Abstract
Human CTR1 is a high-affinity copper transporter that also mediates the uptake of the anticancer drug cisplatin by largely unknown transport mechanisms. Here we report the 6-Å projection structure obtained for human CTR1 by using electron crystallography of 2D protein crystals in a native phospholipid bilayer. The projection of CTR1 reveals a symmetrical trimer that is <40 Å wide. Notably, the center threefold axis of each trimer forms a region of very low electron density likely to be involved in copper translocation. The formation of a putative pore for metal ions at the interface of three identical subunits deviates from the structural design of typical primary and secondary active transporters and reveals that copper uptake transporters have a novel architecture that is structurally more closely related to channel proteins.
Keywords: cryo-electron crystallography, membrane protein, cisplatin, metal transport
Copper is required for the production of ATP in the mitochondria, clearance of toxic-free radicals from the cell, formation of connective tissue during development, production of the neurotransmitter dopamine, and maintenance of proper plasma iron reduction potentials. However, copper also is toxic to the cell because reaction with hydrogen peroxide can result in the formation of dangerous free radicals. Cells must therefore tightly regulate copper homeostasis through an intricate network of membrane transporters and copper chaperones. Concerning transport, two families of membrane proteins are critical for copper homeostasis in mammals: the P-type ATPase copper pumps, ATP7A and ATP7B, that move copper against a concentration gradient during copper secretion or intracellular sequestration, and CTR transporters that are responsible for the initial uptake of copper into cells. Mutations interfering with the function of the ATPases result in debilitating diseases through toxic buildup of copper in the liver (Wilson disease) or a serious deficit of copper in the brain (Menkes disease). In contrast, CTR proteins have not yet been linked with any disease state in humans (1). However, CTR knockouts are embryonic lethal in mice (2, 3), and recent studies showed that human CTR1 (hCTR1) is a major route for the internalization of the anticancer agent cis-platinum(II)diammine dichloride (cisplatin) into yeast, mammalian, and human ovarian carcinoma cells (4–6), attesting to the physiological and clinical importance of this family of transporters.
The observations that hCTR1 is able to transport both copper ions and platinum ions bonded to prosthetic groups is puzzling in light of what is known about structure–function relationships of this protein. At 23 kDa in molecular mass, the hCTR1 monomer has only three transmembrane domains (7, 8), which are insufficient to form an independent translocation pore for metal ions. Accordingly, oligomerization is required to form a copper-permeable pore and has been demonstrated for hCTR1 and yeast CTR1 of which both dimer and trimer models have been proposed (9, 10). Moreover, oligomerization was shown to be functionally important (11), suggesting that interactions between the transmembrane domains of the monomers make critical contributions to the molecular mechanism of copper and/or cisplatin translocation across the plasma membrane. In addition, mutagenesis of residues in the second transmembrane domain identified positions that greatly affect copper uptake in vivo (12, 13). An important implication of these observations is that a copper permeation pathway may be formed at the interface between subunits, a structural design almost exclusively encountered in channel proteins.
Here we present direct structural evidence that hCTR1 has a novel architecture not previously described for any other transporter to our knowledge. Specifically, the projected image of hCTR1 reveals the existence of a copper-permeable pore along the center threefold axis of the trimeric transporter complex, thereby placing the structural design of this transporter family in closer proximity to channel proteins.
Results
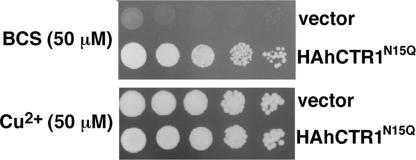
hCTR1 is N-glycosylated at a single site in the N terminus, Asn-15 (14), yet glycosylation is not required for CTR1 to function as a high-affinity copper uptake transporter (8, 15). To increase molecular homogeneity for crystallization experiments we generated mutant N15Q with a hemagglutinin (HA) tag inserted at the N terminus (HAhCTR1N15Q). Fig. 1 demonstrates that HAhCTR1N15Q is functional in copper transport by complementing the growth of yeast deficient in high-affinity copper uptake (Δctr1,3), even under the conditions of copper starvation. Fortuitously, hCTR1 has an extracellular metal binding domain of ≈60 amino acid residues (8), which allowed recombinant protein to be purified on cobalt resin without the need for polyhistidine tagging. Thus, the ability of hCTR1 to bind transition metal ions was not affected by solubilization or the presence of detergent.
Fig. 1.
N-terminal HA-tagged hCTR1N15Q construct is functional in growth complementation under copper starvation. S. cerevisiae yeast deficient in high-affinity copper uptake (Δctr1,3) were transformed with the HA-tagged hCTR1 construct used in this study (labeled as HAhCTR1N15Q) or vector only (vector). (Upper) Copper starvation was achieved with plates that contained the copper chelator bathocuproinedisulfonic acid (BCS) and no additional copper. (Lower) Shown is control growth on plates containing excess copper sulfate (50 μM) without any chelator.
