Abstract
Antibodies are generally thought to be a class of proteins that function without the use of cofactors. However, it is not widely appreciated that antibodies are believed to be the major carrier protein in human circulation for the important riboflavin cofactor that is involved in a host of biological phenomena. A further link between riboflavin and antibodies was discovered 30 years ago when a bright-yellow antibody, IgGGAR, was purified from a patient with multiple myeloma who had turned yellow during the course of her disease. It was subsequently shown that the yellow color of this antibody was due to riboflavin binding. However, it was not known how and where riboflavin was bound to this antibody. We now report the crystal structure of this historically important IgGGAR Fab at 3.0-Å resolution. The riboflavin is located in the antigen-combining site with its isoalloxazine ring stacked between the parallel aromatic moieties of TyrH33, PheH58, and TyrH100A. Together with additional hydrogen bonds, these interactions reveal the structural basis for high-affinity riboflavin binding. The ligand specificity of IgGGAR is compared with another riboflavin-binding antibody, IgGDOT, which was purified from a second patient with multiple myeloma. The crystal structure of IgGGAR provides a starting point for attempts to understand the physiological relevance and chemical functions of cofactor-containing antibodies.
Keywords: cancer, cofactor, human antibody, myeloma, riboflavin-binding
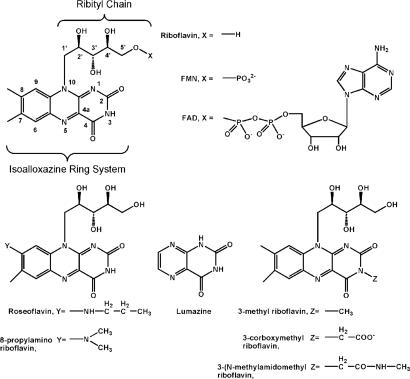
A bright-yellow antibody, IgGGAR, from a patient with multiple myeloma, was discovered and characterized by Farhangi and Osserman in 1976 (1). The patient (“Gar”) was referred to the Francis Delafield Hospital, New York, in May 1973, with a diagnosis of multiple myeloma. Physical examination confirmed that the patient presented with an intense and unusually bright-yellow coloration of the skin (xanthoderma) and hair (xanthotrichia), but normal white sclera, suggesting a non-bilirubin-related abnormality in pigment metabolism. Further studies found that the patient’s serum was bright yellow, and all the xanthochromia was associated with a monoclonal antibody, IgGGAR. The cause of pigmentation was ultimately shown to be riboflavin binding by this antibody. IgGGAR, a human IgG2(λ), is bright-yellow in color and binds riboflavin with a Kd of 1.8 nM (2). Although IgGGAR was isolated with tightly bound riboflavin, other riboflavin derivatives can act as ligands (Fig. 1), such as flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), with Kd values of 2.2 nM and 8 nM, respectively (2). Later, in 1990, Merlini et al. (3) discovered and characterized a second anti-riboflavin antibody, IgGDOT, from another patient with multiple myeloma. IgGDOT was also bright-yellow with bound riboflavin, and showed remarkable similarities to IgGGAR.
Fig. 1.
Chemical structures of riboflavin, FMN, FAD, and six riboflavin analogues: roseoflavin, 8-propylamino riboflavin, lumazine, 3-methyl riboflavin, 3-carboxymethyl riboflavin, and 3-(N-methylamidomethyl) riboflavin.
Although the biological role of that fraction of antibodies that carry riboflavin is unclear, further progress requires an understanding of how riboflavin is bound to antibodies in normal and disease states. Toward this end, the riboflavin-binding myeloma proteins continue to be of key interest because they represent the only available source to investigate the structure and function of natural, human riboflavin-binding antibodies (2). We now report the crystal structure of the myeloma protein, IgGGAR, which had been stored in the freezer for >30 years.
Results and Discussion
Fab Structure of IgGGAR.
