Abstract
The nicotinic acetylcholine receptor (nAChR) is the prototype member of the superfamily of pentameric ligand-gated ion channels. How the extracellular ligand-binding domain coordinates selective binding of ligand molecules to different subtypes of the receptor is unknown at the structural level. Here, we present the 2.2-Å crystal structure of a homolog of the ligand-binding domain of the nAChR, Aplysia californica AChBP (Ac-AChBP), in complex with α-conotoxin ImI. This conotoxin is unique in its selectivity toward the neuronal α3β2 and α7 nAChR, a feature that is reflected in its selective binding to Ac-AChBP compared with other AChBP homologs. We observe a network of interactions between the residues of the ligand-binding site and the toxin, in which ImI Arg-7 and Trp-10 play a key role. The toxin also forms interactions in the ligand-binding site that were not seen in the complex of Ac-AChBP with PnIA(A10L D14K), a conotoxin variant that lacks binding selectivity to AChBP homologs. In combination with electrophysiological recordings obtained by using the wild-type α7 nAChR and L247T mutant, we show that conotoxin ImI inhibits ion conduction by stabilizing the receptor in a desensitized conformation. Comparison of the Ac-AChBP–ImI crystal structure with existing AChBP structures offers structural insight into the extent of flexibility of the interface loops and how their movement may couple ligand binding to channel gating in the context of a nAChR.
Keywords: ligand-gated ion channel, x-ray crystallography, cys-loop receptor, protein structure, acetylcholine binding protein
Acetylcholine binding protein (AChBP) is a water-soluble homolog of the extracellular ligand-binding domain of the nicotinic acetylcholine receptor (nAChR). Its biological role is to modulate synaptic transmission when glial cells release it into cholinergic synapses of the freshwater snail Lymnaea stagnalis (1). AChBP is characterized by pharmacological properties that mostly resemble the neuronal α7 nAChR, which forms functional homopentameric receptors. Determination of the crystal structure of AChBP from L. stagnalis (Ls-AChBP) has revealed the molecular architecture of the ligand-binding domain into atomic detail and has rationalized the results from a vast amount of structure–function studies (2–4). In particular, the structure of AChBP has served as a model for the ligand-binding domain of nAChRs and other members of the family of ligand-gated ion channels, including the 5-HT3, glycine, GABAA, and GABAC receptors.
Recently, AChBPs of two other species were identified, namely Ac-AChBP from Aplysia californica (5) and Bt-AChBP from Bulinus truncatus (6). Crystallization and structure determination of these two novel members of the AChBP family has shown that their overall structural scaffold is strikingly similar despite their low sequence conservation (6, 7). So far, the x-ray structures of all three AChBPs contained buffer molecules, either Hepes (8) or CAPS (6), in their ligand-binding sites when the proteins were crystallized in the absence of known ligands of the nAChR. Cocrystallization of Ls-AChBP with nicotine and carbamylcholine has revealed the atomic interactions required for binding of these two agonists of the nAChR (9). Recently, we also solved the first structure of Ac-AChBP in complex with an α-conotoxin, which acts as an antagonist at the nAChR (7). Comparison with the agonist-bound structures showed that binding of the α-conotoxin to AChBP stabilizes the receptor in a conformation where the C-loop is displaced outward. This feature was also visible in a relatively low-resolution structure of Ls-AChBP in complex with α-cobratoxin (10), a member of the family of the long-chain α-neurotoxins that also inhibits the nAChR (11).
Despite these insights, little is known about the determinants required for binding of subtype-selective ligands to the nAChR at the structural level. Conotoxin ImI, another member of the family of α-conotoxins (12, 13), has initially been described as a specific and high-affinity inhibitor of the α7 nAChR (14) and has been extensively studied using site-directed mutagenesis (15–21), cycle mutant analysis (16), and docking simulations (22, 23). It was recently reported that conotoxin ImI displays even higher affinity for the human α3β2 nAChR, but it has been difficult to generalize the ImI specificity because it was reported not to have any affinity for rat α3β2 nAChR (22, 24). The selective binding of conotoxin ImI to nAChR subtypes is reflected in the specific inhibition of Ac-AChBP compared with Ls-AChBP (5, 7). In this work, we report the crystal structure of Ac-AChBP in complex with conotoxin ImI. By comparison with the structure of Ac-AChBP in complex with conotoxin PnIA(A10L D14K), a conotoxin variant that binds to AChBPs in an unselective manner (7), we dissect the molecular interactions required for subtype-selective ligand binding.
Together with electrophysiological recordings on the α7 nAChR and L247T mutants, we show the relevance of the x-ray structure to the intact nAChR. Based on a comparison of the Ac-AChBP-ImI crystal structure with other AChBP structures, we show the extent of flexibility of the interface loops and how their movement may couple ligand binding to channel gating in the context of a nAChR.
Results and Discussion
Pharmacological and Electrophysiological Properties of α-Conotoxin ImI.
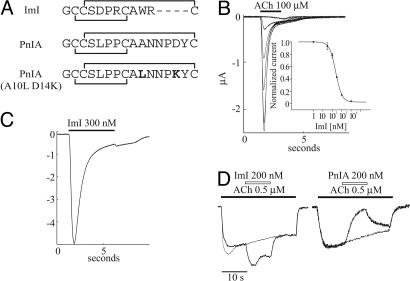
To identify and select members of the α-conotoxin family as suitable candidates for cocrystallization purposes with AChBP, we had characterized the interaction of a series of α-conotoxins with all known AChBPs in a competitive binding assay with 125I α-bungarotoxin and isothermal titration calorimetry (7). We showed that α-conotoxin PnIA, containing mutations A10L and D14K (Fig. 1A), binds with high affinity to both Ls-AChBP (IC50 = 13 ± 2 nM) and Ac-AChBP (IC50 = 28 ± 6 nM). Conversely, α-conotoxin ImI binds in a more discriminative manner and shows high affinity for Ac-AChBP (IC50 = 33 ± 5 nM) but much lower affinity for Ls-AChBP (IC50 = 4,140 ± 400 nM). Strong selectivity has been reported by Hansen et al. (5) using intrinsic Trp fluorescence.