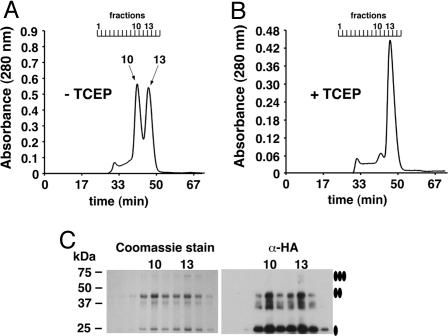
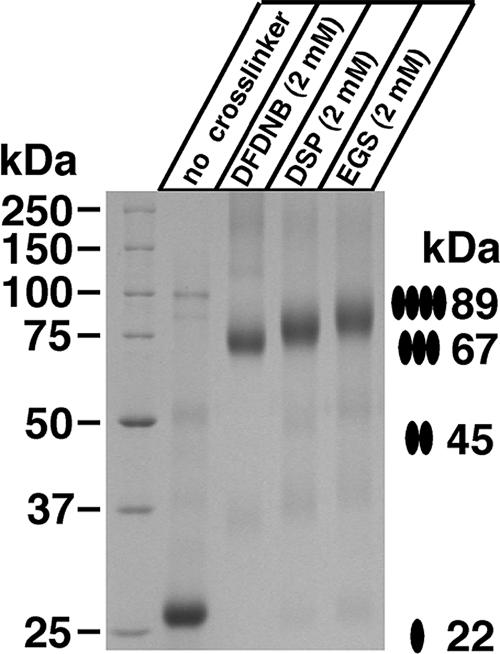
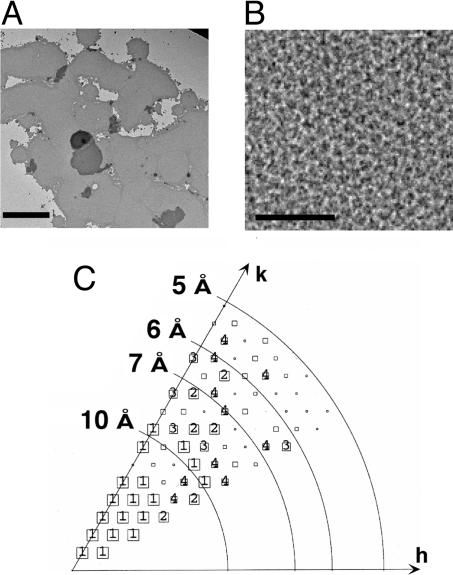
When subjected to gel filtration, HAhCTR1N15Q migrated as two distinct oligomeric states with the first peak (fraction 10, Fig. 2A) representing a molecular mass that was approximately double the size of the second peak (fraction 13). The two species were indistinguishable when subjected to Coomassie staining or anti-HA Western blotting (Fig. 2C). Notably, formation of the larger of the two oligomeric species could be suppressed by the addition of the strong disulfide reducing agent tris(2-carboxyethyl)phosphine (TCEP; Fig. 2B) revealing that changes in the oxidation state of cysteine residues within hCTR1 affected its ability to associate into a larger-order complex. The Kaplan laboratory (8) previously reported that the penultimate residue of hCTR1, Cys-189, mediates disulfide-dependent oligomerization of hCTR1, providing one possible explanation for the observed gel-filtration profiles of HAhCTR1N15Q. To further investigate the oligomeric state of hCTR1, detergent-solubilized purified HAhCTR1N15Q was treated with three lysine-specific chemical crosslinkers that differed by the length of their spacer arms. In each case, crosslinking was extremely efficient (Fig. 3), suggesting the presence of closely spaced lysine residues at the interfaces between monomers. However, crosslinking did not provide a clear answer as to whether hCTR1 is trimeric or tetrameric. Fortunately, protein from fraction 10 formed large, but macroscopically mosaic, 2D crystals (Fig. 4A) whose hexagonal packing ruled out a tetrameric organization (Fig. 4B). Moreover, digital filtering of the transforms revealed trimers, but the resolution was insufficient to reveal details of the hCTR1 structure. Crystals therefore were flash-frozen in liquid ethane, and images were collected by using electron cryomicroscopy.
Fig. 2.
Gel filtration of purified HAhCTR1N15Q reveals two disulfide-dependent oligomerization states. (A) HAhCTR1N15Q in the absence of reducing agents. (B) HAhCTR1N15Q chromatographed after the column was preequilibrated with buffer containing the disulfide-specific reducing agent TCEP (1 mM). In the absence of TCEP, a HAhCTR1N15Q dimer-of-trimer species eluted in fraction 10 and the trimer eluted in fraction 13. (C) Coomassie staining of 10 μl each of gel filtration fractions 7–15 and the same fractions diluted 1:1,000 subjected to anti-HA Western blot. Fractions 10 and 13 are specifically indicated. The smear observed below the band of the dimer most likely reflects anomalous migration caused by flexibility of the dimeric species.
