cochlear progenitor cells was supported by X-gal label in the postnatal day 6 (P6) cochlea occurring both in hair cells and in different types of supporting cells (Fig. 9d) and was also supported by the lack of pRb staining in all cochlear hair cells and many supporting cells in the Pou4f3-pRb−/− mice (Fig. 1j). Therefore, Pou4f3-cre expression seems to follow different patterns: in the vestibular sensory epithelium it is expressed primarily by hair cells, and in the cochlea it is expressed by progenitor cells that give rise to both hair cells and supporting cells.
Fig. 1.
X-gal staining of E13.5 Rosa26 mouse inner ear and Rb deletion in Pou4f3-pRb−/− hair cells. (a–c) In the saccular sensory epithelium, Myo7a-positive hair cells were labeled with X-gal (bracket). Because of the masking effect, Myo7a labeling was weak in the hair cells strongly stained for X-gal, and weak X-gal staining was seen in hair cells with strong Myo7a labeling (a, circled cell). Furthermore, some X-gal-labeled cells were not Myo7a-positive (c, arrows) (see Results), and some were in the stroma region (ST) (b and c). (d) In the P17 control utricle, pRb staining was ubiquitous in all hair cells and supporting cells. (e) In the Pou4f3-pRb−/− utricle, pRb was absent in all hair cells but present in supporting cells (arrows), some of which were displaced from basal lamina (arrowheads). (f–h) X-gal labeling was in cells within the p27kip1-positive ZNPC (bracket) and in the cells outside the ZNPC (between two dashed lines in h) and the mesenchymal cells (MC). (i) In P6 control cochlea, pRb was prominent in hair cells (arrows in Inset) and weaker in supporting cells. (j) In Pou4f3-pRb−/− cochlea, pRb was absent in all hair cells and many supporting cells (arrows) but present in other supporting cells (arrowhead). The cells indicated are likely Deiters’ cells. (Scale bars: 20 μm.)
X-gal staining was also observed in the stroma in the vestibular system (Fig. 1 b and c) and in tissues outside the inner ear, including the brain (data not shown), indicating that the Pou4f3 promoter used in Pou4f3-cre mice drove expression in a manner similar to but not identical to that of the endogenous Pou4f3 gene.
Lack of pRb Leads to Cell-Cycle Reentry of Postmitotic Hair Cells.
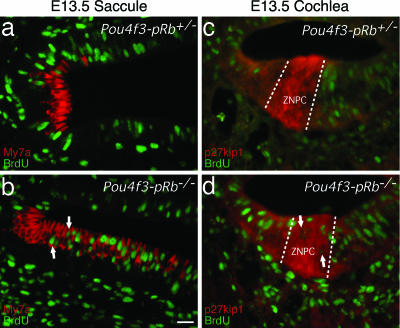
To determine whether pRb is required for cell-cycle reentry of postmitotic hair cells, BrdU was injected into E13.5 pregnant mice, and the embryos were harvested 2 h later. At E13.5, vestibular hair cells and auditory sensory progenitor cells were postmitotic. Labeling by an anti-BrdU antibody showed BrdU incorporation, hence cell-cycle reentry, in the Pou4f3-pRb−/− vestibular hair cells (Fig. 2b, arrows) and cochlear sensory progenitor cells (Fig. 2d, arrows), but not in control cells (Fig. 2 a and c).
Fig. 2.
BrdU labeling in the saccular macula and the primordial organ of Corti at E13.5. (a) In control saccule, no BrdU labeling was seen in hair cells. (b) BrdU labeling was seen in the Pou4f3-pRb−/− hair cells (arrows). (c) No BrdU labeling was in the p27kip1-positive postmitotic ZNPC of control mouse. (d) BrdU labeling was seen in the Pou4f3-pRb−/− ZNPC (arrows). (Scale bar: 20 μm.)
Postnatal Pou4f3-pRb−/− Hair Cells Undergo Cell Division and Can Transduce Mechanical Stimuli.
