Abstract
Chemoattractants like fMet-Leu-Phe (fMLP) induce neutrophils to polarize with phosphatidylinositol 3,4,5-trisphosphate (PIP3) and protrusive F-actin at the front and actomyosin contraction at the sides and back. RhoA and its downstream effector, myosin II, mediate the “backness” response, which locally inhibits the “frontness” response and constrains its location to one part of the cell. In living HL-60 cells, we used a fluorescent PIP3 probe or a single-chain FRET biosensor for RhoA-GTP to assess spatial distribution of frontness or backness responses, respectively, during the first 3 min after exposure to a uniform concentration of fMLP. Increased PIP3 signal or RhoA activity initially localized randomly about the cell’s periphery but progressively redistributed to the front or to the back and sides, respectively. Cells rendered unable to mount the frontness response (by inhibiting actin polymerization or Gi, a trimeric G protein) responded to a micropipette source of attractant by localizing RhoA activity at the up-gradient edge. We infer that protrusive F-actin, induced by the frontness response, constrains the spatial distribution of backness by locally reducing activation of RhoA, thereby reducing its active form at the front. Mutual incompatibility of frontness and backness is responsible for self-organization of neutrophil polarity.
Keywords: actin cytoskeleton, cell polarity, Rho GTPases
Cell polarity regulates and directs functions of many cells, as exemplified by the axons and dendrites of neurons, apical/basal separation in epithelial cells, and distinctive actin assemblies at the leading and trailing edges of migrating cells, such as neutrophils. Chemoattractants like the tripeptide fMet-Leu-Phe (fMLP) induce polarization and migration of differentiated HL-60 (dHL-60) cells, a neutrophil-like cell line (1, 2). The front of a polarized dHL-60 cell shows a protruding pseudopod composed of actively polymerizing F-actin (“frontness”). In contrast, contracting actomyosin complexes induce the back and sides to take on a rounded shape (“backness”), which is sometimes associated with retraction fibers.
Despite morphologic and functional differences, different migratory cells share a conserved set of polarity signals. Phosphoinositide 3-kinases (PI3Ks), Rho GTPases such as RhoA, Rac, and Cdc42, and the actin and microtubule cytoskeletons play key roles in signaling polarity in cells ranging from Dictyostelium discoideum (3–6) to neurons (7–12) and human neutrophils (5, 13–17). Neutrophils and dHL-60 cells show an unusually strong inherent propensity to polarize. Their polarity can be induced (in the absence of spatial cues) by uniform concentrations of diverse stimuli, including chemoattractants, phosphatidylinositol 3,4,5-trisphosphate (PIP3), and drugs that disrupt microtubules or inhibit calpain (1, 16, 18–23). This polarity need not require or induce asymmetry of chemoattractant receptors (24–26).
Stimulating human neutrophils with a homogeneous concentration of chemoattractant induces ruffles to form all over the cell surface; within minutes, the ruffles gradually consolidate into a single pseudopod (27). During this symmetry-breaking process, fMLP induces formation of distinctive actin assemblies, induced by divergent signaling pathways that are activated by a single species of attractant receptor (16). The frontness pathway is mediated by a heterotrimeric G protein, Gi, as well as PIP3, the Rho GTPase Rac, and F-actin (23, 28–30). These components participate in a positive feedback loop to form a protrusive pseudopod (23, 28, 29). The attractant triggers a separate backness pathway by inducing activation of a different heterotrimeric G protein, G12/13, and downstream components including a second Rho GTPase, RhoA, a Rho-dependent kinase, ROCK, and myosin II (16). Activated myosin is thought to form complexes with cortical actin to exert a contractile force parallel to the cell membrane.
How do these disparate signals and cytoskeletal assemblies interact to induce morphologic polarity, with a pseudopod cleanly demarcated from the cell’s sides and back? Evidence that the backness response locally inhibits frontness provided important clues (16). Inhibiting components of the backness pathway caused fMLP to trigger formation of multiple pseudopods and enhanced fMLP-induced frontness signals, including activation of Rac and PIP3 accumulation. Conversely, expressing constitutively active mutant components of the backness pathway dramatically inhibited pseudopod formation and membrane localization of a PIP3 probe, PH-Akt-GFP. From these findings, we inferred that contractile actomyosin somehow leads to inhibition of the signals responsible for frontness and formation of protrusive F-actin in pseudopods (16).
In this article, we ask a reciprocal question: does frontness locally inhibit backness signals and confine morphologic backness to sides and back? We now find, in experiments using a biosensor for RhoA activity, that the answer is yes. From this and other results, we infer that neutrophil symmetry breaking in the absence of spatial cues is a self-organizing process in which distinct frontness and backness responses, initially distributed randomly about the cell surface, mutually and locally inhibit one another to create a polarized cell.
Results
Characterization of a RhoA Biosensor in HL-60 Cells.