Fig. 1.
Functional characterization of ImI. (A) Sequence alignment of α-conotoxins relevant to this work. (B) ImI inhibits nondesensitizing ACh-evoked currents at WT α7 nAChRs. (C) ImI (300 nM) activates a current at α7-L247T receptors. (D) ImI elicits a current in the presence of a low concentration of ACh at the nondesensitizing α7 L247T receptor. The filled bar indicates the application of ACh (0.5 μM), and the open bar indicates the application of toxin. A control current (fine trace) to ACh is shown superimposed; currents were normalized, and control current amplitude was 400 nA.
PnIA containing the A10L mutation dramatically differs from WT toxin in that it preferentially stabilizes a desensitized state of the nAChR (25). Although both α-conotoxins act as inhibitors of WT α7 nAChRs, WT PnIA inhibits the nondesensitizing L247T mutant nAChRs, whereas PnIA (A10L) acts as an agonist at these receptors (25). Because it has been shown that introduction of the mutation L247T in α7 changes the property of this receptor by rendering conductive a desensitized state (26), the easiest explanation for this observation is to assume that PnIA (A10L) stabilizes a desensitized conformation of the nAChR. This unusual behavior is characteristic for the class of curaremimetic compounds (27, 28) and is different from snake α-neurotoxins, which inhibit the nAChR by stabilizing a resting state of the receptor (29, 30). Because the electrophysiological properties of α-conotoxin ImI have not been investigated in this context, we compared the functional effects of ImI on WT and L247T α7 nAChRs. Representative current traces in Fig. 1B show how increasing concentrations of α-conotoxin ImI progressively inhibit acetylcholine (ACh)-evoked currents at the WT α7 nAChR. From the concentration-response relationship, we calculated IC50 = 232 nM and a Hill coefficient of 1.3 (Fig. 1B Inset), which is similar to reported values. Application of conotoxin ImI to oocytes expressing L247T α7 nAChRs evokes currents (Fig. 1C), similar to mutant conotoxin PnIA (A10L), suggesting that these toxins activate the mutant receptor.
Ellison et al. (22) recently demonstrated that conotoxin ImI also displays high affinity for the human α3β2 nAChR (IC50 = 41 nM), which contradicts an earlier report from Johnson et al. (24) showing a lack of affinity for the rat α3β2 nAChR. In an attempt to reconcile these data, we have determined the affinity of conotoxin ImI for the rat α3β2 nAChR and found that ImI inhibits the rat α3β2 nAChR with an IC50 = 67 ± 3.5 nM (n = 5). Our result is very similar to the IC50 value determined by Ellison et al. (22) for the human α3β2 nAChR and suggests that conotoxin ImI should be considered as an α7/α3β2-selective ligand for the nAChRs.
These results have two important implications. First, the WT forms of α-conotoxin ImI and PnIA strikingly differ in their electrophysiological properties (Fig. 1D), and α-conotoxin ImI more closely resembles the mutant variant PnIA (A10L) in that it inhibits nAChR by stabilizing the desensitized conformation of the α7 nAChR. A complex of AChBP with conotoxin ImI is therefore more likely to be similar to the desensitized state, rather than the resting state of the nAChR. Second, binding experiments demonstrate that conotoxin ImI displays high specificity for Ac-AChBP, similar to its specificity toward α3β2 and α7 nAChRs, thereby validating our approach to investigate subtype-specific interactions in the complex of Ac-AChBP with conotoxin ImI.
Crystal Structure of Ac-AChBP in Complex with α-Conotoxin ImI.
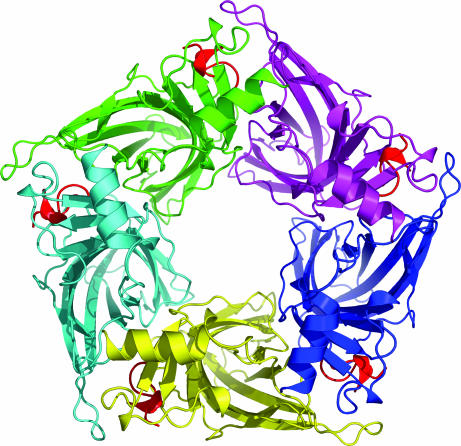
The crystal structure of Ac-AChBP in complex with conotoxin ImI (Ac-AChBP–ImI) was determined at 2.2-Å resolution and solved by molecular replacement. The diffraction data and electron density maps were of excellent quality, although some side chains of the complementary binding site were disordered. The structure of Ac-AChBP–ImI was refined to an Rwork of 17% and an Rfree of 22% (see Table 2, which is published as supporting information on the PNAS web site, for complete statistics). The structure of Ac-AChBP–ImI shows features that are similar to the complex with conotoxin PnIA(A10L D14K), but it also displays some notable differences. First, the pentamer in the Ac-AChBP–ImI structure is highly symmetric, following an exact fivefold symmetry, whereas the monomers in the PnIA(A10L D14K) complex have 3–4° reorientations with respect to each other. The rms deviation (rmsd) between monomers is 0.24 ± 0.03 Å (for 205 Cα atoms) in the ImI complex and 0.53 ± 0.10 Å (for 205 Cα atoms) in the PnIA(A10L D14K) complex. Ac-AChBP–PnIA(A10L D14K) crystals contain one pentamer whereas Ac-AChBP–ImI crystals contain two pentamers in the asymmetric unit, arranged in a head-to-head configuration. In one of the pentamers (subunits A–B–C–D–E), two binding sites, namely at the B–C and D–E interfaces, are characterized by weak electron density for conotoxin ImI. The C-loop at these interfaces forms crystal contacts with neighboring pentamers that push the C-loop in a partially closed conformation. This conformation most likely limits the occupancy of these sites by the toxin. The second pentamer in the asymmetric unit (subunits F–G–H–I–J) is fully occupied by five molecules of conotoxin ImI (Fig. 2). The pentamer of the Ac-AChBP–ImI structure with five toxins bound is very similar to the complex with PnIA(A10L D14K) and has a rmsd of 0.77 Å on 1,021 Cα atoms. At fully occupied binding sites, conotoxin ImI opens the C-loop by a distance of 10.58 ± 0.27 Å as measured between the Cys-188 Cα atom in the Ac-AChBP–ImI and the Ac-AChBP–Hepes structures, which is very similar to the value calculated for the C-loop displacement in the Ac-AChBP–PnIA(A10L D14K) structure (10.40 ± 0.04 Å).