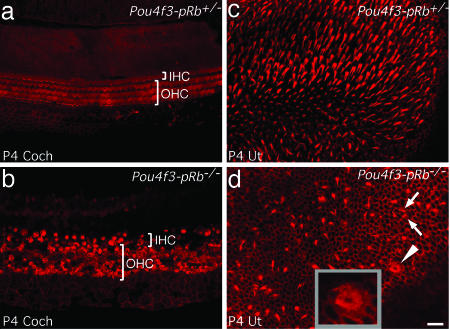
Using confocal microscopy on phalloidin-stained cochleae, we observed hair cells with stereocilia in P4 Pou4f3-pRb−/− mice. In contrast to normal cochleae, which have three rows of outer hair cells (OHCs) and one row of inner hair cells (IHCs), the Pou4f3-pRb−/− cochleae had more IHCs and OHCs, arranged in a disorganized pattern. Many OHCs lacked bundles (Fig. 3b and Fig. 10, which is published as supporting information on the PNAS web site). In the utricle the number of hair bundles was reduced, but many appeared morphologically normal (Fig. 3d). Atypical bundles were also observed, including ring-shaped bundles (Fig. 3d, arrowhead and Inset) and two bundles within one hair cell boundary (Fig. 3d, arrows), indicating ongoing or incomplete cytogenesis.
Fig. 3.
Confocal microscopy of phalloidin-labeled P4 hair bundles. (a) Three rows of OHCs and one row of IHCs were seen in control. (b) Multiple rows of both OHCs and IHCs were seen in the Pou4f3-pRb−/− cochlea; many OHCs lacked hair bundles. (c) Control utricular hair cells with hair bundles. (d) In the Pou4f3-pRb−/− utricle the proportion of hair cells bearing bundles was considerably lower than in control. Abnormal hair bundles included a ring shape (arrowhead and Inset) and two bundles in one hair cell (arrows). (Scale bar: 20 μm.)
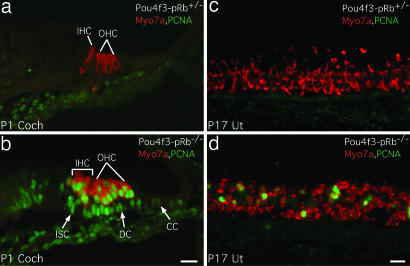
Postnatal Pou4f3-pRb−/− hair cells were assessed for proliferation. Positive antiproliferating cell nuclear antigen (anti-PCNA) antibody labeling was seen in P1 cochlear and P17 utricular hair cells, indicating ongoing cell proliferation (Fig. 4b and d). Many Pou4f3-pRb−/− cochlear supporting cells were also PCNA-positive (Fig. 4b), but no PCNA-positive hair cells or supporting cells were present in control mice (Fig. 4 a and c). Proliferation of hair cells and supporting cells was also observed in P10 Pou4f3-pRb−/− cochlea (data not shown). The number of PCNA-positive utricular hair cells decreased over time: 111 ± 12 (62%) at P4, 77 ± 14 (48%) at P6, and <44 ± 5 (15%) at P17, suggesting that another mechanism compensated for the loss of pRb. BrdU injection and labeling confirmed proliferation at those stages (data not shown).
Fig. 4.
Proliferation of postnatal hair cells. (a) P1 control cochlea lacking PCNA labeling. (b) Pou4f3-pRb−/− cochlea with most hair cells and many supporting cells (arrows) labeled for PCNA. PCNA labeling correlated with X-gal-positive cells in the cochlea (Fig. 9d). (c) No PCNA-positive hair cells in the P17 control utricle. (d) PCNA labeling was colocalized with hair cells but not supporting cells in the Pou4f3-pRb−/− utricle. DC, Deiters’ cell; CC, Claudius cell; ISC, inner sulcus cell. (Scale bars: 20 μm.)
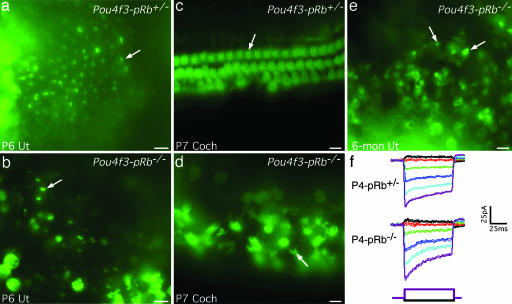
We used FM1-43 dye and transduction current recordings to assay the mechanosensitivity of proliferating Pou4f3-pRb−/− hair cells. FM1-43 passes through open transduction channels and thus marks functional hair cells (26, 27). In mouse utricular hair cells the onset of mechanosensitivity began at E16 (28). At P6, both control and Pou4f3-pRb−/− utricular hair cells with stereocilia accumulated FM1-43 (Fig. 5a and b). At P4, Pou4f3-pRb−/− utricular hair cells had normal transduction currents of ≈100 pA [Fig. 5f; 100.8 ± 23.3 pA for Pou4f3-pRb−/− vs. 98.3 ± 13.3 pA for Pou4f3-pRb+/− (mean ± SEM); n = 4 for each group]. Similarly, P7 Pou4f3-pRb−/− cochlear and 6-month utricular hair cells with bundles took up FM1-43 (Fig. 5 d and e).