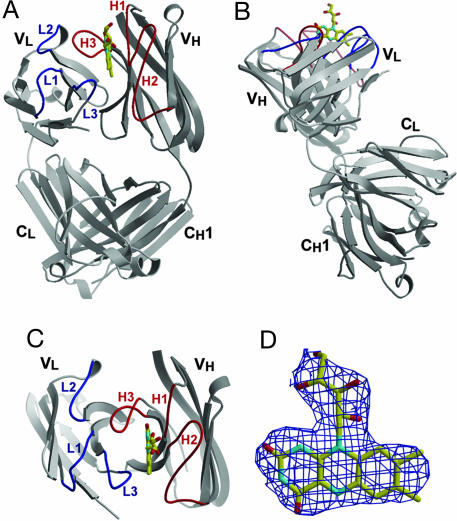
IgGGAR Fab consists of a human Ig γ2 heavy chain and a λ light chain (Fig. 2). In the Fab–riboflavin complex crystals, noncrystallographic symmetry (NCS) results in two pairs of Fabs, corresponding to molecules A (peptide chain ID L, H) and B (peptide chain ID A, B) and to molecules C (peptide chain ID C, D) and D (peptide chain ID E, F) that are related by a pseudotranslation of ≈1/3, 1/2, and 0, respectively. NCS restraints were, therefore, used in the structural refinement. All four Fab–riboflavin complexes are similar and, unless noted otherwise, the discussion will focus only on one representative complex, A.
Fig. 2.
Crystal structure of antibody IgGGAR. (A) Standard view of IgGGAR Fab in its complex with riboflavin, with the light and heavy chains colored in light and dark gray, respectively, a convention followed in all subsequent figures. The loops corresponding to CDRs L1, L2, and L3 are colored blue, whereas CDRs H1, H2, and H3 are colored red. Bound riboflavin is also shown in ball-and-stick presentation with yellow carbons in the combining site. (B) Side view of IgGGAR Fab. The elbow angle of IgGGAR is among one of the largest seen in antibody structures. (C) Top view of IgGGAR Fv fragment, with riboflavin bound between CDRs L3, H1, H2, and H3. (D) Electron density for the riboflavin in IgGGAR Fab. A σA-weighted 2Fo-Fc map was contoured at 1.0 σ. Figs. 2 and 3A were generated in bobscript (26) and rendered in raster3d (27).
The elbow angles, which correspond to the pseudodyad axis between the Fab constant and variable domains, are 231°, 235°, 219°, and 217°, respectively for the four IgGGAR Fab molecules A, B, C, and D (also see Fig. 2 A and B). Antibody flexibility is believed to aid in bivalent recognition of intact pathogens by the two Fab domains of the Ig (4). To date, the reported largest elbow angle in all the other reported antibody structures is 225° (PDB code 1ADQ). Significantly, antibodies with λ light chains have the largest elbow angles and ≈2/3 have elbow angles >180° (5). Fabs A and B and C and D have similar crystal-packing environments that are reflected in similar elbow angles.
Complementarity-determining region (CDR) L1 has no residue insertions after residue 27 and does not belong to any previously classified canonical structures (6, 7). The same lack of any insertion after residue 27 in L1 was also noted in two other λ light-chain Fab structures (PDB codes 1NFD and 8FAB) and may represent another canonical structure. CDR L2 belongs to canonical class λL2-1. CDR L3 has one insertion after residue 95 and is found only in one other λ light-chain Fab structure (PDB code 1AQK), which also does not belong to any previously classified canonical structure. CDRs H1 and H2 belong to canonical classes 1 and 3A according to their length and main-chain torsion angles, respectively. CDR H3 contains three insertions after 100, and has a typical bulged-torso conformation at residue 101 that is stabilized by the canonical salt bridge between ArgH95 and AspH101 (8).
Binding of Riboflavin in the Combining Site.
