Abstract
Group II intron RNAs self-splice in vitro but only at high salt and/or Mg2+ concentrations and have been thought to require proteins to stabilize their active structure for efficient splicing in vivo. Here, we show that a DEAD-box protein, CYT-19, can by itself promote the splicing and reverse splicing of the yeast aI5γ and bI1 group II introns under near-physiological conditions by acting as an ATP-dependent RNA chaperone, whose continued presence is not required after RNA folding. Our results suggest that the folding of some group II introns may be limited by kinetic traps and that their active structures, once formed, do not require proteins or high Mg2+ concentrations for structural stabilization. Thus, during evolution, group II introns could have spliced and transposed by reverse splicing by using ubiquitous RNA chaperones before acquiring more specific protein partners to promote their splicing and mobility. More generally, our results provide additional evidence for the widespread role of RNA chaperones in folding cellular RNAs.
Keywords: catalytic RNA, ribozyme, RNA helicase, RNA structure
Group II introns are mobile catalytic RNAs that splice via a lariat intermediate and may be ancestors of eukaryotic spliceosomal introns (1–3). To catalyze splicing, the intron RNAs fold into a conserved three-dimensional structure, which aligns the splice sites and branch-point nucleotide residue and uses bound Mg2+ ions to activate the appropriate bonds for catalysis. The conserved three-dimensional structure of group II introns consists of six double-helical domains (D1 to D6), which interact with each other via tertiary contacts (3). Some group II introns self-splice in vitro, but only under nonphysiological conditions, including high monovalent salt and Mg2+ concentrations, which were thought essential for RNA folding and structural stabilization. Thus far, most of what is known about group II intron RNA structure, folding, and reactivity has come from studies under such conditions (1–3).
The efficient splicing of group II introns in vivo requires proteins to help the intron RNA fold into the catalytically active structure (reviewed in refs. 2–4). Mobile group II introns encode proteins, which have reverse transcriptase (RT) activity and also promote the splicing of the intron in which they are encoded (“maturase” activity). Other group II introns do not encode maturases and instead rely on host proteins, which appear to differ in different organisms (2–4). Until now, a protein-dependent in vitro splicing system has been developed only for the RT/maturase protein encoded by the mobile Lactococcus lactis Ll.LtrB intron (5, 6). Studies using this system showed that the maturase binds specifically to the intron RNA and promotes its splicing by stabilizing the catalytically active RNA structure (7, 8).
In addition to proteins that stabilize specific RNA structures, cellular RNAs may require RNA chaperones to disrupt stable inactive structure (“kinetic traps”) during RNA folding (9, 10). In the case of group I and II introns, this RNA chaperone function is thought to be fulfilled by specific DExH/D-box proteins (11, 12). DExH/D-box proteins, sometimes referred to as RNA helicases, are a large and ubiquitous protein family, members of which use the energy of ATP hydrolysis to mediate RNA structural rearrangements in a variety of cellular processes, including the splicing of eukaryotic spliceosomal introns (13, 14). Three subfamilies, DExH, DEAH, and DEAD, are distinguished by the amino acid sequence of one of their conserved motifs (14).
In the fungus Neurospora crassa, the efficient splicing of a subset of mitochondrial group I introns requires both the mitochondrial tyrosyl-tRNA synthetase (CYT-18 protein), which stabilizes the active RNA structure, and a DEAD-box protein, CYT-19, which disrupts inactive structures that are rate-limiting for RNA folding (11). The requirement for both proteins for efficient splicing was shown genetically and confirmed by in vitro splicing reactions using purified recombinant CYT-18 and CYT-19. Additionally, RNA structure mapping with the Tetrahymena thermophila LSU-ΔP5abc intron, whose splicing at low Mg2+ concentrations depends on CYT-18, showed that CYT-19 acts to disrupt a predominant misfolded structure containing a nonnative pairing termed Alt-P3 (11). Recent studies have extended these findings by showing that CYT-19 plus ATP can also resolve a variety of other nonnative secondary and tertiary structures of the same intron (S.M. and A.M.L., unpublished data) and misfolded forms of the Tetrahymena L-21 ScaI ribozyme (H. Bhaskaran and R. Russell, personal communication).
Saccharomyces cerevisiae mitochondria contain a DEAD-box protein Mss116p, which is related to CYT-19 and functions in splicing both group I and II introns (12, 15, 16). CYT-19 and Mss116p have 52% similarity in their core regions containing eight conserved motifs, but are more divergent in their C-terminal domains (12). Recently, we showed that disruption of the MSS116 gene inhibits the splicing of all S. cerevisiae mitochondrial group I and II introns, some RNA processing reactions, and translation of a subset of mRNAs, and that all of these defects could be suppressed by the expression of CYT-19 (12). Many of the group I and II introns whose splicing is defective in an MSS116 disruptant additionally require intron-encoded maturases or nuclear gene-encoded splicing factors for structural stabilization. Results for the yeast mobile subgroup IIA intron aI2, which encodes an RT/maturase protein, indicated that DEAD-box protein function is required after maturase binding, as expected for an RNA chaperone that resolves inactive structures formed during maturase-promoted RNA folding (12). Collectively, the above findings suggested that both Mss116p and CYT-19 might act broadly as RNA chaperones on both group I and II introns, as well as other RNAs.
The S. cerevisiae mitochondrial group II introns whose splicing is promoted by Mss116p or CYT-19 in vivo include two small subgroup IIB introns aI5γ and bI1 (12), which have been studied extensively as model systems for group II intron RNA folding and self-splicing (1–3). The in vitro reactions of these introns or ribozymes derived from them are optimal at elevated temperature (42–45°C) with high Mg2+ (50–100 mM) and monovalent salt (≥500 mM) concentrations (17–21). Neither aI5γ nor bI1 encodes a maturase, and their efficient splicing in vivo has been thought to require host proteins to stabilize the catalytically active RNA structure (3).