After exposure to fMLP, RhoA immunofluorescence and transiently expressed RhoA-GFP both localize in cytoplasm at the rear of polarized dHL-60 cells (16). We applied FRET microscopy to assess spatial distribution of active RhoA (RhoA-GTP) during the course of polarization, using a genetically encoded single-chain RhoA biosensor, previously used to detect RhoA activation in fibroblasts (O.P., L. Hodgson, R. Klemke, and K.H., unpublished work). The biosensor is a fusion protein containing RhoA and the RhoA-binding sequence of its effector, rhotekin, as well as cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP), which serve respectively as the FRET donor and acceptor. This biosensor design has the advantage that the C terminus of the RhoA sequence remains intact, thereby preserving normal regulation by guanine dissociation inhibitor and reversible membrane localization. FRET was assessed as the ratio of the FRET signal to the CFP signal in each pixel of the images; details of FRET measurements are described in Supporting Text, which is published as supporting information on the PNAS web site.
To confirm that the FRET/CFP ratio detected transfer of fluorescence resonance energy, we measured FRET and then photobleached the acceptor YFP. As expected, photobleaching the acceptor by >75% increased CFP emission from individual cells, indicating an overall FRET efficiency of ≈20% (see Fig. 5A, which is published as supporting information on the PNAS web site, and Supporting Text). In addition, the spatial distribution of the FRET/CFP signal in individual cells was virtually identical to the relative increase in CFP emission detected after photobleaching (45% YFP bleached; ΔCFP; see Fig. 5B), validating use of the biosensor as a reliable indicator of subcellular location of Rho activation (FRET/CFP; see Fig. 5B).
The response of the biosensor to GTP loading was also validated by coexpressing the probe with a GTPase-activating protein, p50RhoGAP, which inactivates RhoA by catalyzing its GTPase activity (31). Coexpression of p50RhoGAP reduced the average FRET/CFP signal in fMLP-treated cells by 20% (see Fig5C), indicating that the biosensor was regulated by GTP loading. A large fraction of RhoA was not affected by the GTPase-activating protein because it was already in the GDP state, consistent with observations in other cell types (32, 33), indicating that relatively small proportions of the total pools of Rho GTPases are activated and translocated to the membrane to produce biological effects. Extensive characterization of the RhoA biosensor was reported in fibroblasts (O.P., L. Hodgson, R. Klemke, and K.H., unpublished work). Results in both cell types show that the RhoA biosensor can be regulated and accurately reflects cellular RhoA activity.
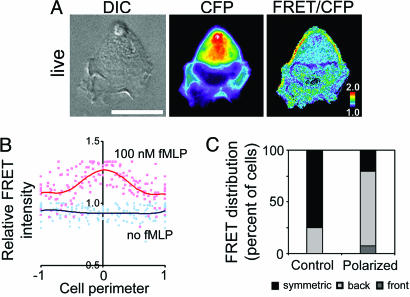
As we expected, the transiently expressed biosensor showed higher RhoA activity at the back of polarized dHL-60 cells; this increase was seen in live (Fig. 1A) as well as fixed cells (not shown). More specifically, the highest FRET/CFP signals in polarized cells were localized predominantly to the cell periphery at the sides and back (Fig. 1A), whereas the biosensor itself, like endogenous RhoA (16), was distributed through the cytoplasm behind the nucleus (CFP image; Fig. 1A); this localization was quantified in 10 cells (Fig. 1B; see Fig. 6B, which is published as supporting information on the PNAS web site, for plots of individual cells). Such an asymmetric distribution of the FRET/CFP signal was detected in the majority of polarized cells examined (73% in the back, 7% in the front; n = 69), whereas only 25% of unstimulated cells (n = 72) showed asymmetrical FRET/CFP signals (Fig. 1C).
Fig. 1.
Characterization of the RhoA biosensor. (A) Distribution of the RhoA biosensor and FRET/CFP in a live polarized dHL-60 cell. CFP and FRET/CFP ratio images are in pseudocolor, with the color indicating the relative value at each pixel. (Scale bar, 10 μm.) (B) Distributions of relative FRET/CFP intensities at the peripheries of 10 unstimulated cells (blue) or 10 cells treated with 100 nM fMLP (red). Peripheral FRET/CFP ratios, assessed as described in Materials and Methods, were significantly increased at the back of stimulated cells. The maximum peripheral FRET/CFP value for stimulated cells is 1.3, reflecting ≈30% greater peripheral FRET/CFP of stimulated cells. Values at the back (origins) of fMLP-treated cells were significantly greater (P < 0.0001) than those at the cell’s front (midpoint), whereas values at the origin and midpoint of unstimulated cells were not different (P = 0.38). Similarly, as indicated by the red and blue regression lines, mean FRET/CFP at the back of stimulated cells was significantly greater (P < 0.0001) than that at the origin of unstimulated cells. Fig. 6 shows how peripheral pixels were identified and analyzed (see also Materials and Methods), as well as pseudocolor images of representative unstimulated or stimulated cells and plots of the corresponding peripheral FRET/CFP values. (C) Relative numbers of cells with asymmetric (gray) or symmetric (black) FRET/CFP distribution in fixed unstimulated cells (n = 72) vs. cells (n = 69) polarized after a 3-min stimulation with 100 nM fMLP. Polarized cells showing asymmetric FRET/CFP distribution are further categorized into front (dark gray) vs. back (light gray) localization.