Fig. 5.
Pou4f3-pRb−/− hair cells are functional, based on FM1-43 loading. Arrows indicate representative labeled cells. (a) P6 control utricular hair cells. (b) P6 Pou4f3-pRb−/− utricular hair cells. (c) P7 cochlear hair cells. (d) P7 Pou4f3-pRb−/− cochlear hair cells. Note the disorganized distribution of hair cells. (e) Six-month-old Pou4f3-pRb−/− utricular hair cells. In all FM1-43 loading experiments, the dye was localized in hair cells bearing bundles that could be identified in differential interference contrast microscopic images (data not shown). (f) Comparable transduction currents, elicited in control (Top) and Pou4f3-pRb−/− (Middle) littermates by step deflections of the hair bundle (Bottom), showed adaptation in response to positive (purple) and negative (black) bundle deflections. Only the largest positive and negative stimuli are shown. (Scale bars: 20 μm.)
Differentiation of Pou4f3-pRb−/− hair cells was studied with antibodies to hair cell markers including Ptprq, espin, myosin VI, and Myo7a. Positive labeling suggested that the Pou4f3-pRb−/− hair cells differentiated like controls (data not shown). Interestingly, in Pou4f3-pRb−/− utricles with multiple layers of hair cells, labeling with antibodies to the hair bundle proteins espin and Ptprq showed that hair cells in all layers could bear bundle-like structures (Fig. 11, which is published as supporting information on the PNAS web site).
pRb Is Required for Auditory Hair Cell Maturation, Survival, and Hearing.
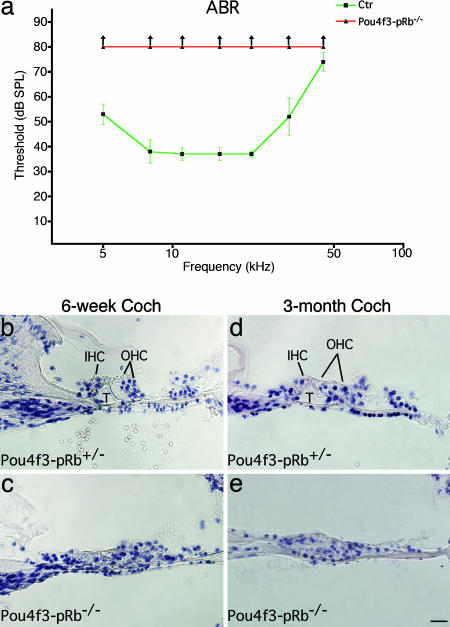
To study functional aspects of the auditory system, auditory brainstem response (ABR) testing was performed on 3-month-old Pou4f3-pRb−/− mice. ABR tests failed to detect any response in Pou4f3-pRb−/− mice, indicating that the mice were profoundly deaf (Fig. 6a).
Fig. 6.
ABR testing and histological analysis of Pou4f3-pRb−/− cochleae. (a) ABR testing. At 3 months, Pou4f3-pRb−/− mice (n = 3) exhibited no response up to at least 80-dB sound pressure level (arrows, thresholds > 80 dB), whereas control mice (n = 5) showed more normal hearing. (b) Hematoxylin and eosin staining of control cochleae at 6 weeks. Morphology, including the tunnel of Corti (T), was normal. The split outside the OHC region was a sectioning artifact. (c) Pou4f3-pRb−/− cochlea at 6 weeks. The tunnel of Corti was absent, and other morphology was abnormal. (d) Control cochlea at 3 months with normal morphology. (e) Pou4f3-pRb−/− cochlea at 3 months showing severe degeneration of organ of Corti with loss of hair cells. (Scale bar: 20 μm.)
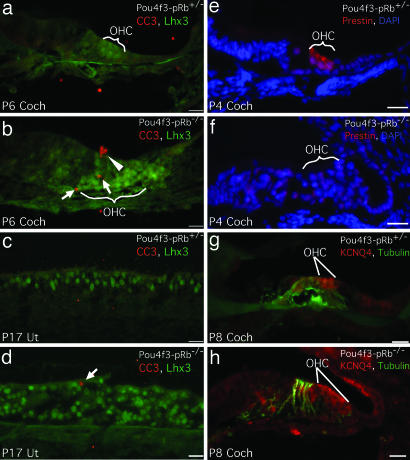
Hematoxylin and eosin staining of Pou4f3-pRb−/− cochleae (29) showed that, in the basal turn, the tunnel of Corti had collapsed in 6-week-old (Fig. 6c) and 3-month-old (Fig. 6e) cochleae. No Myo7a-positive cells were found in 6-week-old or 3-month-old Pou4f3-pRb−/− cochleae, indicating total hair cell loss (Fig. 12, which is published as supporting information on the PNAS web site). Cell death in the Pou4f3-pRb−/− organ of Corti was investigated by using an antibody to activated caspase 3 (CC3). CC3-positive cells were readily identified in the OHC region of the organ of Corti from P6 (Fig. 7b) through P17 (data not shown), starting with OHCs at the base. At P10 OHC death had extended to the apex, whereas IHC death was confined to the base. By 3 months, the Pou4f3-pRb−/− organ of Corti had lost all hair cells (Figs. 6e and 12f). Profound deafness was apparently the consequence of cell death in the organ of Corti.