Here, we show that CYT-19 alone is sufficient to promote the in vitro splicing of both the aI5γ and bI1 group II introns under near-physiological conditions and that it acts as an ATP-dependent RNA chaperone, whose continued presence is not required after RNA folding has occurred. Our results indicate that once formed the active structures of some group II introns do not require proteins or high Mg2+ concentrations for structural stabilization.
Results
CYT-19 Plus ATP Promotes the Splicing of the Yeast aI5γ and bI1 Group II Introns at Near-Physiological Monovalent Salt, Mg2+, and Temperature.
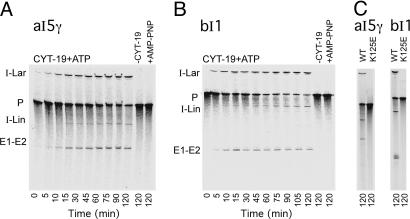
Figs. 1 and 2 show the effect of purified CYT-19 protein on the splicing of aI5γ and bI1 in reaction media containing 100 mM monovalent salt and 5 or 10 mM MgCl2 at 30°C, the standard growth temperature for yeast. As expected, both introns show at most minimal self-splicing under these conditions or in the same reaction media at higher temperatures up to 45°C (Fig. 1 A and B, −CYT-19 lanes, and data not shown). Strikingly, CYT-19 plus ATP induced the splicing of both introns, whereas CYT-19 plus adenylylimidodiphosphate (AMP-PNP), a nonhydrolyzable ATP analog, did not induce splicing (Fig. 1 A and B, +AMP-PNP lanes, and Fig. 2). The CYT-19 mutant K125E, which lacks ATPase activity (11) but still binds aI5γ and bI1 (see below), gave no detectable splicing of either intron (Fig. 1C).
Fig. 1.
CYT-19-dependent splicing of group II introns aI5γ and bI1. (A and B) Splicing time courses were done by incubating 32P-labeled RNA substrate (20 nM) containing the intron and flanking exon sequences with purified CYT-19 protein (500 nM) plus 1 mM ATP in reaction medium containing 100 mM KCl/5 mM MgCl2 at 30°C. Products were analyzed in a denaturing 4% polyacrylamide gel. Control lanes (Right) show RNA substrate incubated without CYT-19 (−CYT-19) or with CYT-19 plus 1 mM AMP-PNP (+AMP-PNP) for 120 min. (C) Splicing reactions with wild-type CYT-19 (WT) and mutant K125E were done as above for 120 min. E1-E2, ligated exons; I-Lar, intron lariat; I-Lin, linear intron; P, precursor RNA. The I-Lin band may contain a mixture of linear intron and broken lariat RNA. The closely spaced doublet for bI1 ligated exons seen previously (21) likely is caused by a premature transcription stop.
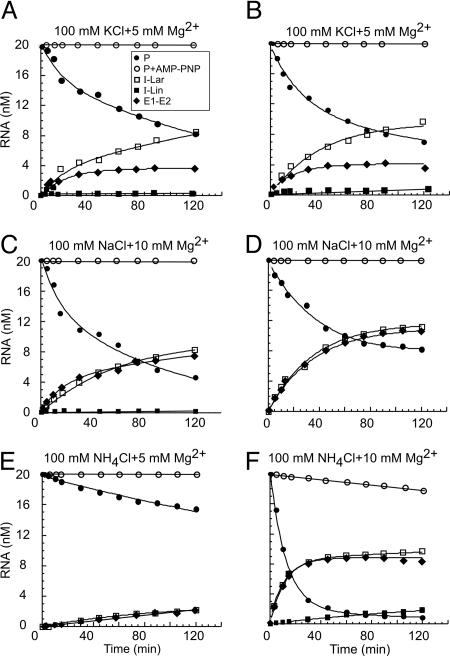
Fig. 2.
Representative kinetic analysis of CYT-19-dependent splicing of aI5γ and bI1. Splicing time courses for aI5γ (A, C, and E) and bI1 (B, D, and F) were done with 500 nM CYT-19, 20 nM 32P-labeled precursor RNA, and 1 mM ATP or AMP-PNP at 30°C in reaction media containing 100 mM KCl (A and B), NaCl (C and D), or NH4Cl (E and F), and 5 or 10 mM MgCl2, as indicated. The plots show the disappearance of precursor RNA and appearance of products as a function of time. In some reactions (e.g., aI5γ in NaCl and bI1 in NH4Cl), precursor RNA degradation at long-time points leads to a 20–30% discrepancy in molar ratios of substrate and products. Differential susceptibility to trace nuclease activity may also account for the finding that in the KCl reactions lariat RNA continues to accumulate at long time points, whereas ligated exons plateau. I-Lin was not detectable in reactions D and E. No CYT-19-promoted splicing was observed for either aI5γ or bI1 in reaction media containing 50 or 100 mM (NH4)2SO4 (an optimal salt for aI5γ self-splicing; ref. 20) and 5 or 10 mM MgCl2 at 30°C (data not shown). Abbreviations are as in Fig. 1.
Previous studies showed that the self-splicing of aI5γ in vitro can occur either by branching, producing lariat intron RNA (I-Lar), or by 5′ splice site hydrolysis, producing linear intron RNA (I-Lin), with the hydrolytic pathway strongly favored in reaction media containing KCl (e.g., 17% I-Lar, 80% I-Lin in 0.5 M KCl/0.1 M MgCl2) (19, 20). Additionally, in KCl, the excised intron RNA efficiently cleaves ligated exons to produce free 5′ and 3′ exons, a reaction known as spliced-exon reopening (SER; refs. 19 and 20). By contrast, in the CYT-19-promoted splicing reactions at 5 mM Mg2+, lariat RNA was the predominant product with all salts tested, and cleaved exons, indicative of SER, were undetectable in KCl or NH4Cl and only a minor fraction (<5% RNA) in NaCl (Table 1 and data not shown).