Fig. 2.
Crystal structure of α-conotoxin ImI bound to Ac-AChBP viewed along the fivefold axis. Conotoxins are in red.
We previously reported that the structure of PnIA(A10L D14K) in the crystal structure of the complex with Ac-AChBP is nearly identical to the crystal structure of free PnIA (rmsd of 0.51 ± 0.08 Å). Of the four conotoxin ImI structures that have been determined by NMR (15, 31–33), the conformation of ImI in our crystal structure closely resembles the NMR structure determined by Lamthanh et al. (15) (Protein Data Bank ID code 1G2G) with a rmsd of 0.46 ± 0.03 Å (on 12 Cα atoms of the average NMR structure). However, in the crystal structure we identified an intramolecular salt bridge between Asp-5 and Arg-7 that was not observed in any of the four NMR structures.
The most notable differences between the two Ac-AChBP complexes with conotoxins, however, are found at the binding site in the number and nature of interactions between Ac-AChBP and the conotoxins.
Molecular Contacts of α-Conotoxin ImI in the Ligand-Binding Site of Ac-AChBP.
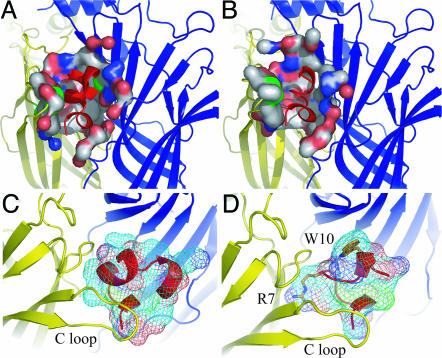
Both conotoxins PnIA(A10L D14K) and ImI, which is smaller, are buried in the ligand-binding site by ≈60% of their surface area. They also share a common orientation with their central helix protruding into the binding site and their N and C termini located at the bottom and top of the binding site, respectively. The rmsd between conotoxin PnIA(A10L D14K) and ImI in the Ac-AChBP-bound structures is 0.82 ± 0.02 Å (on Cα 1–10 of five toxins). By using solvent accessibility calculations, we found that the toxin–receptor interface covers 827 ± 32 Å2 for conotoxin PnIA(A10L D14K) and 679 ± 15 Å2 for the smaller conotoxin ImI (Fig. 3A and B). Both conotoxins also have the Cys-2–Cys-8 disulfide bridge stacked onto the vicinal disulfide bond (Cys-188–Cys-189) in the C-loop of Ac-AChBP (Fig. 4).
Fig. 3.
Surface area at the toxin–receptor interface. (A and B) Comparison of the Ac-AChBP surface area contacted by α-conotoxin PnIA(A10L D14K) (A) and α-conotoxin ImI (B). (C and D) Surface area presentation of α-conotoxins protruding into the binding site. (C) PnIA(A10L D14K) (D) ImI. ImI Arg-7 and Trp-10 are shown in stick presentation.
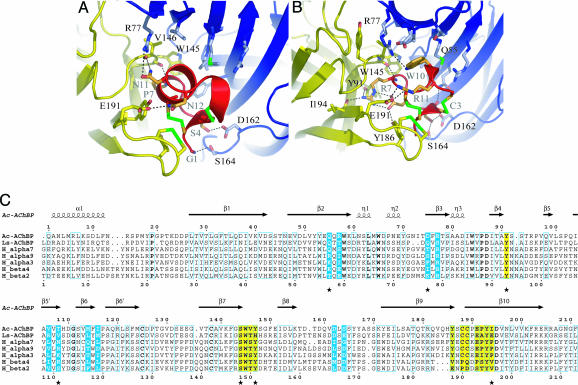
Fig. 4.
Molecular contacts at the toxin–receptor interface. (A and B) Details of molecular contacts between Ac-AChBP and α-conotoxin PnIA(A10L D14K) (A) and α-conotoxin ImI (B). The conotoxin is shown in red, the principal binding side in yellow, and the complementary binding side in blue. Disulfide bridges are green. Dashed lines indicate H-bonds or salt bridges. (C) Sequence alignment of Ac-AChBP, Ls-AChBP, and α7, α9, α3, β4 nAChRs. Sequence numbering at the top is for Ac-AChBP and at the bottom is for α7 nAChR. Residues of the principal binding side that interact with α-conotoxin ImI are shown in yellow; residues of the complementary binding side are in blue. Contacts that are present in the complex with ImI, but not in the complex with PnIA(A10L D14K), are labeled with ★ below the alignment.
As reported, conotoxin PnIA(A10L D14K) contacts highly conserved residues of the principal binding site and residues on the complementary binding side (Table 1 and Fig. 4A).
Table 1.