Fig. 7.
Cell death in the Pou4f3-pRb−/− inner ear and impairment of cochlear hair cell maturation. (a) At P6, no CC3-positive cells were detected in control cochlea. (b) CC3-positive cells (arrows) in the Pou4f3-pRb−/− OHC region (bracket), including a cell extruded from the luminal surface (arrowhead). (c) At P17, no CC3-positive cells were seen in the control utricle. (d) Occasional CC3-positive cells were observed in the Pou4f3-pRb−/− utricle. (e) At P4, control midturn OHCs were stained strongly with an anti-prestin antibody (bracket). (f) In the midturn Pou4f3-pRb−/− cochlea, prestin labeling was completely absent in the OHC region. (g) At P8, control apical OHCs showed labeling for KCNQ4. (h) Similar to control, the apical Pou4f3-pRb−/− OHCs were stained with KCNQ4. Lhx3 (Lim-homeodomain transcription factor 3)-positive hair cells (11) and tubulin-positive nerve fibers are shown. (Scale bars: 20 μm.)
Mouse cochlear hair cells undergo final maturation at P10–P12, as assessed by auditory nerve recordings (30). The electromotility protein prestin and the potassium channel KCNQ4, essential for mature OHC function, are expressed in postnatal OHCs (30–33). In P4 Pou4f3-pRb−/− OHCs we found no labeling with antibodies to prestin, whereas prominent labeling was seen in control littermates (Fig. 7 e and f). Prestin was not detected at later stages (P10), excluding the possibility of delayed expression (data not shown). In contrast, KCNQ4 was detected in P8 Pou4f3-pRb−/− OHCs, similar to controls (Fig. 7 g and h). Similarly, we found using in situ hybridization that the expression of the gene encoding TMC1 (transmembrane channel-like 1), a protein important for postnatal hair cell development (34), was similar in Pou4f3-pRb−/− and control cochlear hair cells (Fig. 13, which is published as supporting information on the PNAS web site). Therefore, the effect of pRb on maturation appears to be gene-specific.
Long-Term Survival of pRb−/− Hair Cells in the Vestibular System.
Unlike auditory hair cells, Pou4f3-pRb−/− vestibular hair cells continued to divide in 6-week-old mice (data not shown). The number of dividing hair cells declined with age, as illustrated by the percentage of PCNA-positive hair cells (from 63% at P4 to <2% at 6 weeks). In 3-month-old Pou4f3-pRb−/− utricles no PCNA-positive hair cells were detected, suggesting that a compensatory mechanism controlled cell division.
Confocal microscopy of phalloidin-labeled 6-month-old Pou4f3-pRb−/− utricles showed abnormalities including thin and disorganized hair bundles in most but not all hair cells (Fig. 14b, which is published as supporting information on the PNAS web site). Therefore, lack of pRb function led to abnormal development of vestibular hair cells.
In Pou4f3-pRb−/− mice the number of vestibular hair cells decreased from early postnatal stages to adult: at P6 there were three to four layers of hair cells in the utricle, whereas at 3 months one to two layers of hair cells remained. Unlike in Pou4f3-pRb−/− cochleae, CC3 antibodies only occasionally labeled Pou4f3-pRb−/− utricular hair cells (Fig. 7 c and d). The survival of hair cells in 6-month-old utricles and the paucity of CC3 labeling, in contrast to rapid cell death in the cochlea, indicated that cell death was a slow process in the utricle and that some hair cells may resist apoptosis. Adult (3- and 6-month-old) Pou4f3-pRb−/− utricular hair cells bearing bundles accumulated FM1-43 dye (Fig. 5e, arrows). Tubulin and synaptophysin immunolabeling showed that hair cells were also surrounded by nerve fibers and apparently formed synapses with ganglion cells (Fig. 15, which is published as supporting information on the PNAS web site).