Table 1.
Kinetic parameters for CYT-19-promoted splicing reactions of group II introns aI5γ and bII
| Intron | Salt | [Mg2+] | Rate constants (×10−3 min−1) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| P |
I-Lar |
I-Lin |
E1–E2 |
||||||
| k1 | k2 | k1 | k2 | kobs | k1 | k2 | |||
| aI5γ | KCl | 5 | 73 (13) | 5.6 (46) | 55 (7) | 2 (36) | 0.1 (2) | 50 (14) | — |
| NaCl | 10 | 76 (23) | 10 (54) | 57 (20) | 4.3 (26) | 0.1 (2) | 60 (20) | 5 (15) | |
| NH4Cl | 5 | 2.2 (18) | — | 1.4 (16) | — | — | 1.4 (14) | — | |
| bI1 | KCl | 5 | 39 (26) | 3.5 (32) | 36 (20) | 3.6 (30) | 0.3 (5) | 51 (14) | — |
| NaCl | 10 | 22 (43) | 2.7 (16) | 21 (30) | 2.1 (25) | — | 20 (30) | 2 (24) | |
| NH4Cl | 10 | 80 (80) | 6.4 (10) | 85 (22) | 8.5 (32) | 6.3 (10) | 78 (21) | 7.8 (30) | |
| aI5γ | KCl + spermidine | 5 | 352 (25) | 3.9 (35) | 286 (16) | 1.4 (24) | 170 (10) | 270 (14) | 2.3 (12) |
| bI1 | KCl + spermidine | 5 | 184 (47) | 9.7 (32) | 145 (14) | 0.2 (40) | 178 (10) | 180 (14) | 0.1 (30) |
Kinetic parameters for the splicing reactions shown in Fig. 2 and additional reactions described in the text are summarized. All reactions were in 100 mM monovalent salt with 500 nM CYT-19, 20 nM precursor RNA, and 1 mM ATP at 30°C. The data were best-fit to equations with one or two exponentials to obtain kobs for the single phase (k1) or for fast and slow phases (k1 and k2 respectively). The minor product I-Lin can be formed either by hydrolytic splicing or breakage of intron lariat RNA, and the rate of its formation is denoted kobs. The numbers in parentheses indicate the percentage of total RNA in each phase or reaction product. For reaction media containing 100 mM KCl/5 mM Mg2+, which we adopted as standard, shown are representative data based on at least 10 time courses for each intron in the absence or presence of 0.1 μM spermidine. For other conditions, the data shown are for time courses that gave maximal rates. The variation in rates was < 2- to 3-fold in replicate experiment with different protein preparations and RNA substrates.
Mg2+ Requirement and Kinetics of the CYT-19-Promoted Splicing Reactions.
The ability of CYT-19 plus ATP to promote aI5γ and bI1 splicing in 100 mM salt was restricted to a relatively narrow range of Mg2+ concentrations, with the Mg2+ requirement lowest in KCl (5 mM Mg2+), higher in NH4Cl (5–10 mM Mg2+), and highest in NaCl (10 mM Mg2+) (Table 1 and data not shown). In reaction medium containing 100 mM KCl, the CYT-19-promoted splicing reactions had broad rate optima between 5 and 15 mM Mg2+, with rates and amplitudes decreasing at higher Mg2+ concentrations, and no reaction at 25 or 50 mM Mg2+ (data not shown). CYT-19’s RNA-stimulated ATPase activity decreased progressively at Mg2+ concentrations >5 mM [assayed with poly(C); data not shown]. Under the most stimulatory conditions for CYT-19, precursor RNA disappearance was frequently biphasic, with rate constants for the fast phase (k1) 73–76 × 10−3 min−1 for aI5γ and 22–80 × 10−3 min−1 for bI1 (Table 1). In 100 mM KCl/5 mM Mg2+, addition of 0.1 μM spermidine did not induce self-splicing, but increased the rate of CYT-19-promoted splicing by 2- to 5-fold (Table 1 and data not shown).
The kinetics of the CYT-19-promoted splicing are comparable to those for self-splicing of aI5γ and bI1 in similar salts, even though the latter reactions are carried out at substantially higher temperatures (e.g., self-splicing of aI5γ at 42°C, 28 × 10−3 min−1 in 500 mM KCl/100 mM Mg2+ and 38 × 10−3 min−1 in 500 mM NH4Cl/100 mM Mg2+; self-spicing of bI1 at 37°C, 120 × 10−3 min−1 in 1.25 M NH4Cl, 60 mM Mg2+, 5 mM spermidine) (20, 21). The self-splicing reactions are also biphasic for precursor RNA disappearance, suggesting at least two RNA conformers (20). In the CYT-19-promoted splicing reactions, an additional fraction of the RNA often (but not always) remained unspliced, possibly reflecting intractable conformations, which could not be refolded by CYT-19 plus ATP.
CYT-19 Alone Cannot Promote Splicing of Maturase-Dependent Group II Introns.
Yeast mitochondria contain two mobile group IIA introns, aI1 and aI2, which encode RT/maturase proteins (see Introduction and refs. 22 and 23). Although CYT-19 can substitute for Mss116p to facilitate the splicing of aI1 and aI2 in vivo (12), we found that the purified protein could not promote their in vitro splicing under any of the conditions tested above for aI5γ or bI1 (data not shown). This lack of effect likely reflects that both introns additionally require the maturase to stabilize the active RNA structure (see below). We showed previously that CYT-19 is similarly unable to promote group I intron splicing in the absence of CYT-18 for RNA structural stabilization (11).