Contacts between Ac-AChBP and α-conotoxins
| Ac-AChBP | Conotoxin PnIA(A10L D14K) | Conotoxin ImI |
|---|---|---|
| Principal side | ||
| Tyr-91* | Arg-7 | |
| Ser-144 | Arg-7 | |
| Trp-145 | Pro-7 | Arg-7, Pro-6 |
| Val-146 | Asn-11 | Arg-7 |
| Tyr-147 | Arg-7 | |
| Tyr-186 | Gly-1, Cys-2, Leu-5 | Gly-1, Cys-2, Asp-5 |
| Cys-188 | Cys-2, Tyr-15 | Cys-2 |
| Cys-189 | Asn-12 | Arg-11 |
| Glu-191 | Asn-12 | Arg-11 |
| Tyr-193 | Cys-8 | Arg-7, Cys-8 |
| Ile-194* | Arg-7 | |
| Complementary side | ||
| Tyr-53 | Ser-4, Pro-6 | — |
| Gln-55* | Cys-3 | |
| Arg-57 | Pro-13 | Cys-12 |
| Asp-75* | Trp-10 | |
| Arg-77 | Asn-11 | Trp-10 |
| Val-106 | Leu-10 | Trp-10 |
| Thr-108* | Trp-10 | |
| Met-114 | Ala-9, Leu-10, Pro-13 | Trp-10, Ala-9 |
| Ile-116 | Ala-9, Leu-10, Pro-13 | Pro-6, Ala-9 |
| Asp-162 | Ser-4 | Ser-4 |
| Ser-164 | Gly-1 | Ser-4 |
H-bonds (2.6–3.4 Å) are shown in bold, a salt bridge is in italics, and van der Waals interactions (3.2–3.8 Å) are in normal font.
*Unique contacts only observed at the Ac-AChBP–ImI interface.
Conotoxin ImI (Fig. 4B) forms a broader range of interactions that partially overlap with those seen in the complex with PnIA(A10L D14K). The nature of those contacts, however, is very different. First, ImI forms a salt bridge between Arg-11 and Glu-191 on the principal binding side (loop C). Four hydrogen bonds (H-bonds) are formed on the principal site, three of which involve ImI Arg-7 and Tyr-91 (loop A), Trp-145 (loop B), and Ile-194 (loop C). ImI Arg-7 further forms an intramolecular salt bridge with ImI Asp-5 and forms extensive van der Waals interactions with Ser-144, Val-146, Tyr-147 (loop B), and Tyr-193 (loop C). Moreover, the four additional residues (Tyr-91, Ser-144, Tyr-147, and Ile-194) of the principal binding side of Ac-AChBP not forming contacts in the complex with PnIA(A10L D14K) are all involved in interactions with Arg-7 (see Table 1 and Fig. 4B). This result seems to suggest a key role of Arg-7 in the binding of conotoxin ImI to the principal binding side. A fourth H-bond at the principal site is formed between ImI Asp-5 and Tyr-186 (loop C).
On the complementary binding side, ImI forms three H-bonds, namely between ImI Cys-3–Gln-55 (loop D) and ImI Ser-4–Asp-162/Ser-164 (loop F). ImI Trp-10 is involved in interactions with two of the three additional residues of the complementary side not contacted in the complex with PnIA(A10L D14K) (Asp-75 and Thr-108). ImI Trp-10 further forms an extended range of van der Waals contacts with Arg-77, Val-106, and Met 114 (loop E). This result seems to suggest an essential contribution of ImI Trp-10 to binding of the toxin on the complementary binding side.
Our results are in excellent agreement with functional data available from cycle mutant analysis on the α7 nAChR and conotoxin ImI, which revealed a predominant contribution of the interaction between ImI Arg-7 and Y195 in the α7 nAChR (Y193 in Ac-AChBP) and multiple weak interactions between ImI Trp-10 and Thr-77 and Asn-111 in the α7 nAChR (16). The importance of Arg-7 and Trp-10 in binding of ImI is further illustrated in Fig. 3D. The contour map of conotoxin ImI shows how both side chains protrude deep into the binding pocket at sites where PnIA(A10L D14K) does not have interactions (Fig. 3C).
Hansen et al. (34) recently reported a crystal structure of Ac-AChBP–ImI in a comprehensive study that also includes complexes of Ac-AChBP with epibatidine, lobeline, and methyllycaconitine. The structure of Ac-AChBP in complex with conotoxin ImI described in their paper is very similar to our structure and has a rmsd of 0.57 Å (1,014 Cα atoms). Small conformational changes can be seen at the N-terminal helix, which contains two additional residues and a FLAG epitope in the structure from Hansen et al. (34). One significant difference is that Asp-195 forms a salt bridge with ImI Arg-7 in the structure from Hansen et al. (34), whereas Asp-195 (197 in the numbering of Hansen et al.) has a different side-chain orientation and forms a H-bond with a water molecule in our structure, possibly due to differences in electrostatic conditions of the crystallization medium. Apparently, there is some flexibility in the detailed interaction at this site. Finally, Hansen et al. (34) did not discuss the salt bridge formed between Glu-191 and ImI Arg-11, although that contact is also present in their structure.
Conotoxin Interactions Related to nAChR Subtypes.