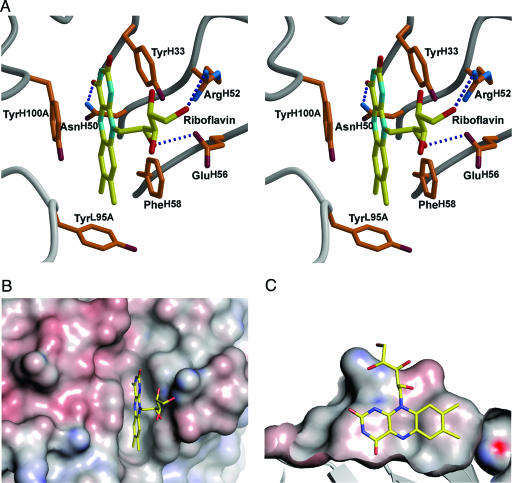
Although no riboflavin was added in the purification and crystallization processes, clear interpretable electron density corresponding to a riboflavin molecule was found in the antibody-combining site (Fig. 2), despite the storage of the antibody for >30 years. The Fab fragment forms a rather shallow binding slot rather than a groove (Fig. 3B and C), which nicely accommodates the riboflavin with a good shape correlation (9) of 0.82. The riboflavin molecule is trapped in the binding site such that the isoalloxazine ring is buried in the narrow slot in the binding pocket, whereas the ribityl side-chain extends out to the pocket rim (Figs. 2 and 3). Upon binding of riboflavin, 258 Å2 of the molecular surface of the Fab fragment is covered, whereas riboflavin itself is 68% buried (229 of 337 Å2) in the binding site. A total of 130 van der Waals’ interactions and four hydrogen bonds are made between antibody and riboflavin, with only 17 of these van der Waals’ interactions from the light chain. Residues from four CDR loops contact the riboflavin: GlyL95 (L3), TyrL95A (L3), ProL96 (L3), TyrH33 (H1), AsnH50 (H2), ArgH52 (H2), GluH56 (H2), PheH58 (H2), ValH95 (H3), and TyrH100A (H3). No contacts are made with CDR loops L1 and L2, although the latter is not surprising, because most haptens are unable to access CDR L2 when bound in the combining site (10).
Fig. 3.
Stereoview of the IgGGAR antigen-binding site. (A) The combining site with bound riboflavin. Hydrogen bonds are shown as dotted blue lines. The isoalloxazine ring is π-stacked between aromatic residues TyrH33, PheH58, and TyrH100A, the N5 atom of the ring hydrogen bonds to AsnH50, and the ribityl side chain contributes two hydrogen bonds to ArgH52 and GluH56. (B and C) Top view (B) and side view (C) of shape complementarity of riboflavin in the antibody IgGGAR-combining site prepared with pymol (http://pymol.sourceforge.net). The isoalloxazine ring is trapped in a narrow slot in the combining site, and the ribityl side chain rests on the outer surface of the combining site. The molecular surface is colored by electrostatic potential (calculated with the program apbs (28) with a 1.4-Å probe radius and contoured between −20 and +20 kT).
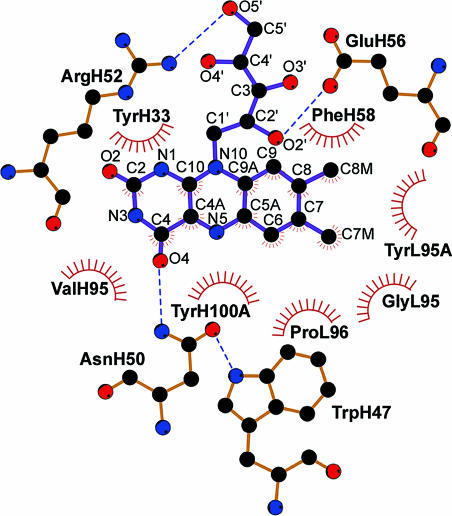
Binding of riboflavin occurs in a narrow cleft, with the vitamin isoalloxazine ring stacked between the parallel aromatic groups of TyrH33 (with the si-face of riboflavin), PheH58 (si-face), and TyrH100A (re-face), with distances between the isoalloxazine ring and the respective aromatic rings from these three residues of ≈3.2, 3.5, and 3.4 Å (Figs. 3A and 4). These π–π stackings presumably contribute to the high-affinity binding of riboflavin. The isoalloxazine ring of flavins is amphipathic, because the xylene portion is hydrophobic, and the pyrimidine moiety is hydrophilic. The isoalloxazine ring of riboflavin in IgGGAR nestles on the floor of the binding site, with xylene and pyrimidine moieties buried. The Nδ2 of AsnH50 is hydrogen bonded to O4 and is only 3.5 Å from N5 of the isoalloxazine ring (Fig. 3). AsnH50 is also oriented by a hydrogen bond from its Oδ1 to Nε1 of TrpH47 (Fig. 4). Hence, AsnH50 is a key residue for flavin binding. The isoalloxazine ring also makes van der Waals’ interactions with GlyL95, TyrL95A, ProL96, and ValH95. According to the Kabat–Wu database (11), the key residues for riboflavin binding, TyrH33, PheH58, TyrH100A, and AsnH50, occur in only ≈21%, 2%, 10%, and 5% of antibody sequences, respectively.