The L. lactis Ll.LtrB intron is closely related to the yeast aI1 and aI2 introns and encodes an RT/maturase (LtrA protein), whose splicing activity can be assayed readily in vitro by using a shortened 0.9-kb intron deleted for the intron ORF (Ll.LtrB-ΔORF; refs. 5 and 6). In this in vitro system, CYT-19 plus ATP did not stimulate the splicing of the Ll.LtrB-ΔORF intron in the presence or absence of the maturase (data not shown). This finding may reflect that the Ll.LtrB-ΔORF intron folds without kinetic traps or that they cannot be resolved by CYT-19 plus ATP.
CYT-19 Binds Nonspecifically to Group I and Group II Intron RNAs.
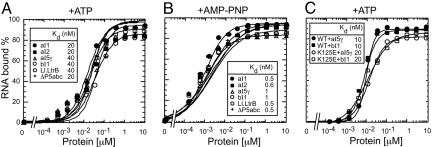
We showed previously that CYT-19 binds nonspecifically to different group I intron RNAs or poly(C) with little, if any, difference in affinity for different RNAs (11). Here, equilibrium-binding assays in 100 mM KCl/5 mM Mg2+ plus 1 mM ATP showed that CYT-19 binds to RNAs containing group II introns aI1, aI2, aI5γ, bI1, and Ll.LtrB-ΔORF and to the T. thermophila LSU-ΔP5abc group I intron, again with no significant difference in its affinity for different RNAs (apparent Kd 10–40 nM; Fig. 3A and C). The nonspecific binding of CYT-19 to group I and II intron RNAs was tighter in the presence of AMP-PNP (apparent Kd 0.5–1 nM), but again with no significant difference for different introns (Fig. 3B; see also ref. 11). In koff assays in 100 mM KCl/5 mM Mg2+ with either 1 mM ATP or AMP-PNP, complexes between CYT-19 and aI5γ or bI1 dissociated rapidly (t1/2 < 5 s; data not shown). We showed previously that the omission of ATP or the addition of ADP, Pi, or ADP plus Pi did not significantly affect the binding of CYT-19 to group I introns (11), and we confirmed that this is also the case for aI5γ and bI1 (data not shown). Importantly, the CYT-19 mutant K125E, which lacks ATPase activity (11) and was shown above to be unable to splice aI5γ and bI1 RNAs (Fig. 1C), binds to both introns with apparent Kd near those of the wild-type protein (Fig. 3C).
Fig. 3.
Equilibrium-binding assays. The indicated 32P-labeled RNAs (5 pM) were incubated with increasing concentrations of CYT-19 protein for 90 min at 30°C, and then filtered through nitrocellulose to bind RNA–protein complexes. A and B compare the binding of a group I intron (T. thermophila LSU-ΔP5abc intron) and the indicated group II introns RNAs in reaction medium containing 100 mM KCl/5 mM MgCl2 with 1 mM ATP or 1 mM AMP-PNP, respectively. C compares the binding of wild-type CYT-19 (WT) and mutant K125E to aI5γ and bI1 in reaction medium containing 100 mM KCl/5 mM MgCl2 plus 1 mM ATP. The plots show the percent of the input RNA bound to a nitrocellulose filter as a function of CYT-19 concentration. Essentially identical binding curves were obtained when RNA and protein were incubated for 10 or 30 min (data not shown).
CYT-19 Concentration Dependence of RNA Splicing.
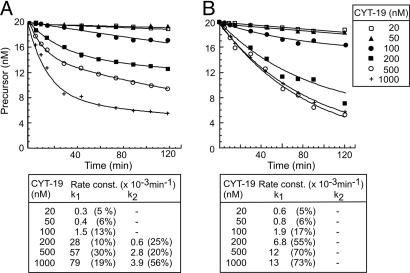
Fig. 4 shows time courses for splicing reactions of aI5γ and bI1 with 20 nM precursor RNA and increasing CYT-19 concentrations from 20 nM to 1 μM. Both the rate constant for the fast phase (k1) and the amount of spliced RNA increased with increasing protein concentration, up to 1 μM CYT-19 for aI5γ and up to 500 nM CYT-19 for bI1, with higher protein concentrations inhibiting splicing for some preparations (Fig. 4 and data not shown). At all protein concentrations tested, no further reaction occurred after 2 h.
Fig. 4.
CYT-19-concentration dependence of group II intron splicing. Splicing time courses with aI5γ (A) and bI1 (B) were done with 20 nM precursor RNA and 20 nM to 1 μM CYT-19 plus 1 mM ATP in reaction media containing 100 mM KCl/5 mM Mg2+ at 30°C. The data were best-fit to equations with one or two exponentials, and calculated rate constants for the single phase (k1) or the fast and slow phases (k1 and k2, respectively) are summarized beneath the plots. The numbers in parentheses indicate the proportion of total RNA spliced in each phase. The I-Lin band, which could contain either linear intron or broken lariat RNA, was not detectable at or below 200 nM CYT-19 for either intron, but increased to ≈4% of the total product at 500 mM CYT-19 and is equimolar with I-lar at 1 μM CYT-19. This increase most likely reflects increased breakage of lariat RNA caused by contaminating nucleases at high protein concentrations.
The increase in k1 with increasing excess CYT-19 is expected if CYT-19 action is inefficient because of its relatively weak nonspecific binding. In the alternative scenario in which CYT-19 binds and acts rapidly and the reaction is limited by the rate of self-splicing, the rate constant would not increase with increasing CYT-19 concentration. The increase in the amount of spliced RNA with increasing protein concentration likely reflects that CYT-19 loses activity during the reaction (as judged by preincubation of 20 nM unlabeled aI5γ substrate with 50 or 500 nM CYT-19 plus 1 mM ATP for 1–2 h, followed by addition of 32P-labeled substrate plus 1 mM ATP; data not shown). This instability, which was not observed for CYT-19 incubated in reaction medium without substrate and ATP (see ref. 11), complicates attempts to demonstrate CYT-19 turnover by carrying out splicing reactions at RNA excess (data not shown). Notably, in the experiment of Fig. 3, there is very little splicing up to 100 nM and then a jump in activity between 100 and 200 nM, possibly reflecting cooperativity between two or more CYT-19 molecules bound to the same RNA. Cooperativity or multimer formation is also consistent with results for group I introns (11).