The front of the pseudopod in most polarized cells contained no detectable amount of probe and, consequently, could not be analyzed for RhoA activation. This very low quantity of Rho in the front of the cell, even if fully activated, would contribute far less activity than that in the back, where Rho is much more abundant. Some polarized cells (20%) exhibited increased Rho activation all around the cell periphery (Fig. 1C), and a few cells showed increased activation behind the nucleus (data not shown). In those cells that did show detectable RhoA activation at the leading edge, activation was low compared to activation at the back (Figs. 1A and 2C; see Fig. 7, which is published as supporting information on the PNAS web site).
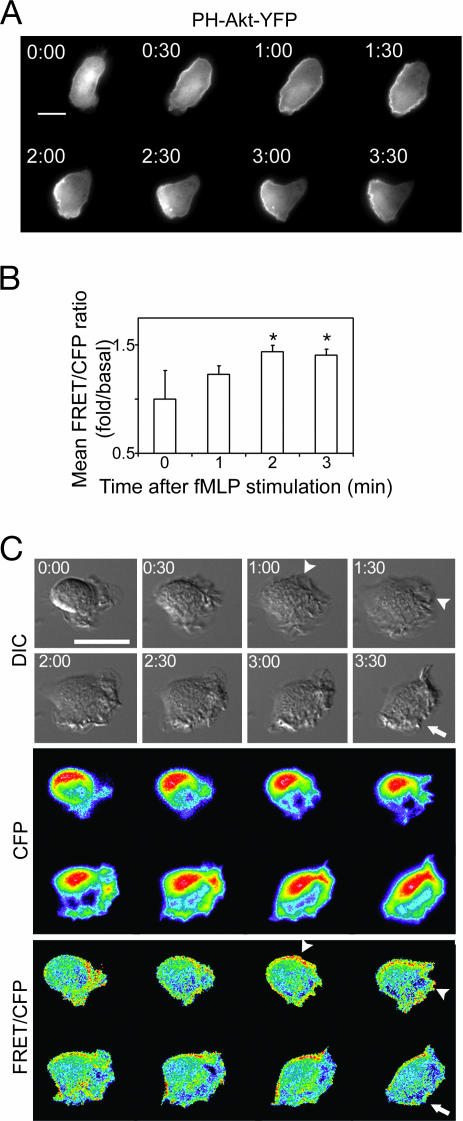
Fig. 2.
Accumulation and localization of PIP3 and RhoA-GTP during dHL-60 cell polarization. (A) Time-lapse microscopy of PH-Akt-YFP recruitment in a live dHL-60 cell. A uniform concentration of fMLP (100 nM) was added at time 0, and fluorescent images of the same cell are shown at the indicated times. (Scale bar, 5 μm.) PH-Akt-YFP translocated from cytoplasm to almost the entire periphery of the cell at 30–60 s and then localized to a clearly demarcated pseudopod at one end of the cell. Similar patterns and timing of these changes were seen in all 10 PH-Akt-YFP-expressing cells tested (data not shown). (B) RhoA biosensor cells were stimulated with 100 nM fMLP for the indicated times, fixed, and imaged for FRET/CFP analysis. fMLP increased the average FRET/CFP ratios, assessed over the footprint of each individual cell, at 2 and 3 min (P ≤ 0.05); the increase at 1 min was not statistically significant (P = 0.3). Data were normalized to the basal FRET/CFP ratio at 0 min (1.0). Shown are representative resu1ts from three individual experiments, each with n ≥ 25 in each condition. Error bars are +2 SEM. (C) Time-lapse microscopy of live dHL-60 cells expressing the RhoA biosensor. Images represent an individual cell before the addition of 100 nM fMLP (time 0) and every 30 s thereafter. Top, Middle, and Bottom represent DIC, CFP, and FRET/CFP ratio images, respectively. Arrows point to ruffles with a low FRET/CFP signal in the front of polarized cells, whereas arrowheads indicate areas that showed both ruffles and high FRET signals before the cell completed morphological polarization. Each image was scaled according to its high and low values at each time point to show relative distributions of RhoA activities at each time point. The dynamic range was 1.5–2.3. Warmer colors correspond to higher values. (Scale bars, 10 μm.)
It is important to stress that spatial distribution of the FRET/CFP signal always differed from that of the probe itself in fMLP-treated polarized cells (Fig. 1A; see also below), indicating that measured increases in FRET/CFP ratios did not mirror local concentrations of the probe itself but instead authentically reflected activation of RhoA. This finding was to be expected because ratiometric measurements of the unimolecular RhoA biosensor cancel out differences in probe distribution.
For collecting data from larger numbers of cells (n ≥ 25) in a single experiment, we used lentiviral-mediated gene transfer to generate a quasistable HL-60 cell line expressing the RhoA biosensor (RhoA biosensor cells). Polarized dHL-60 cells transiently or quasistably expressing the RhoA biosensor showed similar FRET/CFP distributions (compare Fig. 2C vs. controls in Fig. 3A, and see Fig. 6B Lower).
Fig. 3.