Fig. 4.
Schematic presentation of riboflavin binding site in IgGGAR. Residues forming van der Waals’ interactions with the riboflavin are indicated by an arc with radiating spokes toward the ligand atoms they contact; those participating in the hydrogen bonds with the riboflavin are shown in ball-and-stick representations. Hydrogen bonds are illustrated as blue dotted lines. Carbon atoms are colored in black, nitrogen atoms in blue, and oxygen atoms in red. Atom names of the riboflavin are labeled. The figure was generated from the program ligplot (29).
For the ribityl side-chain interactions, two hydrogen bonds are formed between the side chain and the antibody: O2′ to the carboxyl group of GluH56, and O5′ to the guanidinium group of ArgH52 (Figs. 3 and 4). The ribityl moiety also makes van der Waals’ interactions with TyrH33 and PheH58. In the crystallographic asymmetric unit, a side chain from a neighboring molecule hydrogen bonds with the riboflavin; GluH85 in IgGGAR molecules C and D hydrogen bonds to O5′ of riboflavins bound to molecules A and B, respectively.
The oxidized riboflavin in IgGGAR exhibits a planar isoalloxazine-ring configuration (Figs. 2 and 3). IgGGAR loses its yellow color under reducing conditions (data not shown). Although “butterfly” flavin conformations have been observed in a number of crystal structures under reducing conditions (12), other cases have been observed where the reduced flavin is in an almost planar conformation (13). Planar oxidized or puckered reduced flavins have both been observed in the same active site with no large conformational changes in other systems (12). However, whether any structural rearrangements of IgGGAR occur in a reducing environment needs further exploration. In this crystal structure, no water-mediated hydrogen bonds are formed between antibody and riboflavin, as seen for some other flavin-protein structures. However, because this structure is at comparatively modest resolution (3 Å), it is difficult to define all of the bound water molecules.
Structural Basis for Ligand Specificity.
FMN, FAD, and a variety of riboflavin analogues (Fig. 1) bind IgGGAR with various affinities (1, 2, 14). The Kd values for FMN and FAD are 2.2 nM and 8 nM, respectively, as compared with the Kd for riboflavin of 1.8 nM (2). These similar kd values indicate that binding of IgGGAR with flavins is rather insensitive to the relative size and charge of the substituent at the C5′ position of the ribityl moiety. FMN, with a negatively charged phosphate group at C5′, exhibits only a 4-fold reduction in affinity relative to riboflavin, whereas FAD adenine attached to C5′ by a phosphodiester linkage binds IgGGAR with a 10-fold decrease in its kd relative to riboflavin. These findings correlate with our structural results in that the isoalloxazine ring is clearly the most important determinant for recognition and likely contributes most of the binding energy where the ribityl side chain extends out of the binding site toward the antibody surface.
A series of riboflavin analogues have been tested in binding studies with IgGGAR (14). Derivatives with large substituent at the 8-position of the flavin, such as roseoflavin and 8-propylaminoriboflavin (Fig. 1), have only an ≈10-fold decrease in binding affinity. IgGGAR could accommodate such bulky substituents at the 8-position of the flavin where they would point toward the solvent. The altered electronic structure of these analogues was proposed as the possible reason for the observed decrease in binding to IgGGAR (14). Interestingly, lumazine (Fig. 1), which eliminates the xylene ring of the isoalloxazine, has a decreased binding affinity of at least four orders of magnitude relative to riboflavin, indicating the importance of the interactions of the xylene ring, as observed in the crystal structure. A small methyl substituent at N3 decreases binding of flavin to IgGGAR by 10-fold, because this substituent would form close contacts with TyrH33, consistent with the larger substituents not being tolerated, such as 3-carboxymethyl riboflavin or 3-(N-methylamidomethyl) riboflavin (Fig. 1) (14).
Comparison with Other Flavin-Binding Proteins.