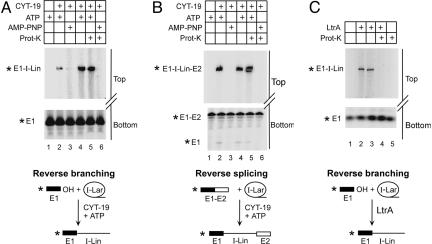
CYT-19 Plus ATP Promotes Reverse Branching and Reverse Splicing of bI1 by Acting as an RNA Chaperone.
To test whether CYT-19 acts as an RNA chaperone, whose continued presence is not required after RNA folding, we used reverse-branching and reverse-splicing assays in which intron lariat reacts in trans with 5′-labeled RNA oligonucleotide substrates corresponding to the 5′ exon (E1) or ligated exons (E1–E2). Reverse branching (reversal of the first splicing step) links the 5′ end of the intron to the 3′ end of E1, whereas reverse splicing (reversal of both steps) results in insertion of the intron between the two exons. Because these reverse reactions of aI5γ are inefficient (see ref. 24), these experiments were carried out only for bI1.
Fig. 5A and B, lanes 1–3, shows that both the reverse-branching and reverse-splicing reactions were promoted by CYT-19 plus ATP, but did not occur without CYT-19 or with CYT-19 plus AMP-PNP. Reaction products were confirmed by sequencing (data not shown). Fig. 5 A and B, lanes 4 and 5, shows reactions with lariat RNA folded in the presence of CYT-19 plus ATP for 1 h, then incubated for 30 min without (lane 4) or with protease K plus 0.1% SDS to remove CYT-19 (lane 5), before adding the RNA oligonucleotide substrate. The results show that after CYT-19-promoted refolding of intron lariat RNA both reverse branching and reverse splicing occurred with undiminished efficiency after the digestion of CYT-19 (2–5% of RNA oligonucleotide and 6–15% lariat RNA reacted in Fig. 5 A and B, lanes 4 and 5). SDS/PAGE showed no detectable CYT-19 fragments after protease K digestion (Fig. 6, which is published as supporting information on the PNAS web site). Further, no reaction was observed after protease digestion for RNA that had been preincubated with CYT-19 plus AMP-PNP (Fig. 5 A and B, lanes 6). This latter control indicates that the reactions require RNA structural rearrangements induced by CYT-19 action and are not promoted simply by CYT-19 binding or residual small peptides remaining after protease digestion.
Fig. 5.
Reverse-branching and reverse-splicing reactions. (A and B) Reactions with bI1. Gel-purified intron lariat RNA was preincubated under the conditions indicated below, before initiating the reactions by adding 5′-labeled (star) RNA oligonucleotide corresponding to the 5′ exon (E1) (reverse branching) or ligated exons (E1–E2) (reverse splicing). Lanes 1–3, lariat RNA preincubated for 90 min without (lane 1) or with CYT-19 plus ATP (lane 2) or CYT-19 plus AMP-PNP (lane 3). Lanes 4 and 5, lariat RNA preincubated with CYT-19 plus ATP for 1 h, then split with halves incubated another 30 min without (lane 4) or with protease K (Prot-K) plus SDS (lane 5). Lane 6, lariat RNA incubated for 1 h with CYT-19 plus AMP-PNP and then for 30 min with protease K plus SDS. (C) Reverse branching of the L. lactis Ll.LtrB intron. Lanes 1 and 2, lariat RNA preincubated for 50 min without (lane 1) or with LtrA maturase (lane 2). Lanes 3 and 4, lariat RNA preincubated with LtrA maturase for 20 min, then split with halves incubated for 30 min without (lane 3) or with protease K (Prot-K) plus SDS (lane 4). Lane 5, lariat RNA preincubated for 20 min and then incubated for 30 min with protease K plus SDS. All incubations were at 30°C. Products were analyzed in denaturing 4% (upper gel) and 10% (lower gel) polyacrylamide gels. Schematics of the reverse-branching and reverse-splicing reactions are shown at the bottom.
Fig. 5C shows that the mode of action of CYT-19 differs fundamentally from that of a splicing factor, the L. lactis LtrA maturase, whose continued presence is required to maintain the active RNA structure. In this case, the bound protein is required for the intron RNA to carry out reverse branching (Fig. 5C, lanes 2 and 3), but the RNA loses this activity if the protein is digested with protease before initiating the reaction (Fig. 5C, lane 4).
Discussion
We show here that the DEAD-box protein CYT-19 is by itself sufficient to promote the splicing and reverse splicing of the yeast aI5γ and bI1 group II introns under near-physiological conditions in vitro. Results for the bI1 intron indicate that CYT-19 functions as a true RNA chaperone, which interacts transiently with the intron RNA and uses the energy of ATP hydrolysis to effect RNA structural rearrangements required for ribozyme activity. Once formed the active RNA structure does not require proteins or high Mg2+ for structural stabilization. The in vitro CYT-19-dependent splicing system developed here should now permit detailed analysis of the mode of action of DEAD-box proteins on natural group II intron RNA substrates.