Effects of inhibiting Gi on RhoA activity. (A) Control or PTX-treated (1 μg/ml; 18 h) RhoA biosensor cells with or without fMLP stimulation (100 nM; 3 min) were fixed and subjected to FRET/CFP imaging. Representative DIC and FRET/CFP ratio images are shown. FRET/CFP images of control and PTX-treated cells were scaled individually and represented in pseudocolor. (Scale bar, 10 μm.) (B) Representative control or PTX-treated RhoA biosensor cells were stimulated with 100 nM fMLP for 3 min and fixed and stained for F-actin by using Alexa Fluor 647/phalloidin. Shown are DIC and the corresponding fluorescence images. (C) RhoA biosensor cells with or without PTX pretreatment were exposed to an fMLP gradient. An asterisk depicts the orientation of an fMLP gradient generated by a micropipette. DIC and FRET/CFP ratio images of representative cells are shown. A warm color represents a high value; a cold color represents a low value. (Scale bar, 10 μm.) (D) FRET/CFP distributions away, toward, or irrespective to the micropipette were categorized as down-gradient, up-gradient, or other, respectively, and the percentage of cells in each group was quantified. The numeral above each bar indicates the number of cells showing the distribution indicated. Distribution was determined as described in Materials and Methods.
Spatial Distributions of PH-Akt-YFP and RhoA Activity During Polarization.
Like human neutrophils (27), dHL-60 cells respond to application of a uniform concentration of fMLP by forming ruffles all around the cell periphery at 1 min and become morphologically polarized with a protruding front and a contracting back 2–3 min after stimulation. Similarly, as shown in Fig. 2A, a marker for the front, PH-Akt-YFP, translocated from cytoplasm to the cell periphery by 1 min after exposure to fMLP; during the next 1–2 min, PH-Akt-YFP fluorescence aggregated to become concentrated at the leading edge. In retrospect, we overlooked the potential importance of our earlier observations of this phenomenon (1, 29).
To assess variations in average cellular RhoA activity during the same period, we determined the FRET/CFP ratio of individual biosensor-expressing cells fixed at various times after exposure to uniform fMLP (Fig. 2B). In keeping with Rho pull-down results reported in ref. 16, fMLP stimulation significantly increased cellular FRET/CFP, confirming that the attractant activates RhoA in dHL-60 cells.
To understand temporal and spatial regulation of RhoA during cell polarization, we monitored distribution of the FRET/CFP signal in live dHL-60 cells transiently expressing the RhoA biosensor. RhoA activity at early times (30 s) after exposure to fMLP was observed in cytoplasm and at the cell periphery but between 1.5 and 3 min progressively increased in peripheral regions that were developing into the back and sides (Fig. 2C; see also Fig. 7). During this period, we often observed a region of the cell periphery simultaneously occupied by protruding ruffles and increased RhoA activation, as indicated by the arrowheads in the differential interference contrast (DIC) and FRET/CFP panels of Fig. 2C. With time, however, a portion of the ruffle often retracted, and RhoA activation persisted so that the region became part of the cell’s back or sides rather than the pseudopod. A few small spots of high RhoA activation were occasionally observed near the front edge of some polarized cells (Fig. 2C).
Gi, RhoA, and Morphologic Backness.
RhoA activation analysis confirmed our previous inference from Rho pull-down assays (16) that fMLP stimulates RhoA by a pathway that is not mediated by Gi and showed, in addition, that fMLP-stimulated activity at the front normally restricts the spatial distribution of activated RhoA to the back. Inhibiting Gi with pertussis toxin (PTX) did not significantly change average RhoA activation in unstimulated cells, but allowed fMLP to do so (data not shown), in keeping with Rho pull-down results (16). Despite its ability to increase RhoA FRET/CFP, a uniform concentration of fMLP did not induce morphologic polarity in PTX-treated cells, and the increased RhoA FRET/CFP was not polarized but instead was randomly distributed (Fig. 3A). PTX did substantially inhibit Gi-dependent pseudopod formation as assessed by its prevention of fMLP-stimulated accumulation of F-actin: 96% of 53 control cells formed distinct F-actin-rich pseudopods, but this staining pattern was observed in only 12% of 86 cells treated with PTX; representative cells are shown in Fig. 3B.
PTX-treated dHL-60 cells respond to a point source of fMLP by forming uropod-like structures at their up-gradient edges (16). In accord with this observation, exposure to a micropipette containing fMLP caused PTX-treated cells to localize the increase in RhoA activation to their up-gradient edges (Fig. 3 C and D). This behavior contrasts with that of control cells, which show elevated RhoA activity distributed to regions of the periphery away from the micropipette (Fig. 3 C and D). We infer that fMLP-stimulated frontness restricts the localization of the RhoA backness response.
Actin Polymerization Suppresses and Localizes RhoA Activity.
Latrunculin B, a toxin that prevents actin polymerization by sequestering monomeric actin (34), increased basal cellular RhoA-GTP in dHL-60 cells as assessed in pull-down assays (16); this finding demonstrated that actin polymerization was essential for suppressing RhoA activity. Experiments with the RhoA biosensor confirmed this inference: latrunculin B substantially increased the average FRET/CFP signal in unstimulated cells, which showed no further increase in response to fMLP (Fig. 4A).
Fig. 4.