The individual interactions between α-conotoxin ImI and Ac-AChBP are shown in a sequence alignment with Ls-AChBP and different subtypes of neuronal nAChRs in Fig. 4C. Contacts made with residues of the principal binding side are colored in yellow; those at the complementary side are in blue. In the sequence alignment (Fig. 4C), the largest variability can be observed at residues of the complementary binding side. The lack of affinity of conotoxin ImI toward Ls-AChBP could, at least in part, be explained by a substitution of Asp-75, which forms a unique interaction with ImI Trp-10, by Gln in Ls-AChBP. ImI Trp-10 also interacts with Val-106 in Ac-AChBP and could sterically clash with Arg at the corresponding position in Ls-AChBP. In addition, Dutertre et al. (35) recently identified a key binding determinant, Leu-119, at the α3β2 nAChR and demonstrated the importance of a hydrophobic environment at this position for binding of α-conotoxins. In Ac-AChBP this residue corresponds to Ile-116, but Ls-AChBP contains a Met-114 at the corresponding position, and this larger side chain may interfere with conotoxin binding. Finally, the substitution of Met-114 in Ac-AChBP by Leu-112 in Ls-AChBP could further contribute to loss of affinity of ImI. The importance of Leu-112 and Met-114 to ligand binding was previously demonstrated in the context of AChBP identified from B. truncatus (6).
Conotoxin ImI shows high affinity toward human α7 nAChR (IC50 = 132 nM) (18) and human α3β2 nAChR (IC50 = 41 nM) (22), but very low affinity toward α9 nAChR (IC50 = 1.8 μM) and no affinity toward α3β4 nAChR (24). The sequence alignment shown in Fig. 4C indicates that low affinity of conotoxin ImI toward α9 and α3β4 nAChRs could be explained by substitution of Gln-55 by Arg or Lys in α9/α3β4 and Thr-108 by Arg or Lys in α9/α3β4. These substitutions by positively charged side chains could cause sterical clashes with ImI Cys-3 and Trp-10, respectively, and are not present at the complementary binding side formed by the β2 subunits. This finding might explain why conotoxin ImI retains high affinity at human α3β2 nAChRs. Substitution of Asp-75 by Phe in α3β4 nAChRs also could cause a sterical clash with Trp-10 and contribute to further loss of affinity at this subtype of nAChR.
Docking simulations of ImI into homology models of the α7 nAChR, restricted by pairwise interactions identified in cycle mutagenesis experiments, have been carried out by two different groups (22, 23). One of the shortcomings in both models is that the C-loop was modeled in a closed conformation and that the conotoxin was incorrectly predicted to bind from above the β8-β9 hairpin. Models from Dutertre et al. (23) also suggested that conotoxin PnIA is located at a different position and protrudes deeper into the ACh-binding pocket compared with conotoxin ImI. However, in the crystal structures of Ac-AChBP, we observe that both conotoxins adopt very similar locations in the binding site, with a relative rotation of less than a few degrees and a translation of ≈0.8 Å. However, the side chains of ImI Arg-7 and Trp-10 protrude deep into the binding site, an effect not seen with PnIA(A10L D14K) (Fig. 3 C and D). Both models (22, 23) correctly predicted some of the interactions of ImI Arg-7, namely those with the highly conserved residues of the principal binding site Tyr-93 (loop A), Trp-149 (loop B), and Tyr-195 (loop C). However, the positive charge of Arg-7 is not stabilized through cation-π interactions with these residues as suggested by the authors, but rather by the intramolecular salt bridge in ImI, H-bonds, and van der Waals interactions with conserved residues of the principal binding site. ImI Arg-11, which has been shown experimentally not to be involved in binding of conotoxin ImI (17, 19), was modeled outside of the binding cavity in the docking simulations (22, 23). However, our x-ray structure shows that ImI Arg-11 forms a salt bridge with Glu-191, a long-ranging interaction that may contribute to the overall orientation of the toxin in the binding site. This result indicates that different binding modes of ImI Arg-11 might exist in Ac-AChBP and the α7 nAChR. Although such details may vary, our crystal structure offers structural insight and identifies a network of interactions that goes beyond the pairwise contacts that were found in docking simulations and cycle mutant analysis (22, 23).
Conformational Changes of Ac-AChBP upon Toxin Binding.
Comparison of the Hepes- (7) and ImI-bound structure of Ac-AChBP shows that toxin binding induces an outward displacement of the C-loop, similar to the movement seen in the complex with PnIA(A10L D14K) (7). We previously reported that binding of PnIA(A10L D14K) to Ac-AChBP induces a reorientation of the monomers by 3–4°, which could have been overestimated because of the variability in the orientation of the monomers in this complex. In the Ac-AChBP–ImI complex, the pentamers are highly symmetric, and individual monomers do not display the rigid body movement seen in the PnIA(A10L D14K)-complex.
Recently, it was demonstrated that functional coupling between the ligand-binding and transmembrane domains of a ligand-gated ion channel requires an interaction of the interface region formed by the β1β2-loop, the Cys-loop, and β8β9-loop with the linker that connects the second and third transmembrane domains (L2–3 linker) (36, 37). It also has been demonstrated that an electrostatic interaction between invariant Arg and Glu residues at the interface of ligand-binding and transmembrane domains of the α-subunit of the nAChR (38), as well as a cis-trans isomerization of a conserved Pro in the L2–3 linker (39), serve as conformational switches during gating of the channel. Coupling between Ls-AChBP and the transmembrane domain of a ligand-gated ion channel further requires substitution of the three interface loops by those of the 5-HT3 receptor (36). Our electrophysiological characterization of conotoxin ImI on WT α7 and L247T nAChRs shows that toxin binding results in channel closing by stabilization of the ligand-binding domain in a desensitized conformation. However, comparison of the Hepes- and ImI-bound Ac-AChBP crystal structures shows no dramatic changes in the conformation of these three interface loops. It is therefore likely that Ac-AChBP, in the absence of a transmembrane domain, is already stabilized in a conformation similar to the desensitized state (40) or that the outward movement of the C-loop upon toxin binding may not be transmitted to the interface loops.