The survival of functional Pou4f3-pRb−/− vestibular hair cells with synaptic connections suggested that the vestibular system might continue to operate. Pou4f3-pRb−/− mice were studied for vestibular function by their capacity to walk and swim. At 6 weeks, Pou4f3-pRb−/− mice showed no circling behavior and could swim with their heads above water. Later, at 3 and 6 months, Pou4f3-pRb−/− mice showed moderate circling behavior and could not consistently swim with their heads above water (data not shown). In contrast, Pou4f3−/− mouse mutants lacking all hair cells exhibited severe circling and were totally unable to swim (35). Despite the hair cell abnormalities and behavioral decline, partial vestibular function remained in Pou4f3-pRb−/− mice even at 6 months.
Discussion
This study reveals essential roles for pRb in auditory hair cell maturation and survival. It demonstrates that postnatal hair cells can be maintained in the cell cycle in the absence of pRb function. Different phenotypes in Pou4f3-pRb−/− cochleae and vestibules likely indicate distinct roles of pRb in the respective systems.
We overcame the neonatal lethality of the previous Rb knockout mouse model by creating a conditional pRb knockout mouse in which the Rb gene is primarily deleted in the inner ear. In the vestibular system Rb is primarily deleted in the hair cells, whereas in the cochlea Rb is deleted in sensory progenitor cells. Despite the difference in expression patterns of Pou4f3-cre and the endogenous Pou4f3 gene, including the expression of Pou4f3-cre in nonsensory cells of inner ear, the survival of the Pou4f3-pRb−/− mice allowed us to study the effects of pRb in postnatal development and function of the inner ear.
We found that postnatal Pou4f3-pRb−/− auditory hair cells and many supporting cells undergo cell division. The proliferating cells are correlated with Pou4f3-cre-expressing (X-gal-positive) and pRb-negative cells. Therefore, pRb function is essential for blocking cell-cycle reentry of postnatal hair cells and supporting cells.
pRb is also required for auditory hair cell maturation, as shown by the lack of prestin expression in Pou4f3-pRb−/− OHCs. Failure of OHCs to mature results in cell death (36, 37). In prestin knockout mice cell death starts with OHCs at the cochlear base at P28 and subsequently extends to the apex and to IHCs (37). Pou4f3-pRb−/− hair cell loss follows a similar pattern but with more rapid progression, suggesting that pRb may regulate expression of additional maturation genes. However, it is likely that the effect of pRb on maturation gene expression is gene-specific rather than global, as demonstrated by the expression of KCNQ4 and TMC1 in the Pou4f3-pRb−/− cochlear hair cells.
How pRb regulates prestin expression is unknown. pRb can interact with the cell-specific transcription factors to transactivate cell-specific genes in skeletal muscle and osteoblasts (38, 39) and can also antagonize the inhibitory role of Id2, which in turn activates a master regulator of macrophage differentiation, PU.1 (40). It is possible that pRb interacts with unknown hair cell transcription factors to regulate expression of maturation genes such as prestin.
Pou4f3-pRb−/− IHCs die later, after the death of OHCs, consistent with previous observations that OHCs are required for IHC survival (37). It is unknown whether pRb function is directly required for IHC survival. Experiments using transgenic mice with Rb specifically deleted in IHCs and OHCs could answer this question. As a consequence of cell death, adult Pou4f3-pRb−/− mice are profoundly deaf. Although pRb is well known for its role in inhibition of apoptosis through a p53-dependent pathway, we did not detect p53 expression in the postnatal Pou4f3-pRb−/− inner ear (data not shown), suggesting that hair cell death may be p53-independent.
In the utricle, postnatal Pou4f3-pRb−/− hair cells continue to divide in 6-week-old mice. Proliferation in postnatal Pou4f3-pRb−/− utricles is associated with hair cells, rather than the supporting cells that are also pRb-positive. pRb, therefore, appears to have a cell-autonomous role in regulating cell-cycle exit in vestibular hair cells. Supernumerary hair cells are primarily derived from hair cell division and not from supporting cell division or conversion.
The differentiation of Pou4f3-pRb−/− utricular hair cells seems relatively normal, judging by labeling with various cellular markers and synapse formation with ganglion neurons. Significantly, Pou4f3-pRb−/− hair cells with hair bundles are able to transduce mechanical stimuli by P4 and load with FM1-43 until at least 6 months. Therefore, the newly generated hair cells appear functional. It is notable that hair bundle morphology is relatively normal during early postnatal stages, but by 6 months many hair bundles have abnormally thin stereocilia. The continued absence of pRb function may ultimately contribute to abnormal vestibular hair cells.