Actin polymerization restricts the distribution of active RhoA. (A) Quantitation of average FRET/CFP ratios in control or latrunculin B-treated (LatB; 20 μg/ml; 10 min) RhoA biosensor cells with or without fMLP stimulation (100 nM; 3 min). Data were obtained from >25 cells for each condition and were normalized to the FRET/CFP level of unstimulated controls (1.0). Similar results were observed in three independent experiments. Error bars show + 2 SEM. Asterisks indicate statistical significances of the differences (∗, P < 0.05; ∗∗, P < 0.001) from control cells not treated with fMLP (Student’s t test). Cells pretreated with latrunculin B showed no statistically significant effect of further stimulation with fMLP (P ≥ 0.3). (B) RhoA biosensor cells treated with LatB (20 μg/ml) for 5 min were subjected to an fMLP gradient generated by a micropipette filled with 10 μM fMLP. Shown are DIC and FRET/CFP images of a representative cell before and after the lowering of a micropipette or of a representative cell treated with uniform fMLP (100 nM) for 3 min and fixed. Each ratio image was scaled independently according to its high and low values to show relative distributions of RhoA activities. (Scale bar, 5 μm.) A warm color represents a high value; a cold color represents a low value. (C) Plots of relative RhoA FRET/CFP level along the peripheries of the cells shown in B. The origin of these peripheral measurements, marked as “0” in B, has a value of 0 on the abscissa, in which the diametrically opposite region of the periphery corresponds to −1 or +1. The origins are either arbitrary (untreated cell and cells treated with uniform fMLP) or indicate the orientation of the micropipette. Each data point was normalized by the maximum (1.0) for both axes (see Materials and Methods). (D) Number of LatB-treated cells that fell into down-gradient, up-gradient, or other categories. Ninety percent of control cells (9 of 10) showed high FRET/CFP at the back (away from the pipette), whereas 3 of 13 LatB-treated cells (23%) showed down-gradient localization (see Materials and Methods).
The effects of Latrunculin B on the distribution of RhoA FRET/CFP showed, in addition, that the F-actin polymerized in response to fMLP confines RhoA activity primarily to regions of the cell periphery outside F-actin-rich pseudopods. Fig. 4 B and C compare, in representative cells, the patchy distributions of RhoA activity in the absence or presence of uniform fMLP vs. its peripheral, relatively up-gradient distribution in response to fMLP supplied by the micropipette. Latrunculin B-treated cells adopted a rounded morphology, with an increased FRET/CFP signal, which in some cases did not localize at the cell periphery, either in the absence of fMLP or after addition of a uniform fMLP concentration (Figs. 3A and 4B). Exposure of latrunculin B-treated cells to a point source of fMLP did not induce the RhoA activation signal to distribute predominantly at the cells’ down-gradient edges as seen in normal cells; instead, activated RhoA was greater at the up-gradient edges in most cells (10 of 13, 77%; Fig. 4D). A plot of FRET/CFP in pixels at the cell periphery shows this up-gradient distribution in a representative cell (relative to the micropipette, located at position 0) (Fig. 4C); this distribution contrasted sharply with the down-gradient localization of RhoA activation in normal cells (9 of 10 cells, 90%; Fig. 4D)
Discussion
The present evidence, in combination with previous observations (16), lead us to propose a model for the mechanism of self-organizing polarity in dHL-60 cells. We hypothesize that a uniform concentration of fMLP induces a break in symmetry by stimulating formation of two different actin assemblies, protrusive F-actin and contractile actomyosin complexes, and that each assembly locally inhibits signals necessary for promoting the other. The two assemblies initially compete for the entire cell periphery but soon segregate into two clearly demarcated domains, controlled by different signals. Acting downstream of Gi, a trimeric G protein, PIP3 and Rac promote formation of pseudopods rich in F-actin (i.e., frontness). Myosin II-dependent contraction (i.e., backness) is activated by a Rho/ROCK signaling pathway downstream of different trimeric G proteins, G12 and G13.
This symmetry-breaking mechanism is based on a unifying principle: actin assemblies at the front and back serve not only as downstream “readouts” of signaling pathways but also as regulators of upstream signals. Polarity is initiated and stabilized by the ability of each actin assembly to inhibit locally the signals responsible for the other. In this way, frontness confines backness signals to regions of the cell periphery outside the pseudopod, whereas backness confines frontness signals to the pseudopod. The same principle underlies the positive feedback circuits responsible for robust pseudopods at the leading edge of neutrophils and dHL-60 cells: in this case, protruding actin polymers enhance upstream signals, including PIP3 and Rac, that are responsible for their formation (22, 23, 28, 35).
Evidence for the Proposed Symmetry-Breaking Mechanism.
As we reported in ref. 16, Rho-dependent backness strongly inhibits frontness and constrains it to a single region at the front of the cell: global activation of backness by expression of constitutively active mutant G12, G13, RhoA, or myosin light chains prevented accumulation of F-actin and PH-Akt-GFP at the cell membrane and blocked pseudopod formation (16). In the opposite direction, cells treated with backness inhibitors (dominant negative mutants or pharmacologic inhibitors of p160-ROCK or myosin activities) responded to fMLP with enhanced increases in Rac activity and association of PH-Akt-GFP with particulate cell fractions and by forming multiple pseudopods unconstrained by competition from backness (16, 36). In both directions, perturbing myosin activity produced effects identical to those caused by perturbing signals upstream of myosin, indicating that the final cytoskeletal readout of the backness response suffices to negatively regulate and confine the location of frontness signals and the pseudopods they promote.