Flavins can participate in both one- and two-electron transfer processes and play a pivotal role in a host of biological phenomena. These flavoprotein enzymes catalyze a vast number of biochemical reactions, from oxidases to dehydrogenases and monooxygenases (15). The isoalloxazine ring is the redox-active center of the flavin, where its flavin redox potential is regulated by its protein environment. In a typical redox reaction, when electrons are transferred into the isoalloxazine ring through the electron-deficient N5 atom (16), the negative charge is dispersed over C4A, N5, N10, and N1 (17). Stabilization of this negative charge by the protein is an important factor for the redox potential; a positively charged environment around the pyrimidine ring will increase its potential, whereas a negatively charged or hydrophobic environment will decrease it (15). In the available crystal structures of >40 flavoproteins, the majority of the flavin–protein interactions are made with the ribityl side chains of riboflavin, FMN, or FAD (18). In IgGGAR, the pyrimidine ring of riboflavin is buried in a mostly hydrophobic environment, where the N5 atom is positioned inside the binding pocket. Thus, N5 could be difficult to access by most large potential redox substrates, but molecular oxygen and its redox derivatives would not be excluded.
IgGDOT is another natural riboflavin-binding antibody that was purified from a patient also with multiple myeloma, xanthoderma, and xanthotrichia (3). Purified IgGDOT is also bright yellow, and its ligand was identified as riboflavin. IgGDOT shares an almost identical affinity for flavins with IgGGAR, with kd values for riboflavin, FMN, and FAD of 1.7, 6.6, and 18 nM, respectively (2). Comparison of their amino acid sequences (2) indicates that the key residues for riboflavin binding in IgGGAR, TyrL95A, TyrH33, and TyrH100A, are conserved in both IgGGAR and IgGDOT antibodies. The sequence similarity (51% and 45% identity between the light and heavy chains, respectively), together with the similar affinity for flavins, suggests that riboflavin might adopt a similar binding mode with IgGDOT as with IgGGAR. It is noticeable, however, that some other key binding residues in IgGGAR, such as AsnH50, ArgH52, and PheH58, are not conserved and are substituted by IleH50, AsnH52, and SerH58, respectively, in IgGDOT, suggesting a slightly different or modified flavin-binding mode.
Discussion.
Immunoglobulins and albumin are the main riboflavin-carrier proteins in the plasma (19). Albumin binds riboflavin only very weakly, with kd values of 3.8 to 10.4 mM (19), whereas high-affinity binding was detected in the normal human plasma Ig fraction with kd values of 2.43 and 0.07 nM for two proposed binding sites (20). Any assumptions about the physiological role of riboflavin-binding immunoglobulins must center on whether one considers the bound riboflavin an antigen or whether the riboflavin-binding myeloma proteins simply reflect clonal expansion during disease of that subclass of antibodies that normally bind riboflavin. If, in our studies, riboflavin had been found to bind outside of the combining site, one might have simply concluded that, although it is curious that immunoglobulins are the major carrier of riboflavin, it has no role in antibody function. However, the fact that riboflavin located deep within the combining site makes the situation more intriguing and complex. If one considers riboflavin simply an antigen, this immediately invites speculation as to the consequences of having an immune system that attempts to remove an exogenous antigen that is essential to life. In essence, this situation reduces to antigenic drive from which there is no escape. One might expect such a system to drive to oncogenesis, as might have happened in the two reported patients whose myeloma proteins bound riboflavin. If this is the case, myeloma proteins that bind other essential nutrients may have been missed because the nutrients lack the striking color of riboflavin or they bind substochiometric amounts of ligands because their physiological concentration is much lower than that of riboflavin. An alternative view is that, although riboflavin is bound in the combining site, it is not necessarily an antigen. For this notion, riboflavin may function as a cofactor for a subclass of antibodies and, thus, endow that class of antibodies that carry it with unique chemical properties in much the same way as it does for so many enzymes. Here, it would be likely that any oxidant would be derived from redox-active leukocytes that have responded to the antigen–antibody union. Alternatively, the bound riboflavin acts as a sensitizer that is activated by light through the skin in the same way that porphyrins cause skin damage when they are present in excess quantities in the disease porphyria. Although many years ago this idea may have seemed somewhat heretical, we now know that antibodies are capable of catalyzing a wide variety of chemical reactions. Indeed, we have found that, in the presence of white light and riboflavin, antibodies are capable of carrying out complex redox reactions that destroy bound antigens (data not shown).
Materials and Methods
Purification, Crystallization, and Data Collection.