Fig. 3.
Crosslinking of purified HAhCTR1N15Q. Gel filtration fraction 10 was incubated in the indicated concentrations of primary-amine reactive crosslinkers of varying spacer lengths (1,5-difluoro-2,4-dinitrobenzene, DFDNB, 3 Å; dithiobis-[succinimidylpropionate], DSP, 12 Å; ethylene glycolbis[succinimidylsuccinate], EGS, 16 Å) for 30 min before quenching the reaction and subjection to nonreducing SDS/PAGE and Coomassie staining. The apparent increase in weight likely reflects the differences in molecular mass of the crosslinking agents themselves, but could also be related to slight differences in the compactness of the crosslinked oligomer.
Fig. 4.
2D crystals of purified HAhCTR1N15Q. (A) Low-magnification view of flat sheets of HAhCTR1N15Q crystals stained with 2% phosphotungstic acid. (Scale bar corresponds to 1.5 μm.) (B) High-magnification view (×37,000) of the hexagonal lattice. (Scale bar corresponds to 500 Å.) (C) Phase error of unique reflections to 5-Å resolution. The size of the boxes in the plot correspond to the phase error associated with each measurement after averaging data from five images and rounding to 0° or 180° as dictated by the centrosymmetric constraint (1, <8°; 2, <14°; 3, <20°; 4, <30°; 5, <40°; 6, <50°; 7, <80°; 8,<90°, where 90° is random). Boxes are shown as decreasing in size depending on the phase error, and sizes 1–4 are individually labeled.
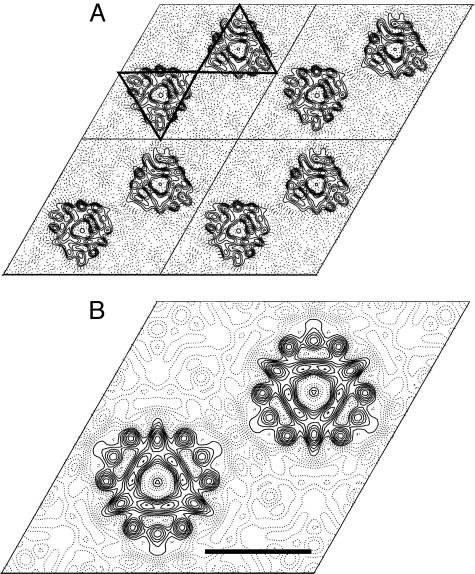
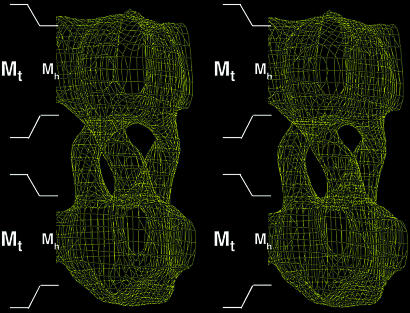
Analysis of the data obtained from frozen hydrated samples confirmed that hCTR1 is trimeric, even without any symmetry imposed, as can be seen by the triangular molecular boundary apparent in Fig. 5A. Moreover, phase relationships in the projection data revealed that the crystals belong to the two-sided plane group p622, indicating a double-layered crystal form (see Fig. 4C for a plot of the combined phase error after processing and merging of the data and Tables 1 and 2 for image statistics). The double-layered nature of the crystals was confirmed independently by determining a low-resolution 3D structure from single-axis tilt series (0–55° tilt) of three hCTR1 crystals that had been preserved in negative stain. Merged in symmetry p6, this reconstruction clearly showed two ordered layers that were related by a twofold axis of rotation between the two layers (Fig. 6 and Table 3). Based on the phase constraints in the projection data of the vitrified sample and the 3D negative stain reconstruction, imposition of p622 plane group symmetry therefore appeared justified. The physiological significance of the crystal packing contacts between the two layers remains unknown, and clarification of the exact mechanism underlying the formation of the dimer of trimers will need detailed information about the 3D structure. A contour plot of one unit cell, calculated with p622 symmetry imposed, shows two groups of superimposed trimers. Several strong circular densities are indicative of α-helical secondary structure (Fig. 5B). However, the complicated double-layered topology of the crystals precludes assignment of these density features to individual membrane-spanning helices or extramembraneous regions of the molecules. In contrast, both layers of the crystal define a circular region of low protein density about the center threefold axis. The ≈9-Å size of this hole is well within the resolution limit of the data and is compatible with the presence of a copper translocation pathway along the center threefold axis of the trimer. The exact dimensions of such a pore cannot yet be established, partly because individual amino acid side chains are not resolved.
Fig. 5.