Experiments presented in this article used an intramolecular FRET biosensor for RhoA activation to test the reciprocal element of the proposed symmetry-breaking mechanism, negative regulation of backness signals by frontness (F-actin). The biosensor revealed that frontness normally confines a key upstream backness signal to the portion of the cell periphery not occupied by the pseudopod. In cells exposed to a point source of fMLP, loss of frontness, produced either by inhibiting upstream signals with a Gi inhibitor PTX (Fig. 3) or by inhibiting actin polymerization with latrunculin B (Fig. 4), produced cells in which RhoA was activated predominantly at the up-gradient edge.
The precise biochemical mechanism(s) by which frontness locally inhibits RhoA activation are unknown. We suspect, however, that this negative regulation is mediated primarily by protrusive actin polymers or, perhaps more likely, by as yet unidentified signals or RhoGAPs and other proteins that may bind to such polymers and depend on them for their activation. In keeping with this idea, latrunculin B, a frontness inhibitor that prevents morphologic polarity by sequestering monomeric actin and inhibiting actin polymerization, markedly elevates RhoA activity even in the absence of fMLP, as assessed either by the RhoA biosensor (Fig. 4) or by a RhoA-GTP pull-down assay (16). This finding suggests that some process dependent on actin polymers suppresses basal RhoA activity even in unstimulated dHL-60 cells. Relative to the elevated baseline of latrunculin-treated cells, uniformly applied fMLP does not further increase RhoA activity (Fig. 4), although an fMLP gradient does cause RhoA activity to localize predominantly at the up-gradient edge. Gi-dependent signals upstream of actin polymerization are less likely to play essential roles in locally inhibiting RhoA activity because such signals are at least partly intact in latrunculin-treated cells. Thus, latrunculin treatment allows fMLP-induced accumulation of PH-Akt-GFP at the up-gradient edge (1, 28), as well as substantial fMLP-dependent activation of Cdc42, PAK1, Rac, and phosphorylated Akt (A. Van Keymeulen, K.W., Z. Knight, C. Govaerts, K.H., K. Shokat, and H.B., unpublished data).
A critical prediction of the proposed symmetry-breaking mechanism is that the cell’s first response to uniform application of fMLP is to activate backness and frontness signals transiently at multiple random locations on the cell periphery; later, however, protrusive F-actin and myosin-dependent contraction cause these signals to segregate into cleanly demarcated domains as each actin assembly succeeds in locally inhibiting upstream signals responsible for the other. In keeping with this prediction, at early times after fMLP treatment, both PH-Akt-YFP and the RhoA FRET/CFP appear at multiple regions of the cell periphery (Fig. 2 A and C); RhoA activity is often observed at regions that simultaneously exhibit ruffles (Fig. 2C). Over the course of the next 3 min, however, PH-Akt-YFP or RhoA FRET/CFP become confined to the front or the back and sides, respectively, of the polarized cell (Fig. 2 A and C). The time course of these responses and of their segregation into separate domains of the plasma membrane are in keeping with a self-organizing polarization process that depends on mutual inhibition of frontness by backness and vice versa.
Perspective.
Our symmetry-breaking hypothesis states that mutual local inhibition and competition between frontness and backness responses creates asymmetry. In the form presented here, however, the simple competition hypothesis may not fully account for the stable asymmetry of neutrophils and dHL-60 cells, which remain completely polarized, with a single pseudopod as long as fMLP is present. The competition might result in triumph of one response over the other unless the cell can preserve a precise, potentially delicate balance between frontness and backness signals. Observations reported in a separate paper (A. Van Keymeulen, K.W., Z. Knight, C. Govaerts, K. H., K. Shokat, and H.B., unpublished data) identify a mechanism for preserving stable polarity that depends on the ability of some frontness signals to reinforce RhoA activity and actomyosin contraction at the trailing edge.
The spatiotemporal pattern of RhoA activation in randomly migrating fibroblasts, assessed with the same RhoA biosensor (O.P., L. Hodgson, R. Klemke, and K.H., unpublished work), differs strikingly from that of polarized dHL-60 cells. Unlike polarized dHL-60 cells, the fibroblasts show only transient activation of RhoA at the back during tail retraction but persistently high RhoA activity at their leading edges. This difference presumably reflects marked differences in migration speeds and roles of the leading and trailing edges between the two cell types. Neutrophils migrate dramatically faster than fibroblasts, continuously contracting their backs to restrict location of the pseudopod and to allow rapid forward movement of the trailing edge. The pseudopod of the neutrophil moves more or less continuously forward, adhering loosely to the substratum, in contrast to the fibroblast’s leading edge, where slower forward motion is accomplished by tight adhesion to the substratum, whereas many peripheral ruffles extend and retract without sticking. We suspect that the persisting RhoA activation at the fibroblast leading edge plays a key role in forming adhesive structures regulated by F-actin (37) and that these structures are absent or much less important in neutrophils. Thus, persistent RhoA activation at the fibroblast’s front allows it to probe and enhance structural integrity of connective tissue, whereas predominant RhoA activation at the rear makes the neutrophil stick less tightly to extracellular matrix so that it can more rapidly penetrate and explore damaged or inflamed tissues.