The CYT-19-promoted splicing reactions at near-physiological KCl and Mg2+ concentrations yield predominantly intron lariat RNA and ligated-exon products. By contrast, the self-splicing reactions of aI5γ and other group II introns at high monovalent salt and Mg2+ concentrations produce a high proportion of linear intron RNA via hydrolysis and cleaved exons via spliced-exon reopening, particularly in reaction media containing KCl (19, 20). These differences suggest that the active structures of aI5γ and bI1 at physiological KCl and Mg2+ concentrations may differ in significant ways from those formed under self-splicing conditions. A similar conclusion was reached for the maturase-promoted and self-splicing reactions of the Ll.LtrB intron based on chemical structure mapping and analysis of UV-induced RNA–RNA cross-links (7, 8). We suggested previously that inactive structures stabilized by high Mg2+ concentrations might contribute to the strongly decreased rate of self-splicing compared with maturase-dependent splicing of that intron (7, 8).
Fully understanding the mechanism by which CYT-19 promotes group II intron splicing will require detailed RNA structural analysis. However, given its demonstrated mode of action in group I intron splicing (see Introduction), it is likely that CYT-19 promotes group II intron splicing at least in part by disrupting stable inactive or intermediate structures that are kinetic traps in RNA folding. This mode of action is also suggested by the findings that CYT-19 binds RNAs nonspecifically, that it promotes the splicing of diverse group I and II introns, and that its ability to splice both group I and II introns requires ATP hydrolysis in addition to RNA binding (this work and refs. 11 and 12). It is also possible that the disruption of stable inactive structures is coupled to the formation of critical duplexes required for catalysis by a strand annealing activity, which has been demonstrated for some DEAD-box proteins (ref. 25 and references therein).
Pyle and coworkers have extensively studied the folding of a ribozyme derivative of aI5γ referred to as D135 RNA, which consists of aI5γ domains D1, D3, and D5 (26). In reaction medium containing 500 mM KCl, 100 mM MgCl2 at 42°C, D135 RNA efficiently cleaves an RNA substrate corresponding to the 5′ exon-intron junction in an efficient multiturnover reaction. By correlating the acquisition of ribozyme activity with tertiary-structure formation detected by hydroxyl-radical footprinting and other methods as a function of time and Mg2+ concentration, they concluded that the RNA folds slowly but directly to the native state without detectable kinetic traps (27, 28). The relevance of D135 folding to that of the full-length intron is unclear, however, because D135 lacks intron domains 2, 4, and 6, as well as 5′ and 3′ exons. The folding of these additional RNA segments or their interactions with other RNA segments could lead to kinetic traps. Indeed, previous studies of aI5γ splicing and reverse branching by Nolte et al. (29) provided direct evidence that 5′ exon sequences induce inhibitory structures that are rate-limiting for these reactions. Additionally, the high salt concentrations and temperatures used in the D135 ribozyme reaction may alleviate kinetic traps.
Our results suggest that group II introns may differ in their folding pathways and their degree of dependence on different types of proteins. Thus, the RNA chaperone CYT-19 is by itself sufficient to promote the in vitro splicing of aI5γ and bI1, indicating that their active structures are stable relative to inactive structures under near-physiological conditions. Nevertheless, the splicing reactions remain slower than in vivo, likely reflecting that additional, as yet unidentified, host proteins are needed for optimal RNA folding. The yeast aI1 and aI2 introns differ by encoding maturases, which stabilize their active structures (22, 23). CYT-19 contributes to the splicing of these introns in vivo (12), but cannot by itself promote their splicing in vitro because the active structure is not sufficiently stable in the absence of the maturase. The Ll.LtrB-ΔORF intron may belong to a third class, which requires a maturase but can fold without CYT-19-resolvable kinetic traps. It remains to be determined whether the full-length Ll.LtrB intron in its natural exon context in vivo requires an RNA chaperone in addition to the maturase.
Finally, with respect to evolution, the behavior of aI5γ and bI1 suggests that ancestral group II introns could have spliced and been mobile by reverse splicing into RNA and possibly DNA (30), by taking advantage of ubiquitous DExH/D-box proteins or other RNA chaperones. Such ancestral group II introns may have acquired RTs to enhance mobility and then become progressively more dependent on the encoded protein and/or host proteins for structural stabilization. The alternative “retroelement ancestor hypothesis” posits that all extant group II introns evolved from ancestral mobile introns, which used intron-encoded RT/maturases for splicing and mobility, and that many of these introns subsequently lost their ORFs and substituted host proteins for structural stabilization (31). The ability of an RNA chaperone alone to promote the splicing and reverse splicing of the ORF-less aI5γ and bI1 introns is not expected if they evolved from ORF-containing introns, like yeast aI1 and aI2 or Ll.LtrB, which were already structurally debilitated as result of their dependence on an RT/maturase (see Fig. 5). A scenario in which ancestral group II introns were self-sufficient catalytic RNAs would parallel the situation for group I introns, some of which acquired site-specific DNA endonucleases that promote mobility and then became dependent on them and/or host proteins to promote splicing (32). Spliceosomal introns continue to rely heavily on DExH/D-box proteins, which may retain the same RNA chaperone function found here now manifested in structural rearrangements of small nuclear RNAs thought to have evolved from group II intron domains.
Materials and Methods
RNA Substrates.
RNA splicing substrates containing an intron and flanking exons were synthesized by transcribing the following restriction enzyme-digested recombinant plasmids with phage T3 (aI1 and aI2) or T7 RNA polymerase (all other introns; Megascript kit; Ambion, Austin, TX): aI5γ, pJD20/HindIII (33), 1,491-nt RNA containing 288-nt 5′ exon, 887-nt intron, and 316-nt 3′ exon; bI1, pHrH154#4/EcoRI (12), 1,143-nt RNA containing 138-nt 5′ exon, 768-nt intron, and 239-nt 3′ exon; aI1, pSHΔAC/EcoRI (34); aI2, pSZD1/BstEII (35); Ll.LtrB-ΔORF, pGMΔORF/BamHI (5); and T. thermophila LSU-ΔP5abc, pBlueΔP5abc/XhoI (S.M. and A.M.L., unpublished work).