Projection views of hCTR1. (A) Projection density map of HAhCTR1N15Q at 6-Å resolution with no symmetry enforced (p1). Four unit cells (a = b = 87.9 Å) are shown. Triangles are drawn around two of the superimposed dimers of trimers in a single unit cell to emphasize the triangular shape of the complex. (B) A single unit cell is displayed with p622 symmetry imposed. The double-layered crystals have four hCTR1 trimers in each unit cell. Strong circular density features are indicative of α-helical secondary structure but cannot be unambiguously attributed to the three transmembrane domains. A region of very low electron density at the center axis of each hCTR1 trimer marks the presence of a putative copper permeable pore within the complex. (Scale bar corresponds to a distance of 30 Å.)
Table 1.
Image statistics (projection structure)
| Plane group symmetry* | p622 |
| No. of images | 5 |
| Unit cell parameters | a = b = 87.9 ± 0.4 Å (n = 10); γ = 119.9 ± 0.2 (n = 5) |
| Range of defocus | 4,723 − 13,626 Å |
| Total no. of observations† | 578 |
| Total no. of unique reflections† | 57 |
| Range of independent observations for each structure factor | Minimum: 1 [(0,6), (3,10), and (4,8)] Maximum: 30 [(1,3)] |
| Overall phase residual compared to 0°/180° (45° is random)‡ | 14.8° |
*Symmetry was determined by using allspace (46).
†Including reflections with IQ ≤ 6 to 6 Å.
‡Amplitude weighted vectorially averaged phase residual that shows the phase deviation from theoretical 0°/180° (where 45° is random).
Table 2.
Resolution shell statistics (projection structure)
| Resolution range, Å | No. of unique reflections | Phase residual (0°/180°) (45° is random) |
|---|---|---|
| 88–12 | 17 | 7.7 |
| 11.9–8.5 | 14 | 11.3 |
| 8.4–6.9 | 13 | 24.0 |
| 6.8–6.0 | 13 | 18.8 |
Fig. 6.
3D reconstruction of HAhCTR1N15Q. A stereo side view of half a unit cell (two stacked trimers) is shown with p6 symmetry imposed. Because p6 symmetry cannot impose a second layer during data processing, the two layers are truly crystallographic. Thus, enforcing p622 symmetry for the calculated projection structure (Fig. 5) is justified. A distance of 30 Å for the hydrophobic acyl chains of each phospholipid bilayer is marked to scale as Mh. The total distance of each phospholipid bilayer including lipid head groups (≈45 Å) is marked Mt. The placement of these boundaries is an approximation. The plot is contoured at 1.2 σ.
Table 3.
Data analysis summary (3D reconstruction)
| No. of images | 36 |
| Unit cell parameters | a = b = 84.6 ± 0.6 Å (n = 6); c =200 Å, γ = 120.2 ± 0.3 (n = 3) |
| Two-sided plane group | p6 |
| Range of crystal tilts | 0° to 55° |
| Total no. of measurements | 728 |
| Total no. of fitted unique reflections | 86 |
| Overall weighted phase error | 10.5° |
| Effective resolution cutoffs | 15 Å (in plane) |
| 22 Å (vertical) |
Discussion
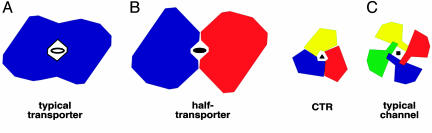
Here we present evidence that hCTR1, a membrane protein thought to act as transporter for copper ions, structurally displays the type of radial symmetry that is observed in the majority of known channel structures. Transporters (primary and secondary active) comprise an enormous superfamily of α-helical integral membrane proteins with very diverse hydrophobic, hydrophilic, and charged substrates. Almost every known transporter structure to date reveals a monomeric unit that is capable of forming an independent pore for the transport of substrates across the lipid bilayer. So far, the only exceptions to this general rule are the “half-transporter” architecture particular to EmrE and the ATP binding cassette transporters MsbA and BtuCD in which the substrate is transported at the interface between two subunits (16–18). In the majority of cases, (the bacterial multidrug efflux transporter AcrB, the ammonia transporter AmtB, the Escherichia coli H+/Cl− exchanger, the glycerol-3-phosphate transporter GlpT, the lactose permease LacY, the oxalate transporter OxlT, the leucine transporter LeuT, and the p-glycoprotein P-gp) an internal repeat in the primary amino acid sequence corresponds to a pseudo twofold symmetry within each monomer (19–26). This is a key design principle for these transporters in which a pathway for substrates is formed along the axis of pseudo twofold symmetry. In the few examples where a pseudo twofold symmetry is not present in the 3D structure (the Na+/H+ antiporter NhaA, the Ca2+ ATPase, and the glutamate transporter homologue Glt) an independent pathway for substrates is still formed within the monomer of the protein (27–29). In contrast, the function of the hCTR1 copper transporter depends on the trimeric organization of hCTR1 and differs from the design of canonical transporters. This observation is summarized in Fig. 7, which also illustrates how EmrE/MsbA/BtuCD and hCTR1 are structural “hybrids” between classical transporters and bona fide channel proteins such as the potassium channels, mechanosensitive channels, acetylcholine receptor ion channel, cardiac gap junction channel, and bacterial-type three secretion systems (30–35).