Materials and Methods
Materials and methods used in cell culture, transient expression of exogenous cDNA, micropipette assays, and fixation and staining for actin, drugs, and toxins have been described in detail (1, 16, 28, 29). All methods are described in further detail in Supporting Text.
DNA Constructs and Lentiviruses Expressing the RhoA Biosensor and PH-Akt-YFP.
To generate lentiviruses, pVSV.G, pCMVD8.9 (gifts from Todd Brennan, University of California, San Francisco), and the FuPw vector containing either the RhoA biosensor or PH-Akt-YFP were cotransfected into human embryonic kidney (HEK) cells using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s protocol. The concentrated solution of lentiviral particles was added to undifferentiated HL-60 cells.
Image Acquisition and Data Processing.
For time-lapse and fixed cell imaging, FRET, CFP, and YFP images were captured using filter sets S436/10 and S535/30, S436/10 and S470/30, and S500/20 and S535/30, respectively (Chroma Technology, Rockingham, VT). DIC images were acquired for each cell and binning was set at 2 × 2.
For FRET/CFP ratiometric processing, CFP and FRET images were background-subtracted. (Note that the initial analysis showed that applying a shading correction was not necessary; that is, higher activity on one side of the cell was consistent regardless of cell orientation and not attributable to uneven illumination across the image field.) The resulting background-corrected FRET image was divided by that of the CFP image to obtain a pixel-to-pixel FRET/CFP ratio image. Cells were thresholded to discard any portions of the image with insufficient intensity to provide reasonable signal/noise. Unless stated otherwise, the final FRET images were displayed in pseudocolors scaled linearly from the lowest to the highest signal within each cell.
Assessing Polarity and Peripheral Distribution of RhoA Activity.
Cells were defined as polarized on the basis of morphology under DIC microscopy. Polarized cells showed a broad front containing a pseudopod on one side and a round, narrow back at the other side. Cells normally became fully polarized after a 2- to 3-min treatment with 100 nM fMLP. Symmetry or asymmetry of FRET distribution in a cell was determined by examining the profile of relative FRET intensity along the cell’s periphery, in pseudocolor images. Similarly, cells exposed to an fMLP gradient with the highest FRET facing toward or away from the micropipette were categorized as “up-gradient” or “down-gradient,” respectively. Cells that showed a relatively even distribution around the periphery were assigned as “other.”
Statistical Analysis.
To compare FRET/CFP intensities of cells treated under different conditions, the average FRET/CFP ratio of each cell was determined by dividing the cell’s total FRET/CFP ratio by its entire cell area. Results were normalized against the control unstimulated sample (set to 1.0). Unless stated otherwise, values from at least 25 cells were used to assess mean FRET/CFP ratios for each condition, and the effect of each condition was compared with the unstimulated control by using Student’s t test. Each set of experiments was repeated at least three times.
Supplementary Material
Acknowledgments
We thank members of the Bourne Laboratory, Max Krummel, Orion Weiner, Dyche Mullins, and Mark von Zastrow for advice and useful comments on the manuscript. This work was supported by National Institutes of Health Grants GM 27800 (to H.B.) and GM 32089 and GM 57824 (to K.H.). K.W. was a postdoctoral fellow supported by the American Heart Association. O.P. is supported by grants from the Swiss National Science Foundation, Roche Research Foundation, Novartis, and Philip Morris.
Abbreviations
- fMLP
fMet-Leu-Phe
- dHL-60
differentiated HL-60
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
- PTX
pertussis toxin
- DIC
differential interference contrast.
Footnotes
Conflict of interest statement: K.H. is a paid consultant of Genospectra (Fremont, CA).