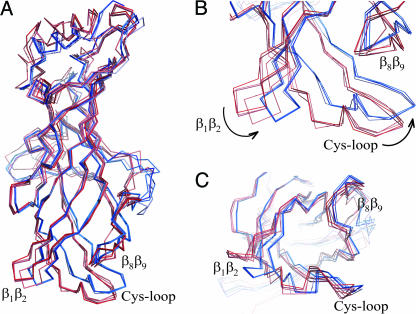
However, the availability of crystal structures of AChBP from different species in complex with different ligands offers us structural insights into how ligand binding may be coupled to channel gating in the intact nAChR. Upon superposition of all AChBP structures available, we surprisingly find that the structures separate into two groups. The first group is formed by Ls-AChBP in complex with Hepes, carbamylcholine, nicotine, and cobratoxin (colored in shades of blue in Fig. 5). The second group is formed by Bt-AChBP in complex with CAPS, Ac-AChBP in complex with Hepes, conotoxin PnIA(A10L D14K), and conotoxin ImI (shades of red in Fig. 5). Comparison between the two groups shows significant rigid body movements of the β1β2-loop and the Cys-loop, and a smaller movement of the β8β9-loop (Fig. 5 B and C). Although some of these differences are clearly species-related (6), they offer structural insight into the extent of flexibility of the interface loops and how they may alter their conformation during channel gating. Movie 1, which is published as supporting information on the PNAS web site, shows the transition between the Ls-AChBP–nicotine and the Ac-AChBP–ImI structure, two of the extreme conformations in the superposition.
Fig. 5.
Superposition of AChBP crystal structures using secondary-structure matching (41) in coot (42). The Ls-AChBP–Hepes structure was used as a reference, and the rmsds for pairwise superpositions are as follows: Ls-AChBP–carbamylcholine 0.52 Å (1,025 Cα atoms), Ls-AChBP–nicotine 0.58 Å (1,028 Cα atoms), Ls-AChBP–cobratoxin 0.96 Å (991 Cα atoms), Ac-AChBP–Hepes 1.68 Å (943 Cα atoms), Ac-AChBP–ImI 1.78 Å (926 Cα atoms), Ac-AChBP–PnIA(A10L D14K) 1.90 Å (925 Cα atoms), and Bt-AChBP–CAPS 1.78 Å (950 Cα atoms). Structures of Ac-AChBP and Bt-AChBP form one group and are colored in shades of red; structures of Ls-AChBP form another group and are in shades of blue. (A) Side view of one monomer, with the N terminus at the top and interface loops at the bottom. (B) Magnified view of the interface loops. (C) Magnified view of the interface loops seen parallel to the fivefold axis. The extreme C terminus was omitted for clarity.
In this work, we show that two naturally occurring conotoxins have evolutionarily diverged to target distinct conformational states of the nAChR. Determination of the crystal structure of the complex of conotoxin ImI with Ac-AChBP demonstrates the structural determinants required for binding of an α3β2/α7-selective ligand. Comparison of this crystal structure with other AChBP structures sheds light on the flexibility of the interface loops and how their conformational change may couple ligand binding to channel gating in the context of the nAChR.
Materials and Methods
Electrophysiology.
Expression and electrophysiological recordings from nAChRs were carried out as described in ref. 25. Briefly, oocytes were injected intranuclearly with 2 ng of cDNA that encoded the human α7, chick α7 L247T, or rat α3β2 nAChR [cDNAs were a gift from M. Ballivet (University of Geneva, Geneva) and J. P. Changeux (Institute Pasteur, Paris)]. All recordings were performed in OR2 medium that contained 82.5 mM NaCl, 2.5 mM KCl, 5 mM CaCl2, and 2 mM Hepes (pH 7.4) adjusted with NaOH. ACh and conotoxins were dissolved in the solution just before use. To prevent contamination by the activation of calcium-dependent chloride channels, oocytes were incubated for at least 3 h in the presence of the chelating agent 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate-acetoxymethyl ester (100 μM). Unless indicated, all recordings were performed at −100 mV; data were digitized online, stored on a Macintosh computer, and analyzed off-line using matlab (Mathworks, Natick, MA).
Protein Expression and Purification.
Untagged Ac-AChBP was expressed from baculovirus in SF9 insect cells and purified from medium as described in ref. 9. Synthesis of α-conotoxin ImI was carried out as described in ref. 33. An excess amount of α-conotoxin ImI was added to Ac-AChBP, and the complex was equilibrated at room temperature for 2 h before crystallization trials.
Crystallization.
Crystals in spacegroup P21 were grown at 20°C by using the hanging-drop or sitting-drop vapor diffusion technique. Cell constants were as follows: a = 113.21 Å, b = 123.13 Å, c = 118.75 Å, α = γ = 90°, β = 117.47°, and two AChBP pentamers in the asymmetric unit. For each drop, the protein complex was mixed with an equal volume of reservoir solution containing 100 mM sodium acetate (pH 5.5) and polyethylene glycol 5000 monomethylether (PEG 5000 MME) at 12.5%. Cryoprotection was achieved by increasing the PEG 5000 MME concentration in the reservoir solution to 30% in 5% increments. All crystals were flash-cooled by immersion in liquid nitrogen.
Structure Determination.
Diffraction data were collected at beamline ID14EH2 of the European Synchrotron Radiation Facility, Grenoble. mosflm (43) was used for indexing, designing the optimal data collection strategy, and data processing. scala was used for data reduction and scaling in the ccp4 program suite (43). The structure of Ac-AChBP in complex with α-conotoxin ImI was solved by molecular replacement using molrep (44) and the published structure of Ac-AChBP–PnIA (D14K A10L) as the search model (Protein Data Bank ID code 2BR8) (7). The atomic coordinates for the α-conotoxin were removed from the search model to reduce model bias. Automated model building was carried out by using arp/warp (45). The structure was rebuilt in coot (42) and further refined using refmac (46) with translation, liberation, and screw (TLS) parameters (47). A multimeric Gly peptide was automatically built by arp/warp in some of the ligand-binding sites, which was mutated according to the amino acid sequence of α-conotoxin ImI (14) and manually rebuilt. Conotoxins in the other binding sites could be placed in electron density by applying rotation and translation operations according to the underlying 52-symmetry of the AChBP pentamer. The C-terminal residues 206–216 were not visible in the electron density map. The final model has rmsds of 0.017 Å for bond lengths and 1.594° for bond angles and has no Ramachandran outliers. The final model contains 17,092 protein atoms and 1,515 water molecules.