Fig. 7.
hCTR1 is organized like a transporter-channel hybrid. (A) In the majority of known transporter structures, the permeation pathway is contained within a single monomer that displays pseudo twofold symmetry. This architecture is seen for the structures of the multidrug efflux transporter AcrB, the ammonia transporter AmtB, the E. coli H+/Cl− exchange transporter, the glycerol-3-phosphate transporter GlpT, the lactose permease LacY, the Na+/Cl−-dependent leucine transporter LeuT, and the oxalate transporter OxlT. In three cases, even though the pseudo twofold axis of symmetry is not evident, the monomeric unit still forms an independent pore (glutamate transporter Glt, NhaA, and the calcium pump of sarcoplasmic reticulum). (B) Half-transporters of the ATP-binding cassette family, such as MsbA and BtuCD, have 6–10 transmembrane domains. Although these helical bundles would be large enough to accommodate a pore within the monomer, these transporters homodimerize, forming a pore for substrates at the interface of monomers. In contrast, hCTR1 monomers are too small to form a pore, thus making oligomerization mandatory for function. (C) Our data support that the pore for copper ions is formed at the interface of three identical subunits that oligomerize in a manner much like traditional channel proteins.
The combination of a channel-like architecture with uniporter-like throughput of copper ions raises several important questions about the mechanism by which CTR1 transports copper. First, why can’t hCTR1 be a high-flux channel in mechanism? The toxic nature of copper ions requires this metal to be tightly bound to proteins at all times to prevent the formation of dangerous free radicals in the cell or the formation of metallic copper by disproportionation. Not surprisingly, proteins involved in copper homeostasis have very high affinities for the metal such that the probability of finding free copper ions in the cell approaches zero (36). Neither does copper exist in free form in the blood circulation, with most coordinated to ceruloplasmin and the remainder bound by albumin and amino acids. Thus, controlled coordination of copper ions immediately before, during, and after passage through the hCTR1 is most compatible with a slow transport cycle governed by exchange of copper ions with delivery proteins, intracellular chaperones, and/or low molecular weight chelators such as glutathione. This view is supported by recent work showing that the isolated C terminus of the yeast CTR1 can donate copper ions into the copper chaperone Atx1 in vitro (37) and work by Eisses and Kaplan (8, 38) demonstrated that hCTR1 transports copper with high affinity and a relatively low turnover. Yet copper transport through hCTR1 is passive because neither inhibitors of ATP synthesis (antimycin, oligomycin, and sodium azide) nor ouabain, an inhibitor of Na+/K+-ATPase, affect copper uptake (9).
Consistent with a passive transport mechanism, a seemingly unobstructed pore is present in our projection density map. However, the lack of any obvious barriers raises the question of which molecular determinants within hCTR1 are critical for controlled copper translocation across the membrane and how they contribute to the mechanism of copper transport. Although the final answer to this question has to await information about the 3D structure, the amino acid sequences of CTR proteins and several mutagenesis studies point toward critical contributions of the N and C termini, as well as TM2 and TM3. Specifically, nearly every member of the CTR family has potential metal binding motifs both in the extracellular N terminus and the intracellular C terminus where they are ideally positioned to coordinate receipt and metal exchange with chaperones during a putative transport cycle of hCTR1 in vivo. However, in the absence of copper these sites may be mostly unstructured and thus would not contribute to our projection density map.
Although the N and C termini may contribute significantly to the overall mechanism of copper transport, additional determinants are required to allow for a controlled translocation across the bilayer. For instance, the importance of an MxxxM motif at the extracellular end of TM2 has been established (12). More recently, the Kaplan laboratory (38) showed that additional residues on TM2 also affect transport in vivo, and a study of TM3 revealed the importance of a GxxxG (GG4) motif for CTR oligomerization and function (11). The exact interplay between these different elements needs to be established. However, GG4 motifs have been shown to play critical roles in the structure of several membrane proteins. Such roles include intramonomer interhelical packing as seen in the aquaporin-1 water channel (39), and the formation of intersubunit oligomerization as in glycophorin A (40), and via the related AG4 motif as in the mechanosensitive channel MscS (31). Glycine residues also play essential roles in the design of the selectivity filter in potassium channels and serve as gating hinges in KcsA and MthK (41). Notably, the glycines of the GYG motif in the selectivity filter of potassium channels are within 5.4 Å of each other and their symmetry-related mates in the functional tetramer. Contemplating these spatial relationships, a similar set of constraints could, in principle, be created by orienting the GG4 motif on TM3 toward the packing interface along the central threefold axis of the trimer. In such a model, the GG4 motif on TM3 would contribute to both selectivity and “gating” while the state of the gate could be regulated by the occupancy of extracellular and intracellular metal binding sites. In addition, this model suggests that the trimeric nature of hCTR1 reflects an adaptation to its physiological substrate (copper ions), which at a radius of between 0.68 and 0.91 Å in coordination, is significantly smaller than potassium ions whose flux is mediated by channels that are tetrameric in architecture.