The presence of hair cells in 6-month-old mice, with a majority of hair cells ceasing to divide by 6 weeks, suggests that compensatory mechanisms replace pRb function. p107, well known to compensate for pRb loss in other systems, was not up-regulated in Pou4f3-pRb−/− hair cells (data not shown). The long-term survival suggests that some vestibular hair cells may be resistant to cell death. In addition, we found no sign of tumor formation in multiple animals studied over time; therefore, the pRb−/− vestibular hair cells are unlikely to be tumorigenic.
Is it possible to regenerate functional hair cells by manipulating the pRb pathway? Permanent deletion of Rb is not optimal because it causes cell loss and hair bundle abnormalities. However, the effects of transient and reversible block of pRb function in mature hair cells are not known. Knockdown of pRb function with short hairpin RNA in cultured cells resulted in increased proliferation and failure of contact inhibition (P.W.H., unpublished data). Proliferation was subsequently blocked and contact inhibition was reestablished upon reexpression of Rb after degradation of short hairpin RNA. Similar manipulations may allow cell-cycle reentry of hair cells, and the subsequent return of pRb function may lead to expression of maturation genes (such as prestin) and inhibition of apoptosis. In the vestibular system many hair cells lacking pRb continued to divide up to 6 weeks, survived for 6 months, and remained functional at both the cellular and system levels. Therefore, temporary block of pRb function in vestibular hair cells may lead to regeneration for recovery of vestibular function.
Methods
Generation of Pou4f3-cre Transgenic Mice.
A 9-kb portion of the 5′ upstream region of the Pou4f3 gene in pBluescript was obtained (L. Erkman and G. Rosenfeld, University of California at San Diego, La Jolla) and used to drive Cre expression. A β-globin intronic sequence of 800 bp was blunt-ligated to the 5′ end of the Cre insert of pOG231 (S. O’Gorman, The Salk Institute, La Jolla, CA). The pOG231 Cre-containing plasmid had a nuclear localization signal peptide sequence (CCCAAGAAGAAGAGGAAGGTG) of the SV40 large T antigen immediately 3′ of the ATG initiation sequence. A human growth hormone polyA initiation sequence was ligated to the 3′ end of the Cre insert. Additionally, the β-globin/Cre/human growth hormone polyA construct went through two rounds of mutagenesis with a QuikChange mutagenesis kit (Stratagene) to introduce a consensus Kozak site (GCCGCCACC) at the 5′ end of the Cre nuclear localization signal sequence ATG codon. The construct was verified by sequencing. The construct was then ligated to the 5′ upstream Pou4f3 clone. Pou4f3Cre transgene was released by Cla digestion and was used to generate the Pou4f3Cre transgenic mouse lines by using standard pronuclear injection techniques. Mouse lines (with background 129/SvEv) were successfully created. All animal work was conducted by using procedures reviewed and approved by the institutional animal care and use committees of Massachusetts General Hospital, The Salk Institute for Biological Studies (Pou4f3Cre transgenic mouse line generation), and Tufts University School of Medicine (Pou4f3Cre line maintenance and breeding) and were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Research Animals.
Genotyping.
For Cre genotyping, forward (CCAATTTACTGACCGTACACC) and reverse (CGGCTATACGTAACAGGGTG) primers were used. PCR was performed by using 95°C for 2 min followed by 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min for 30 cycles. The final annealing was 72°C for 10 min. A band of 500 bp indicated the presence of the Cre allele. Rbflox/flox genotyping was conducted as described (11).
BrdU Labeling.
Timed pregnant mice at E13.5 were injected once with BrdU (Sigma) (prepared in 1× PBS, pH 7.0), and embryos were harvested 2 h later. For postnatal mice, BrdU was injected once per day for 2 days before killing (final: 50 μg of BrdU per gram of body weight).
Immunohistochemistry, FM1-43 Loading, and Whole-Cell Patch-Clamping.
Frozen sections of the inner ear tissues were prepared for immunolabeling by using the protocol as described (11). For sources of antibodies used in the study see Table 1, which is published as supporting information on the PNAS web site. DAPI and rhodamine–phalloidin were from Molecular Probes. FM1-43 loading and whole-cell patch-clamping followed previous protocols (11).
ABR Testing.
ABR was measured essentially as previously described (29), with the exception that the highest threshold tested was 80-dB sound pressure level.
Vestibular Tests.