Structure Analysis.
Structure validation was carried out by using what if (48) and molprobity (49). areaimol and contact were used to analyze interaction surface areas and contacts (43). Interactions between residues of conotoxins and Ac-AChBP were only considered if present in at least three of five binding sites. Secondary-structure matching (41) was carried out by using the webservice on the European Bioinformatics Institute macromolecular structure database web site (www.ebi.ac.uk/msd). espript was used for sequence alignment and secondary structure analysis (50). Figures were prepared by using pymol (DeLano Scientific, San Carlos, CA).
Supplementary Material
Acknowledgments
We thank beamline staff at the European Synchrotron Radiation Facility, Grenoble, for assistance with data collection, Puck Knipscheer and Ganesh Natrajan for advice during data collection and processing, Tassos Perrakis for advice on model building and refinement, members of the T.K.S. and A. Perrakis laboratory for comments and suggestions, and Baldomero Olivera (University of Utah, Salt Lake City) for a sample of α-conotoxin ImI. C.U. is supported by a long-term fellowship from the European Molecular Biology Organization. This work was supported by Netherlands Technology Foundation Grant BBC6035 and Netherlands Organization for Scientific Research–Russian Foundation for Basic Research Grant 047.015.016 (to A.B.S. and V.T.).
Abbreviations
- ACh
acetylcholine
- AChBP
ACh binding protein
- Ac-AChBP
AChBP from Aplysia californica
- Bt-AChBP
AChBP from Bulinus truncatus
- Ls-AChBP
AChBP from L. stagnalis
- nAChR
nicotinic acetylcholine receptor
- rmsd
rms deviation.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2C9T).
References
- 1.Smit A. B., Syed N. I., Schaap D., van Minnen J., Klumperman J., Kits K. S., Lodder H., van der Schors R. C., van Elk R., Sorgedrager B., et al. Nature. 2001;411:261–268. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- 2.Karlin A. Nat. Rev. Neurosci. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 3.Sixma T. K., Smit A. B. Annu. Rev. Biophys. Biomol. Struct. 2003;32:311–334. doi: 10.1146/annurev.biophys.32.110601.142536. [DOI] [PubMed] [Google Scholar]
- 4.Smit A. B., Brejc K., Syed N., Sixma T. K. Ann N.Y. Acad. Sci. 2003;998:81–92. doi: 10.1196/annals.1254.010. [DOI] [PubMed] [Google Scholar]
- 5.Hansen S. B., Talley T. T., Radic Z., Taylor P. J. Biol. Chem. 2004;279:24197–24202. doi: 10.1074/jbc.M402452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celie P. H., Klaassen R. V., van Rossum-Fikkert S. E., van Elk R., van Nierop P., Smit A. B., Sixma T. K. J. Biol. Chem. 2005;280:26457–26466. doi: 10.1074/jbc.M414476200. [DOI] [PubMed] [Google Scholar]
- 7.Celie P. H., Kasheverov I. E., Mordvintsev D. Y., Hogg R. C., van Nierop P., van Elk R., van Rossum-Fikkert S. E., Zhmak M. N., Bertrand D., Tsetlin V., et al. Nat. Struct. Mol. Biol. 2005;12:582–588. doi: 10.1038/nsmb951. [DOI] [PubMed] [Google Scholar]
- 8.Brejc K., van Dijk W. J., Klaassen R. V., Schuurmans M., van Der Oost J., Smit A. B., Sixma T. K. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 9.Celie P. H., van Rossum-Fikkert S. E., van Dijk W. J., Brejc K., Smit A. B., Sixma T. K. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 10.Bourne Y., Talley T. T., Hansen S. B., Taylor P., Marchot P. EMBO J. 2005;24:1512–1522. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsetlin V. I., Hucho F. FEBS Lett. 2004;557:9–13. doi: 10.1016/s0014-5793(03)01454-6. [DOI] [PubMed] [Google Scholar]
- 12.McIntosh J. M., Santos A. D., Olivera B. M. Annu. Rev. Biochem. 1999;68:59–88. doi: 10.1146/annurev.biochem.68.1.59. [DOI] [PubMed] [Google Scholar]
- 13.Terlau H., Olivera B. M. Physiol. Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 14.McIntosh J. M., Yoshikami D., Mahe E., Nielsen D. B., Rivier J. E., Gray W. R., Olivera B. M. J. Biol. Chem. 1994;269:16733–16739. [PubMed] [Google Scholar]
- 15.Lamthanh H., Jegou-Matheron C., Servent D., Menez A., Lancelin J. M. FEBS Lett. 1999;454:293–298. doi: 10.1016/s0014-5793(99)00831-5. [DOI] [PubMed] [Google Scholar]
- 16.Quiram P. A., Jones J. J., Sine S. M. J. Biol. Chem. 1999;274:19517–19524. doi: 10.1074/jbc.274.28.19517. [DOI] [PubMed] [Google Scholar]
- 17.Quiram P. A., Sine S. M. J. Biol. Chem. 1998;273:11001–11006. doi: 10.1074/jbc.273.18.11001. [DOI] [PubMed] [Google Scholar]
- 18.Rogers J. P., Luginbuhl P., Pemberton K., Harty P., Wemmer D. E., Stevens R. C. J. Mol. Biol. 2000;304:911–926. doi: 10.1006/jmbi.2000.4247. [DOI] [PubMed] [Google Scholar]