The structure of hCTR1 reported here represents a glimpse of a fold for a transporter of extreme importance in mammalian development and human chemotherapy. A region of low electron density is clearly visualized at the center of the hCTR1 trimer and represents direct visual evidence of a pore for copper ions. A 3D reconstruction of hCTR1 from a tilt series of frozen hydrated crystals will allow a more detailed look into transmembrane helical packing and assignment. Additionally, high-resolution structural data on hCTR1 will help resolve how this protein is able to transport copper ions versus the cisplatin anticancer compound that is coordinated to prosthetic groups.
Materials and Methods
Expression Constructs and Yeast Transformation.
hCTR1 cDNA and Saccharomyces cerevisiae strain Δctr1,3 were obtained from Jane Gitschier (University of California, San Francisco). The initiation methionine of hCTR1 was replaced with a HA tag (MEYPYDVPDYA) to achieve HAhCTR1 as an ORF. N-linked glycosylation of hCTR1 was eliminated (N15Q). HAhCTR1N15Q was ligated into the pDB20 vector as described for complementation studies and into the Pichia pastoris plasmid pPIC3.5K for overexpression and purification. The Δctr1,3 yeast strain was transformed by using lithium acetate, and cells were selected for uracil complementation by plating on minimal dextrose plates containing dextrose (2%), adenine, histidine, lysine, and tryptophan (all 20 mg/liter) but lacking uracil. Pichia strain SMD1163 (Invitrogen) was electroporated (BioRad micropulser) and subjected to one round of selection on regeneration dextrose base plates lacking histidine and a second selection on yeast extract/peptone/dextrose plates containing 0.75 μg/liter G418, all according to the manufacturer’s instructions (MultiCopy Pichia Expression Kit; Invitrogen).
Functional Complementation.
Assays were set up essentially as described (11). Briefly, yeast were washed once with sterile distilled water and adjusted to OD600 of 1.0 in water. Five serial dilutions (1:10 in succession) were spotted onto yeast/peptone/glycerol plates containing glycerol as the only carbon source [1% (wt/vol) yeast extract, 2% (wt/vol) bactopeptone, 3% (vol/vol) glycerol] supplemented with either 50 μM bathocuproinedisulfonic acid (Sigma) for copper depletion or 50 μM copper sulfate for excess copper.
Purification of HAhCTR1N15Q.
Two-liter cultures of P. pastoris expressing HAhCTR1N15Q were inoculated from overnight cultures and grown in a biofermenter [Inceltech (Toulouse, France) LH Series 210] for 50–60 h with 3% glycerol as the sole carbon source. HAhCTR1N15Q expression was induced after complete consumption of the glycerol by the addition of methanol (3.6 ml/h per liter of culture volume) for 4–12 h in the presence of at least 30% dissolved oxygen. Cells were lysed by a single pass through a cell disrupter (Z Plus Series; Constant Cell Disruption Systems, Daventry, U.K.) at 40,000 psi. Cell wall and debris were removed by low-speed centrifugation (600 × g; Beckman JA-10 rotor, 20 min, 4°C), and total membranes were obtained by a second spin at 35,000 rpm in a Beckman type 45 Ti rotor for 15 min at 12°C. Membranes were resuspended in buffer (280 mM NaCl/10 mM Mops, pH 7.4) and homogenized in a dounce glass homogenizer on ice. The membrane suspension was solubilized by the addition of 3% Triton X-100 (Anatrace, Maumee, OH) with stirring for 20 min at room temperature. Unsolubilized material was removed by spinning at 40,000 rpm (Beckman type 45 Ti rotor, 22 min at 22°C), and 10 mM imidazole was added to the supernatant before metal affinity chromatography. HAhCTR1N15Q was purified by direct binding to cobalt metal affinity resin (Talon Superflow; BD Biosciences). Detergent was exchanged by washing the beads with buffer containing 10 mM imidazole and 4 mM decyl-β-maltoside (Anatrace). Protein was eluted with buffer containing 30 mM imidazole and concentrated (Centricon YM-100; Millipore), and batches were subjected to gel filtration chromatography (Superdex 200 10/300 GL; Amersham Pharmacia Biosciences) at a flow rate of 0.3 ml/min in buffer (280 mM NaCl/10 mM Mops/4 mM decylmaltoside, pH 7.4). Bio-Rad gel filtration standards were used to calibrate the gel filtration column.
2D Crystallization of HAhCTR1N15Q.