Human monoclonal antibody IgGGAR Fab was produced by standard protocols. The yellow native IgGGAR was purified from material received from the late Elliott Osserman (Columbia University, New York). IgGGAR, a human IgG2(λ) Ig, was isolated from an 80-year-old patient, as reported in ref. 1. In brief, IgGGAR was recovered from the patient’s plasma by ammonium sulfate precipitation, followed by gel filtration, as described in ref. 21. The purified IgG from the 30-year-old frozen sample was digested to Fab fragments with 1% (wt/wt) papain for 20 h at 37°C, followed by size-exclusion (Superdex-200 column; Amersham Pharmacia) and protein A-affinity chromatography.
IgGGAR Fab was concentrated to 10.7 mg/ml in 0.025 M Tris (pH 7.5) and 100 mM NaCl. Yellow Fab crystals were grown by the sitting-drop vapor-diffusion method from 23% (wt/vol) PEG 4000, 0.2 M diammonium hydrogen citrate, and 0.05 M Tris (pH 7.5). A 3.0-Å data set was collected at the Stanford Synchrotron Radiation Laboratory (SSRL) on beamline 11–1 [Area Detection Systems Corporation (Poway, CA) Quantum 315 charge-coupled device detector] from a single crystal with 25% (vol/vol) glycerol as cryoprotectant. The crystal space group is P212121, with four Fab/riboflavin complexes per asymmetric unit (Vm = 2.8 Å3/Da, 57% solvent). Data were integrated and scaled with HKL2000 (22) (see Table 1).
Table 1.
Data collection and refinement statistics of IgGGAR Fab
| Space group | P212121 |
| Unit cell, Å | a = 100.6, b = 111.1, c = 188.5 |
| Resolution, Å | 50.0–3.00 (3.07–3.00)* |
| X-ray source | SSRL 11–1 |
| No unique reflections | 42,610 |
| Redundancy | 3.8 (3.7) |
| Average I/σ(I) | 15.5 (2.1)* |
| Completeness | 98.9 (99.3)* |
| Rmerge† | 0.10 (0.81)* |
| Refined residues | 1,716 |
| Refined waters‡ | 144 |
| Rcryst§ | 0.240 |
| Rfree¶ | 0.290 |
| Average B values, Å2 | |
| Protein | 61.4 |
| Waters | 33.9 |
| Ligands | 67.5 |
| Ramachandran plot, %∥ | 83.0, 16.5, 0.3, 0.3 |
| rmsd Bond lengths, Å | 0.009 |
| rmsd Bond angles, ° | 1.6 |
rmsd, rms deviation.
*Parentheses denote outer-shell statistics.
†Rmerge = ΣhΣi |Ii(h) − 〈I(h)〉|/ΣhΣi Ii (h), where 〈I(h)〉 is the average intensity of i symmetry-related observations of reflections with Bragg index h.
‡Although it is unusual to refine many waters at this resolution, many were unambiguous and refined well. The 144 waters bind to four Fab molecules in the asymmetric unit.
§Rcryst = Σhkl |Fo − Fc|/Σhkl |Fo|, where Fo and Fc are the observed and calculated structure factors.
¶Rfree was calculated as for Rcryst but on 5% of data excluded before refinement.
∥The values are percentages of residues in the most favored, additional allowed, generously allowed, and disallowed regions. AspL51, as expected, shows main-chain torsion angles in the disallowed regions but in a well defined γ-turn that is observed in most antibody structures (6).
Structure Determination.
The IgGGAR Fab structure was determined by molecular replacement methods by using the program molrep (23). A Fab fragment from human Ig Hil (IgG1, λ) (PDB code 8fab; molecule 2) was used as the search model for rotation and translation function calculations (correlation coefficient = 0.38, Rcryst = 0.54 for the resolution range 40.0–4.0 Å). A pseudotranslation relates pairs of Fabs by the vector (0.33, 0.5, 0) or by a/3 + b/2, as deduced from native Patterson maps. Structural refinement was completed to 3.0-Å resolution by using the program cns (24) to a final Rcryst = 0.24 and Rfree = 0.29 for all data (see Table 1). Model rebuilding was performed by using the program o (25). Data collection and refinement statistics are summarized in Table 1.
Acknowledgments
We remember with gratitude and affection the late Dr. Elliott Osserman, who provided the sample of the native antibody IgGGAR. We thank the staff of the Stanford Synchrotron Radiation Laboratory BL11-1 and several laboratory members, especially Dr. Xiaoping Dai, for assistance with data collection. This work was supported by National Institutes of Health Grants CA38273 (to I.A.W.) and CA27489 (to I.A.W. and R.A.L.) and The Skaggs Institute for Chemical Biology.