References
- 1.Servant G., Weiner O. D., Herzmark P., Balla T., Sedat J. W., Bourne H. R. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauert A. B., Martinelli S., Marone C., Niggli V. Int. J. Biochem. Cell Biol. 2002;34:838–854. doi: 10.1016/s1357-2725(02)00010-9. [DOI] [PubMed] [Google Scholar]
- 3.Merlot S., Firtel R. A. J. Cell Sci. 2003;116:3471–3478. doi: 10.1242/jcs.00703. [DOI] [PubMed] [Google Scholar]
- 4.Chung C. Y., Funamoto S., Firtel R. A. Trends Biochem. Sci. 2001;26:557–566. doi: 10.1016/s0968-0004(01)01934-x. [DOI] [PubMed] [Google Scholar]
- 5.Raftopoulou M., Hall A. Dev. Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Devreotes P., Janetopoulos C. J. Biol. Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 7.Andersen S. S., Bi G. Q. BioEssays. 2000;22:172–179. doi: 10.1002/(SICI)1521-1878(200002)22:2<172::AID-BIES8>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 8.Menager C., Arimura N., Fukata Y., Kaibuchi K. J. Neurochem. 2004;89:109–118. doi: 10.1046/j.1471-4159.2004.02302.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhou F.-Q., Zhou J., Dedhar S., Wu Y.-H., Snider W. D. Neuron. 2004;42:897–912. doi: 10.1016/j.neuron.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Wong K., Ren X.-R., Huang Y.-Z., Xie Y., Liu G., Saito H., Tang H., Wen L., Brady-Kalnay S. M., Mei L., et al. Cell. 2001;107:209–221. doi: 10.1016/s0092-8674(01)00530-x. [DOI] [PubMed] [Google Scholar]
- 11.Shi S.-H., Jan L. Y., Jan Y.-N. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- 12.Fukata M., Nakagawa M., Kaibuchi K. Curr. Opin. Cell Biol. 2003;15:590–597. doi: 10.1016/s0955-0674(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 13.Niggli V. Int. J. Bioch. Cell Biol. 2003;35:1619–1638. doi: 10.1016/s1357-2725(03)00144-4. [DOI] [PubMed] [Google Scholar]
- 14.Rickert P., Weiner O. D., Wang F., Bourne H. R., Servant G. Trends Cell Biol. 2000;10:466–473. doi: 10.1016/s0962-8924(00)01841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiner O. D. Curr. Opin. Cell Biol. 2002;14:196–202. doi: 10.1016/s0955-0674(02)00310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J., Wang F., Van Keymeulen A., Herzmark P., Straight A., Kelly K., Takuwa Y., Sugimoto N., Mitchison T., Bourne H. R. Cell. 2003;114:201–214. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- 17.Bokoch G. M. Immunol. Res. 2000;21:139–148. doi: 10.1385/IR:21:2-3:139. [DOI] [PubMed] [Google Scholar]
- 18.Keller H. U., Naef A., Zimmermann A. Exp. Cell Res. 1984;153:173–185. doi: 10.1016/0014-4827(84)90459-2. [DOI] [PubMed] [Google Scholar]
- 19.Niggli V. J. Cell Sci. 2003;116:813–822. doi: 10.1242/jcs.00306. [DOI] [PubMed] [Google Scholar]
- 20.Shields J. M., Haston W. S. J. Cell Sci. 1985;74:75–93. doi: 10.1242/jcs.74.1.75. [DOI] [PubMed] [Google Scholar]
- 21.Lokuta M. A., Nuzzi P. A., Huttenlocher A. Proc. Natl. Acad. Sci. USA. 2003;100:4006–4011. doi: 10.1073/pnas.0636533100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niggli V. FEBS Lett. 2000;473:217–221. doi: 10.1016/s0014-5793(00)01534-9. [DOI] [PubMed] [Google Scholar]
- 23.Weiner O. D., Neilsen P. O., Prestwich G. D., Kirschner M. W., Cantley L. C., Bourne H. R. Nat. Cell Biol. 2002;4:509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Servant G., Weiner O. D., Neptune E. R., Sedat J. W., Bourne H. R. Mol. Biol. Cell. 1999;10:1163–1178. doi: 10.1091/mbc.10.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson B., Wymann M., Holmgren-Peterson K., Magnusson K. J. Cell Biol. 1993;121:1281–1289. doi: 10.1083/jcb.121.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loitto V.-M., Rasmusson B., Magnusson K.-E. J. Leukoc. Biol. 2001;69:762–771. [PubMed] [Google Scholar]
- 27.Zigmond S. H., Sullivan S. J. J. Cell Biol. 1979;82:517–527. doi: 10.1083/jcb.82.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F., Herzmark P., Weiner O. D., Srinivasan S., Servant G., Bourne H. R. Nat. Cell Biol. 2002;4:513–518. doi: 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- 29.Srinivasan S., Wang F., Glavas S., Ott A., Hofmann F., Aktories K., Kalman D., Bourne H. R. J. Cell Biol. 2003;160:375–385. doi: 10.1083/jcb.200208179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z., Hannigan M., Mo Z., Liu B., Lu W., Wu Y., Smrcka A. V., Wu G., Li L., Liu M. Cell. 2003;114:215–227. doi: 10.1016/s0092-8674(03)00559-2. [DOI] [PubMed] [Google Scholar]
- 31.Souchet M., Poupon A., Callebaut I., Leger I., Mornon J., Bril A., Calmels T. P. FEBS Lett. 2000;477:99–105. doi: 10.1016/s0014-5793(00)01769-5. [DOI] [PubMed] [Google Scholar]
- 32.Ren X. D., Kiosses W. B., Schwartz M. A. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraynov V. S., Chamberlain C., Bokoch G. M., Schwartz M. A., Slabaugh S., Hahn K. M. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 34.Spector I., Shochet N. R., Kashman Y., Groweiss A. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 36.Weiner O. D., Rentel M. C., Ott A., Brown G. E., Jedrychowski M., Yaffe M. B., Gygi S. P., Cantley L. C., Bourne H. R., Kirschner M. W. PLoS Biol. 2006 doi: 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J., Wang F., Van Keymeulen A., Rentel M., Bourne H. R. Proc. Natl. Acad. Sci. USA. 2005;102:6884–6889. doi: 10.1073/pnas.0502106102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burridge K., Wennerberg K. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.