Protein preparations of HAhCTR1N15Q from gel filtration fraction 10 were diluted to a final concentration of 0.3–0.5 mg/ml in buffer at a final concentration of 280 mM NaCl, 10 mM Mops, 2 mM EDTA, 4 mM decylmaltoside, pH 7.4 in a total volume of 100 μl. Dioleoyl phosphatidylcholine was added to a final lipid-to-protein ratio of 0.7 (mass/mass), and the solution was allowed to mix for 1 h with shaking at 26°C. Crystals were grown by transferring the solution to minidialysis units (10-kDa cutoff; Pierce Slide-A-Lyzer) and dialyzed against the same buffer without detergent for 24–48 h at 26°C.
Electron Microscopy.
Crystals were screened, and tilt series were taken by using phosphotungstic acid-stained HAhCTR1N15Q crystals with a Philips Technai 12 electron microscope operating at 120 kV. The best adherence of the crystals to the continuous carbon film was achieved by applying the mixture to the carbon-coated grids after a 45-s glow discharge in the presence of amylamine by using a sputter coater (SCD005; BAL-TEC, Tucson, AZ) set at a 45-mA discharge current. To freeze the crystals in fully hydrated form, grids were similarly glow discharged, sample was applied to the grid in the cold room, and excess liquid was blotted off for 6–7 s, then rapidly plunged into liquid ethane. Images were taken at a sample temperature (−172°C) under low-dose conditions (≈10 e−/A2) on a Philips Technai F20 microscope equipped with a field emission gun, operating at an accelerating voltage of 200 kV, a nominal magnification of 50,000, and defocus values of −4,500 to −12,500 Å. Images were recorded on Kodak SO-163 film and developed for 12 min in a full-strength Kodak D19 developer.
Image Processing.
Electron micrographs were digitized by using a precision scanner (SCAI; Zeiss) with a scanning pixel size of 7 μm (1.4 Å at the level of the specimen). Well ordered areas of 3,0002 to 4,0002 pixels were identified by calculated forward Fourier transform by using the program ximdisp (42). Images were corrected for lattice distortions, effects of the contrast transfer function, and astigmatism by using the mrc image-processing software package (43). Two passes of unbending were used, with a reference area of 10% of total pixel area for the first pass and 5% for the second. Five images were merged in plane group p622, and an inverse temperature factor B = −320 was found to be sufficient to compensate for resolution-dependent attenuation in the amplitudes (44, 45). The final projection density map is contoured to 0.266 × the SD of the density histogram.
Acknowledgments
We thank Jane Gitschier for the gift of hCTR1 cDNA and Δctr1,3 yeast strain. This work is based on S.G.A.’s dissertation submitted to fulfill in part the requirements for the degree of Doctor of Philosophy at Yale University. This work was supported by National Institutes of Health Grant T32 GM07223 (to S.G.A.), National Research Service Award Predoctoral Fellowship F32 NS45550 (to S.G.A.), a Hellman Family Fellowship, and Public Health Service Grant GM071590 (to V.M.U.).
Abbreviations
- hCTR1
human CTR1
- HA
hemagglutinin
- TCEP
tris(2-carboxyethyl)phosphine.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fu X., Rinaldo P., Hahn S. H., Kodama H., Packman S. J. Inherited Metab. Dis. 2003;26:55–66. doi: 10.1023/a:1024027630589. [DOI] [PubMed] [Google Scholar]
- 2.Lee J., Prohaska J. R., Thiele D. J. Proc. Natl. Acad. Sci. USA. 2001;98:6842–6847. doi: 10.1073/pnas.111058698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuo Y. M., Zhou B., Cosco D., Gitschier J. Proc. Natl. Acad. Sci. USA. 2001;98:6836–6841. doi: 10.1073/pnas.111057298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishida S., Lee J., Thiele D. J., Herskowitz I. Proc. Natl. Acad. Sci. USA. 2002;99:14298–14302. doi: 10.1073/pnas.162491399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin X., Okuda T., Holzer A., Howell S. B. Mol. Pharmacol. 2002;62:1154–1159. doi: 10.1124/mol.62.5.1154. [DOI] [PubMed] [Google Scholar]
- 6.Holzer A. K., Samimi G., Katano K., Naerdemann W., Lin X., Safaei R., Howell S. B. Mol. Pharmacol. 2004;66:817–823. doi: 10.1124/mol.104.001198. [DOI] [PubMed] [Google Scholar]
- 7.Zhou B., Gitschier J. Proc. Natl. Acad. Sci. USA. 1997;94:7481–7486. doi: 10.1073/pnas.94.14.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisses J. F., Kaplan J. H. J. Biol. Chem. 2002;277:29162–29171. doi: 10.1074/jbc.M203652200. [DOI] [PubMed] [Google Scholar]
- 9.Lee J., Pena M. M., Nose Y., Thiele D. J. J. Biol. Chem. 2002;277:4380–4387. doi: 10.1074/jbc.M104728200. [DOI] [PubMed] [Google Scholar]