Abbreviations
- CDR
complementarity-determining region
- FAD
flavin adenine dinucleotide
- FMN
flavin mononucleotide
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: Atomic coordinates and diffraction data for IgGGAR Fab have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2FL5).
References
- 1.Farhangi M., Osserman E. F. N. Engl. J. Med. 1976;294:177–183. doi: 10.1056/NEJM197601222940401. [DOI] [PubMed] [Google Scholar]
- 2.Stoppini M., Bellotti V., Negri A., Merlini G., Garver F., Ferri G. Eur. J. Biochem. 1995;228:886–893. doi: 10.1111/j.1432-1033.1995.tb20336.x. [DOI] [PubMed] [Google Scholar]
- 3.Merlini G., Bruening R., Kyle R. A., Osserman E. F. Mol. Immunol. 1990;27:385–394. doi: 10.1016/0161-5890(90)90162-s. [DOI] [PubMed] [Google Scholar]
- 4.Wilson I. A., Stanfield R. L. Nat. Immunol. 2001;2:579–581. doi: 10.1038/89718. [DOI] [PubMed] [Google Scholar]
- 5.Stanfield R. L., Zemla A., Wilson I. A., Rupp B. J. Mol. Biol. 2006 doi: 10.1016/j.jmb.2006.01.023. in press. [DOI] [PubMed] [Google Scholar]
- 6.Al-Lazikani B., Lesk A. M., Chothia C. J. Mol. Biol. 1997;273:927–948. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- 7.Martin A. C., Thornton J. M. J. Mol. Biol. 1996;263:800–815. doi: 10.1006/jmbi.1996.0617. [DOI] [PubMed] [Google Scholar]
- 8.Morea V., Tramontano A., Rustici M., Chothia C., Lesk A. M. J. Mol. Biol. 1998;275:269–294. doi: 10.1006/jmbi.1997.1442. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence M. C., Colman P. M. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 10.Wilson I. A., Stanfield R. L. Curr. Opin. Struct. Biol. 1994;4:857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 11.Johnson G., Wu T. T. Nucleic Acids Res. 2001;29:205–206. doi: 10.1093/nar/29.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lennon B. W., Williams C. H., Jr., Ludwig M. L. Protein Sci. 1999;8:2366–2379. doi: 10.1110/ps.8.11.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyte J. Mechanism in Protein Chemistry. New York: Garland; 1995. pp. 72–90. [Google Scholar]
- 14.Pologe L. G., Goyal A., Greer J. Mol. Immunol. 1982;19:1499–1507. doi: 10.1016/0161-5890(82)90196-1. [DOI] [PubMed] [Google Scholar]
- 15.Bruggeman Y. E., Honegger A., Kreuwel H., Visser A. J., Laane C., Schots A., Hilhorst R. Eur. J. Biochem. 1997;249:393–400. doi: 10.1111/j.1432-1033.1997.00393.x. [DOI] [PubMed] [Google Scholar]
- 16.Platenkamp R. J., Palmer M. H., Visser A. J. W. G. Eur. Biophys. J. 1987;14:393–402. [Google Scholar]
- 17.Hall L. H., Orchard B. J., Tripathy S. K. Int. J. Quantum Chem. 1987;31:217–242. [Google Scholar]
- 18.Massey V. Biochem. Soc. Trans. 2000;28:283–296. [PubMed] [Google Scholar]
- 19.Innis W. S., McCormick D. B., Merrill A. H., Jr. Biochem. Med. 1985;34:151–165. doi: 10.1016/0006-2944(85)90106-1. [DOI] [PubMed] [Google Scholar]
- 20.Watson C. D., Ford H. C. Biochem. Int. 1988;16:1067–1074. [PubMed] [Google Scholar]
- 21.Chang M. Y., Friedman F. K., Beychok S., Shyong J. S., Osserman E. F. Biochemistry. 1981;20:2916–2921. doi: 10.1021/bi00513a031. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Vagin A., Teplyakov A. J. Appl. Crystallog. 1997;30:1022–1025. [Google Scholar]
- 24.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Esnouf R. M. Acta Crystallogr. D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 27.Merritt E. A., Murphy M. E. P. Acta Crystallogr. D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 28.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace A. C., Laskowski R. A., Thornton J. M. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]