Mice were observed for circling and head-tossing behavior, an indication of vestibular dysfunction. Mice were placed into a water tank, and their ability to swim with heads above water was assessed. Pou4f3 knockout mice (which lack functional hair cells) were used as positive controls, and WT littermates were used as negative controls.
Supplementary Material
Acknowledgments
We thank Dr. Sang Goo Lee for his help with X-gal staining. The project was supported by National Institutes of Health Grants DC-04546 and DC-06908 (to Z.-Y.C.), DC-AG20208 (to P.W.H.), DC-02281 (to D.P.C.), DC-01089 (to M.C.B.), and DC- 06258 (to D.E.V.). D.P.C. is an Investigator of the Howard Hughes Medical Institute. The research was conducted while Z.-Y.C. was a Pfizer/American Federation for Aging Research Innovation in Aging Research Grant recipient.
Abbreviations
- OHC
outer hair cell
- IHC
inner hair cell
- Myo7a
myosin VIIa
- PCNA
proliferating cell nuclear antigen
- ZNPC
zone of nonproliferating cells
- ABR
auditory brainstem response
- En
embryonic day n
- Pn
postnatal day n
- CC3
caspase 3.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bermingham N. A., Hassan B. A., Price S. D., Vollrath M. A., Ben-Arie N., Eatock R. A., Bellen H. J., Lysakowski A., Zoghbi H. Y. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 2.Zheng J. L., Gao W. Q. Nat. Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- 3.Kiernan A. E., Ahituv N., Fuchs H., Balling R., Avraham K. B., Steel K. P., Hrabe de Angelis M. Proc. Natl. Acad. Sci. USA. 2001;98:3873–3878. doi: 10.1073/pnas.071496998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanford P. J., Lan Y., Jiang R., Lindsell C., Weinmaster G., Gridley T., Kelley M. W. Nat. Genet. 1999;21:289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- 5.Raz Y., Kelley M. W. Dev. Biol. 1999;213:180–193. doi: 10.1006/dbio.1999.9364. [DOI] [PubMed] [Google Scholar]
- 6.Forge A., Li L., Corwin J. T., Nevill G. Science. 1993;259:1616–1619. doi: 10.1126/science.8456284. [DOI] [PubMed] [Google Scholar]
- 7.Warchol M. E., Lambert P. R., Goldstein B. J., Forge A., Corwin J. T. Science. 1993;259:1619–1622. doi: 10.1126/science.8456285. [DOI] [PubMed] [Google Scholar]
- 8.Chen P., Segil N. Development (Cambridge, U.K.) 1999;126:1581–1590. doi: 10.1242/dev.126.8.1581. [DOI] [PubMed] [Google Scholar]
- 9.Lowenheim H., Furness D. N., Kil J., Zinn C., Gultig K., Fero M. L., Frost D., Gummer A. W., Roberts J. M., Rubel E. W., et al. Proc. Natl. Acad. Sci. USA. 1999;96:4084–4088. doi: 10.1073/pnas.96.7.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen P., Zindy F., Abdala C., Liu F., Li X., Roussel M. F., Segil N. Nat. Cell Biol. 2003;5:422–426. doi: 10.1038/ncb976. [DOI] [PubMed] [Google Scholar]
- 11.Sage C., Huang M., Karimi K., Gutierrez G., Vollrath M. A., Zhang D. S., Garcia-Anoveros J., Hinds P. W., Corwin J. T., Corey D. P., Chen Z. Y. Science. 2005;307:1114–1118. doi: 10.1126/science.1106642. [DOI] [PubMed] [Google Scholar]
- 12.Mantela J., Jiang Z., Ylikoski J., Fritzsch B., Zacksenhaus E., Pirvola U. Development (Cambridge, U.K.) 2005;132:2377–2388. doi: 10.1242/dev.01834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Classon M., Harlow E. Nat. Rev. Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 14.Lipinski M. M., Jacks T. Oncogene. 1999;18:7873–7882. doi: 10.1038/sj.onc.1203244. [DOI] [PubMed] [Google Scholar]