- 10.Klomp A. E., Juijn J. A., van der Gun L. T., van den Berg I. E., Berger R., Klomp L. W. Biochem. J. 2003;370:881–889. doi: 10.1042/BJ20021128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aller S. G., Eng E. T., De Feo C. J., Unger V. M. J. Biol. Chem. 2004;279:53435–53441. doi: 10.1074/jbc.M409421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Y., Smith K., Lee J., Thiele D. J., Petris M. J. J. Biol. Chem. 2004;279:17428–17433. doi: 10.1074/jbc.M401493200. [DOI] [PubMed] [Google Scholar]
- 13.Eisses J. F., Chi Y., Kaplan J. H. J. Biol. Chem. 2005;280:9635–9639. doi: 10.1074/jbc.M500116200. [DOI] [PubMed] [Google Scholar]
- 14.Klomp A. E., Tops B. B., Van Denberg I. E., Berger R., Klomp L. W. Biochem. J. 2002;364:497–505. doi: 10.1042/BJ20011803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puig S., Lee J., Lau M., Thiele D. J. J. Biol. Chem. 2002;277:26021–26030. doi: 10.1074/jbc.M202547200. [DOI] [PubMed] [Google Scholar]
- 16.Reyes C. L., Chang G. Science. 2005;308:1028–1031. doi: 10.1126/science.1107733. [DOI] [PubMed] [Google Scholar]
- 17.Ma C., Chang G. Proc. Natl. Acad. Sci. USA. 2004;101:2852–2857. doi: 10.1073/pnas.0400137101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Locher K. P., Lee A. T., Rees D. C. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 20.Murakami S., Nakashima R., Yamashita E., Yamaguchi A. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 21.Khademi S., O’Connell J., 3rd, Remis J., Robles-Colmenares Y., Miercke L. J., Stroud R. M. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 22.Dutzler R., Campbell E. B., Cadene M., Chait B. T., MacKinnon R. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 24.Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 25.Hirai T., Heymann J. A., Shi D., Sarker R., Maloney P. C., Subramaniam S. Nat. Struct. Biol. 2002;9:597–600. doi: 10.1038/nsb821. [DOI] [PubMed] [Google Scholar]
- 26.Rosenberg M. F., Callaghan R., Modok S., Higgins C. F., Ford R. C. J. Biol. Chem. 2005;280:2857–2862. doi: 10.1074/jbc.M410296200. [DOI] [PubMed] [Google Scholar]
- 27.Williams K. A. Nature. 2000;403:112–115. doi: 10.1038/47534. [DOI] [PubMed] [Google Scholar]
- 28.Yernool D., Boudker O., Jin Y., Gouaux E. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 29.Toyoshima C., Nakasako M., Nomura H., Ogawa H. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 30.Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 31.Bass R. B., Strop P., Barclay M., Rees D. C. Science. 2002;298:1582–1587. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- 32.Chang G., Spencer R. H., Lee A. T., Barclay M. T., Rees D. C. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 33.Miyazawa A., Fujiyoshi Y., Unwin N. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 34.Unger V. M., Kumar N. M., Gilula N. B., Yeager M. Science. 1999;283:1176–1180. doi: 10.1126/science.283.5405.1176. [DOI] [PubMed] [Google Scholar]
- 35.Marlovits T. C., Kubori T., Sukhan A., Thomas D. R., Galan J. E., Unger V. M. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rae T. D., Schmidt P. J., Pufahl R. A., Culotta V. C., O’Halloran T. V. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 37.Xiao Z., Wedd A. G. Chem. Commun. 2002;6:588–589. doi: 10.1039/b111180a. [DOI] [PubMed] [Google Scholar]
- 38.Eisses J. F., Kaplan J. H. J. Biol. Chem. 2005;280:37159–37168. doi: 10.1074/jbc.M508822200. [DOI] [PubMed] [Google Scholar]
- 39.Murata K., Mitsuoka K., Hirai T., Walz T., Agre P., Heymann J. B., Engel A., Fujiyoshi Y. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 40.MacKenzie K. R., Prestegard J. H., Engelman D. M. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 41.Jiang Y., Lee A., Chen J., Cadene M., Chait B. T., MacKinnon R. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 42.Smith J. M. J. Struct. Biol. 1999;125:223–228. doi: 10.1006/jsbi.1998.4073. [DOI] [PubMed] [Google Scholar]
- 43.Crowther R. A., Henderson R., Smith J. M. J. Struct. Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 44.Williams K. A., Geldmacher-Kaufer U., Padan E., Schuldiner S., Kuhlbrandt W. EMBO J. 1999;18:3558–3563. doi: 10.1093/emboj/18.13.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Unger V. M., Kumar N. M., Gilula N. B., Yeager M. Nat. Struct. Biol. 1997;4:39–43. doi: 10.1038/nsb0197-39. [DOI] [PubMed] [Google Scholar]
- 46.Valpuesta J. M., Carrascosa J. L., Henderson R. J. Mol. Biol. 1994;240:281–287. doi: 10.1006/jmbi.1994.1445. [DOI] [PubMed] [Google Scholar]