32P-labeled RNAs used for RNA splicing assays were transcribed in the presence of 50 μCi [α-32P]UTP (3,000 Ci/mmol; Amersham Pharmacia). Higher specific activity RNAs for equilibrium-binding assays were synthesized by using a Maxiscript kit (Ambion) with 500 μM ATP, GTP, and CTP, plus 50 μM UTP and 50 μCi [α-32P]UTP (3,000 Ci/mmol). After transcription, the DNA template was digested with DNase I (Invitrogen or Ambion), and RNAs were purified through three consecutive Sephadex G-50 spin columns. bI1 and Ll.LtrB lariat RNAs, used for reverse-branching and reverse-splicing reactions, were generated by self-splicing of precursor RNAs [bI1: 0.5 M (NH4)SO4, 0.1 M MgCl2, 40 mM Mops, pH 6.5, 2 h at 45°C; Ll.LtrB: 1.25 M NH4Cl, 50 mM MgCl2, 50 mM Tris·HCl, pH 7.5, 3 h, 37°C] and purified in denaturing 4% polyacrylamide gels. Before use, RNAs were dissolved in water, renatured by heating to 92°C for 2 min, 50°C for 5 min (lariat RNA only), and equilibrated in reaction medium at 30°C for 5 min. RNAs were left at room temperature and used for experiments within 2 h of preparation; RNAs stored frozen or on ice for longer periods gave inconsistent results.
RNA oligonucleotides used for reverse-branching and reverse-splicing assays were bI1 E1 (5′-GUGUUUAUGGACAGA), bI1 E1-E2 (5′-GUGUUUAUGGACAGAUGUCACAUUUG), and Ll.LtrB E1 (5′-CGUGAACACAUCCAUAAC). They were synthesized by Dharmacon Research (Lafayette, CO) and 5′ 32P-labeled with T4 polynucleotide kinase (Invitrogen).
Protein Purification.
CYT-19 protein was expressed in Escherichia coli HMS174(DE3) from plasmid pTWC19 and purified as described (11), except that proteins were precipitated with 0.01% polyethylenimine after cell lysis to ensure complete removal of nucleic acids. The purified protein was stored in 50% glycerol at 4°C. The LtrA protein encoded by the L. lactis Ll.LtrB intron was expressed in E. coli BL21(DE3) from plasmid pImp-1P and purified as described (6).
Biochemical Assays.
In vitro splicing reactions were at 30°C with specified amounts of 32P-labeled RNA and CYT-19 protein in 100 μl of reaction medium containing 100 mM NaCl, KCl, or NH4Cl, different MgCl2 concentrations, 10 mM Tris·HCl, pH 7.5 (plus 10 mM Hepes-KOH for reactions with NaCl) (16), and 10% glycerol, plus 1 mM ATP or AMP-PNP (both prepared with equimolar MgCl2). MgCl2 was purchased as a 1 M stock solution (Molecular Biology Grade; RNase-free; Sigma-Aldrich). Reactions were initiated by adding CYT-19, portions were removed at different times, and reactions were terminated by adding 0.1 M EDTA, 0.2 M Tris·HCl, pH 7.5, followed by extraction with phenol/chloroform/isoamyl alcohol (25:24:1) and ethanol precipitation. Products were analyzed in denaturing 4% polyacrylamide gels, which were dried and quantified with a PhosphorImager. PhosphorImager counts were corrected for background and normalized based on the number of U residues to calculate the molar fraction of total RNA in each band. The data from splicing time courses were best-fit to equations with one or two exponentials to obtain kobs for the single phase (k1) or for fast and slow phases (k1 and k2, respectively).
Equilibrium-binding assays were done by incubating 5 pM 32P-labeled RNA with increasing concentrations of CYT-19 at 30°C, and then filtering through nitrocellulose backed by nylon (11). The reaction medium for equilibrium-binding experiments was supplemented with BSA (0.1 mg/ml) to stabilize CYT-19 at low concentration. For koff experiments, 200 nM RNA was incubated with 100 nM CYT-19 in reaction medium containing 100 mM KCl/5 mM MgCl2 plus 1 mM ATP or AMP-PNP for 1 h at 30°C, then diluted 10-fold into reaction medium containing 600 nM unlabeled RNA and filtered through nitrocellulose as above.
Reverse-branching and reverse-splicing reactions were done with gel-purified lariat RNA (100 nM) and 5′ 32P-labeled RNA oligonucleotides (300 nM) corresponding to the 5′ exon (E1) or ligated exons (E1–E2) (see above). For bI1, reactions were at 30°C in 30 μl of 100 mM NaCl, 10 mM MgCl2, 10 mM Tris·HCl, 10 mM Hepes-KOH, pH 7.5 plus 1 mM ATP or AMP-PNP at 30°C. After preincubating lariat RNA with 500 nM CYT-19 plus 1 mM ATP or AMP-PNP, reactions were initiated by adding the RNA oligonucleotide and incubated for 10 min (reverse branching) or 60 min (reverse splicing). Reverse-branching reactions with the Ll.LtrB intron were done in 100 mM NH4Cl, 5 mM MgCl2, 40 mM Tris·HCl, pH 7.5 at 30°C. Similar results were obtained with 500 mM NH4Cl, which is optimal for maturase-promoted forward splicing (6, 7). In some incubations, 20 mg/ml protease K and 0.1% SDS were added to lariat RNA after preincubation with CYT-19 or LtrA. Products of reverse-branching and reverse-splicing reactions were analyzed in denaturing 4% and 10% polyacrylamide gels to detect both high molecular weight products and RNA oligonucleotides.
Supplementary Material
Acknowledgments
We thank Marlene Belfort (Wadsworth Center, Albany, NY), Dan Herschlag (Stanford University, Stanford, CA), Anna Marie Pyle (Yale University, New Haven, CT), and Rick Russell (University of Texas) for comments at different stages of the manuscript. This work was supported by National Institutes of Health Grant GM37951.