- 19.Servent D., Thanh H. L., Antil S., Bertrand D., Corringer P. J., Changeux J. P., Menez A. J. Physiol. (Paris) 1998;92:107–111. doi: 10.1016/s0928-4257(98)80146-0. [DOI] [PubMed] [Google Scholar]
- 20.Sine S. M., Bren N., Quiram P. A. J. Physiol. (Paris) 1998;92:101–105. doi: 10.1016/S0928-4257(98)80145-9. [DOI] [PubMed] [Google Scholar]
- 21.Utkin Y. N., Zhmak M. N., Methfessel C., Tsetlin V. I. Toxicon. 1999;37:1683–1695. doi: 10.1016/s0041-0101(99)00111-7. [DOI] [PubMed] [Google Scholar]
- 22.Ellison M., Gao F., Wang H. L., Sine S. M., McIntosh J. M., Olivera B. M. Biochemistry. 2004;43:16019–16026. doi: 10.1021/bi048918g. [DOI] [PubMed] [Google Scholar]
- 23.Dutertre S., Nicke A., Tyndall J. D., Lewis R. J. J. Mol. Recognit. 2004;17:339–347. doi: 10.1002/jmr.683. [DOI] [PubMed] [Google Scholar]
- 24.Johnson D. S., Martinez J., Elgoyhen A. B., Heinemann S. F., McIntosh J. M. Mol. Pharmacol. 1995;48:194–199. [PubMed] [Google Scholar]
- 25.Hogg R. C., Hopping G., Alewood P. F., Adams D. J., Bertrand D. J. Biol. Chem. 2003;278:26908–26914. doi: 10.1074/jbc.M212628200. [DOI] [PubMed] [Google Scholar]
- 26.Revah F., Bertrand D., Galzi J. L., Devillers-Thiery A., Mulle C., Hussy N., Bertrand S., Ballivet M., Changeux J. P. Nature. 1991;353:846–849. doi: 10.1038/353846a0. [DOI] [PubMed] [Google Scholar]
- 27.Bertrand D., Devillers-Thiery A., Revah F., Galzi J. L., Hussy N., Mulle C., Bertrand S., Ballivet M., Changeux J. P. Proc. Natl. Acad. Sci. USA. 1992;89:1261–1265. doi: 10.1073/pnas.89.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palma E., Mileo A. M., Eusebi F., Miledi R. Proc. Natl. Acad. Sci. USA. 1996;93:11231–11235. doi: 10.1073/pnas.93.20.11231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertrand S., Devillers-Thiery A., Palma E., Buisson B., Edelstein S. J., Corringer P. J., Changeux J. P., Bertrand D. NeuroReport. 1997;8:3591–3596. doi: 10.1097/00001756-199711100-00034. [DOI] [PubMed] [Google Scholar]
- 30.Fruchart-Gaillard C., Gilquin B., Antil-Delbeke S., Le Novere N., Tamiya T., Corringer P. J., Changeux J. P., Menez A., Servent D. Proc. Natl. Acad. Sci. USA. 2002;99:3216–3221. doi: 10.1073/pnas.042699899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gehrmann J., Daly N. L., Alewood P. F., Craik D. J. J. Med. Chem. 1999;42:2364–2372. doi: 10.1021/jm990114p. [DOI] [PubMed] [Google Scholar]
- 32.Rogers J. P., Luginbuhl P., Shen G. S., McCabe R. T., Stevens R. C., Wemmer D. E. Biochemistry. 1999;38:3874–3882. doi: 10.1021/bi9826254. [DOI] [PubMed] [Google Scholar]
- 33.Maslennikov I. V., Shenkarev Z. O., Zhmak M. N., Ivanov V. T., Methfessel C., Tsetlin V. I., Arseniev A. S. FEBS Lett. 1999;444:275–280. doi: 10.1016/s0014-5793(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 34.Hansen S. B., Sulzenbacher G., Huxford T., Marchot P., Taylor P., Bourne Y. EMBO J. 2005;24:3635–3646. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dutertre S., Nicke A., Lewis R. J. J. Biol. Chem. 2005;280:30460–30468. doi: 10.1074/jbc.M504229200. [DOI] [PubMed] [Google Scholar]
- 36.Bouzat C., Gumilar F., Spitzmaul G., Wang H. L., Rayes D., Hansen S. B., Taylor P., Sine S. M. Nature. 2004;430:896–900. doi: 10.1038/nature02753. [DOI] [PubMed] [Google Scholar]
- 37.Chakrapani S., Bailey T. D., Auerbach A. J. Gen. Physiol. 2004;123:341–356. doi: 10.1085/jgp.200309004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee W. Y., Sine S. M. Nature. 2005;438:243–247. doi: 10.1038/nature04156. [DOI] [PubMed] [Google Scholar]
- 39.Lummis S. C., Beene D. L., Lee L. W., Lester H. A., Broadhurst R. W., Dougherty D. A. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 40.Grutter T., Prado de Carvalho L., Virginie D., Taly A., Fischer M., Changeux J. P. C. R. Biol. 2005;328:223–234. doi: 10.1016/j.crvi.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 41.Krissinel E., Henrick K. Acta Crystallogr. D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P., Cowtan K. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Collaborative Computational Project, No. 4. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 44.Vagin A., Teplyakov A. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 45.Perrakis A., Morris R., Lamzin V. S. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov G. N., Vagin A. A., Dodson E. J. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Winn M. D., Isupov M. N., Murshudov G. N. Acta Crystallogr. D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 48.Hooft R. W., Vriend G., Sander C., Abola E. E. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 49.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gouet P., Courcelle E., Stuart D. I., Metoz F. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.