- 15.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 16.Slack R. S., El-Bizri H., Wong J., Belliveau D. J., Miller F. D. J. Cell Biol. 1998;140:1497–1509. doi: 10.1083/jcb.140.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gloster A., El-Bizri H., Bamji S. X., Rogers D., Miller F. D. J. Comp. Neurol. 1999;405:45–60. doi: 10.1002/(sici)1096-9861(19990301)405:1<45::aid-cne4>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson K. L., Vanderluit J. L., Hebert J. M., McIntosh W. C., Tibbo E., MacLaurin J. G., Park D. S., Wallace V. A., Vooijs M., McConnell S. K., Slack R. S. EMBO J. 2002;21:3337–3346. doi: 10.1093/emboj/cdf338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacPherson D., Sage J., Crowley D., Trumpp A., Bronson R. T., Jacks T. Mol. Cell. Biol. 2003;23:1044–1053. doi: 10.1128/MCB.23.3.1044-1053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marino S., Hoogervoorst D., Brandner S., Berns A. Development (Cambridge, U.K.) 2003;130:3359–3368. doi: 10.1242/dev.00553. [DOI] [PubMed] [Google Scholar]
- 21.Vooijs M., te Riele H., van der Valk M., Berns A. Oncogene. 2002;21:4635–4645. doi: 10.1038/sj.onc.1205575. [DOI] [PubMed] [Google Scholar]
- 22.Friedrich G., Soriano P. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 23.Xiang M., Gao W. Q., Hasson T., Shin J. J. Development (Cambridge, U.K.) 1998;125:3935–3946. doi: 10.1242/dev.125.20.3935. [DOI] [PubMed] [Google Scholar]
- 24.Radde-Gallwitz K., Pan L., Gan L., Lin X., Segil N., Chen P. J. Comp. Neurol. 2004;477:412–421. doi: 10.1002/cne.20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen P., Johnson J. E., Zoghbi H. Y., Segil N. Development (Cambridge, U.K.) 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- 26.Meyers J. R., MacDonald R. B., Duggan A., Lenzi D., Standaert D. G., Corwin J. T., Corey D. P. J. Neurosci. 2003;23:4054–4065. doi: 10.1523/JNEUROSCI.23-10-04054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gale J. E., Marcotti W., Kennedy H. J., Kros C. J., Richardson G. P. J. Neurosci. 2001;21:7013–7025. doi: 10.1523/JNEUROSCI.21-18-07013.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geleoc G. S., Holt J. R. Nat. Neurosci. 2003;6:1019–1020. doi: 10.1038/nn1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rehm H. L., Zhang D. S., Brown M. C., Burgess B., Halpin C., Berger W., Morton C. C., Corey D. P., Chen Z. Y. J. Neurosci. 2002;22:4286–4292. doi: 10.1523/JNEUROSCI.22-11-04286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kros C. J., Ruppersberg J. P., Rusch A. Nature. 1998;394:281–284. doi: 10.1038/28401. [DOI] [PubMed] [Google Scholar]
- 31.Zheng J., Shen W., He D. Z., Long K. B., Madison L. D., Dallos P. Nature. 2000;405:149–155. doi: 10.1038/35012009. [DOI] [PubMed] [Google Scholar]
- 32.Kharkovets T., Hardelin J. P., Safieddine S., Schweizer M., El-Amraoui A., Petit C., Jentsch T. J. Proc. Natl. Acad. Sci. USA. 2000;97:4333–4338. doi: 10.1073/pnas.97.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belyantseva I. A., Adler H. J., Curi R., Frolenkov G. I., Kachar B. J. Neurosci. 2000;20, RC116:1–5. doi: 10.1523/JNEUROSCI.20-24-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vreugde S., Erven A., Kros C. J., Marcotti W., Fuchs H., Kurima K., Wilcox E. R., Friedman T. B., Griffith A. J., Balling R., et al. Nat. Genet. 2002;30:257–258. doi: 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
- 35.Erkman L., McEvilly R. J., Luo L., Ryan A. K., Hooshmand F., O’Connell S. M., Keithley E. M., Rapaport D. H., Ryan A. F., Rosenfeld M. G. Nature. 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- 36.Nouvian R., Ruel J., Wang J., Guitton M. J., Pujol R., Puel J. L. Eur. J. Neurosci. 2003;17:2553–2562. doi: 10.1046/j.1460-9568.2003.02715.x. [DOI] [PubMed] [Google Scholar]
- 37.Wu X., Gao J., Guo Y., Zuo J. Brain Res. Mol. Brain Res. 2004;126:30–37. doi: 10.1016/j.molbrainres.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 38.Novitch B. G., Spicer D. B., Kim P. S., Cheung W. L., Lassar A. B. Curr. Biol. 1999;9:449–459. doi: 10.1016/s0960-9822(99)80210-3. [DOI] [PubMed] [Google Scholar]
- 39.Thomas D. M., Carty S. A., Piscopo D. M., Lee J. S., Wang W. F., Forrester W. C., Hinds P. W. Mol. Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 40.Iavarone A., King E. R., Dai X. M., Leone G., Stanley E. R., Lasorella A. Nature. 2004;432:1040–1045. doi: 10.1038/nature03068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.