Abbreviations
- AMP-PNP
adenylylimidodiphosphate
- RT
reverse transcriptase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Michel M., Ferat J. L. Biochemistry. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann K., Schmidt U. Crit. Rev. Biochem. Mol. Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- 3.Pyle A. M., Lambowitz A. M. In: The RNA World. 3rd Ed. Gesteland R. F., Atkins J. F., Cech T. R., editors. Plainview, NY: Cold Spring Harbor Lab Press; 2006. pp. 469–505. [Google Scholar]
- 4.Lambowitz A. M., Caprara M. G., Zimmerly S., Perlman P. S. In: The RNA World. 2nd Ed. Gesteland R. F., Atkins J. F., Cech T. R., editors. Plainview, NY: Cold Spring Harbor Lab Press; 1999. pp. 451–485. [Google Scholar]
- 5.Matsuura M., Saldanha R., Ma H., Wank H., Yang J., Mohr G., Cavanagh S., Dunny G. M., Belfort M., Lambowitz A. M. Genes Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saldanha R., Chen B., Wank H., Matsuura M., Edwards J., Lambowitz A. M. Biochemistry. 1999;38:9069–9083. doi: 10.1021/bi982799l. [DOI] [PubMed] [Google Scholar]
- 7.Matsuura M., Noah J. W., Lambowitz A. M. EMBO J. 2001;20:7259–7270. doi: 10.1093/emboj/20.24.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noah J. W., Lambowitz A. M. Biochemistry. 2003;42:12466–12480. doi: 10.1021/bi035339n. [DOI] [PubMed] [Google Scholar]
- 9.Karpel R. L., Miller N. S., Fresco J. R. Biochemistry. 1982;21:2102–2108. doi: 10.1021/bi00538a019. [DOI] [PubMed] [Google Scholar]
- 10.Herschlag D. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 11.Mohr S., Stryker J., Lambowitz A. M. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 12.Huang H. R., Rowe C. E., Mohr S., Jiang Y., Lambowitz A. M., Perlman P. S. Proc. Natl. Acad. Sci. USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanner N. K., Linder P. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 14.Rocak S., Linder P. Nat. Rev. Mol. Cell. Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 15.Séraphin B., Simon M., Boulet A., Faye G. Nature. 1989;337:84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- 16.Niemer I., Schmelzer C., Börner G. V. Nucleic Acids Res. 1995;23:2966–2972. [PubMed] [Google Scholar]
- 17.Peebles C. L., Perlman P. S., Mecklenburg K. L., Petrillo M. L., Tabor J. H., Jarrell K. A., Cheng H. L. Cell. 1986;44:213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- 18.Schmelzer C., Schweyen R. J. Cell. 1986;46:557–565. doi: 10.1016/0092-8674(86)90881-0. [DOI] [PubMed] [Google Scholar]
- 19.Jarrell K. A., Peebles C. L., Dietrich R. C., Romiti S. L., Perlman P. S. J. Biol. Chem. 1988;263:3432–3439. [PubMed] [Google Scholar]
- 20.Daniels D. L., Michels W. J., Jr., Pyle A. M. J. Mol. Biol. 1996;256:31–49. doi: 10.1006/jmbi.1996.0066. [DOI] [PubMed] [Google Scholar]
- 21.Hiller R., Hetzer M., Schweyen R. J., Mueller M. W. J. Mol. Biol. 2000;297:301–308. doi: 10.1006/jmbi.2000.3582. [DOI] [PubMed] [Google Scholar]
- 22.Carignani G., Groudinsky O., Frezza D., Schiavon E., Bergantino E., Slonimski P. P. Cell. 1983;35:733–742. doi: 10.1016/0092-8674(83)90106-x. [DOI] [PubMed] [Google Scholar]
- 23.Moran J. V., Mecklenburg K. L., Sass P., Belcher S. M., Mahnke D., Lewin A., Perlman P. Nucleic Acids Res. 1994;22:2057–2064. doi: 10.1093/nar/22.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dème E., Nolte A., Jacquier A. Biochemistry. 1999;38:3157–3167. doi: 10.1021/bi982462j. [DOI] [PubMed] [Google Scholar]
- 25.Yang Q., Jankowsky E. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 26.Qin P. Z., Pyle A. M. Biochemistry. 1997;36:4718–4730. doi: 10.1021/bi962665c. [DOI] [PubMed] [Google Scholar]
- 27.Swisher J. F., Su L. J., Brenowitz M., Anderson V. E., Pyle A. M. J. Mol. Biol. 2002;315:297–310. doi: 10.1006/jmbi.2001.5233. [DOI] [PubMed] [Google Scholar]
- 28.Su L. J., Brenowitz M., Pyle A. M. J. Mol. Biol. 2003;334:639–652. doi: 10.1016/j.jmb.2003.09.071. [DOI] [PubMed] [Google Scholar]
- 29.Nolte A., Chanfreau G., Jacquier A. RNA. 1998;4:694–708. doi: 10.1017/s1355838298980165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambowitz A. M., Zimmerly S. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 31.Toor N., Hausner G., Zimmerly S. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belfort M., Derbyshire V., Parker M., Cousineau B., Lambowitz A. M. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol; 2002. pp. 761–783. [Google Scholar]
- 33.Jarrell K. A., Dietrich R. C., Perlman P. S. Mol. Cell. Biol. 1988;8:2361–2366. doi: 10.1128/mcb.8.6.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hebbar S. K., Belcher S. M., Perlman P. S. Nucleic Acids Res. 1992;20:1747–1754. doi: 10.1093/nar/20.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmerly S., Guo H., Eskes R., Yang J., Perlman P. S., Lambowitz A. M. Cell. 1995;83:529–538. doi: 10.1016/0092-8674(95)90092-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.