Abstract
The division of the living world into three cellular domains, Archaea, Bacteria, and Eukarya, is now generally accepted. However, there is no consensus about the evolutionary relationships among these domains, because all of the proposed models have a number of more or less severe pitfalls. Another drawback of current models for the universal tree of life is the exclusion of viruses, otherwise a major component of the biosphere. Recently, it was suggested that the transition from RNA to DNA genomes occurred in the viral world, and that cellular DNA and its replication machineries originated via transfers from DNA viruses to RNA cells. Here, I explore the possibility that three such independent transfers were at the origin of Archaea, Bacteria, and Eukarya, respectively. The reduction of evolutionary rates following the transition from RNA to DNA genomes would have stabilized the three canonical versions of proteins involved in translation, whereas the existence of three different founder DNA viruses explains why each domain has its specific DNA replication apparatus. In that model, plasmids can be viewed as transitional forms between DNA viruses and cellular chromosomes, and the formation of different levels of cellular organization (prokaryote or eukaryote) could be traced back to the nature of the founder DNA viruses and RNA cells.
The main achievement of evolutionary biology in the last century has been the unification of cellular life by the recognition that all cells share a common mechanism for protein synthesis with the same genetic code and thus originated from a common ancestor [here called the Last Universal Cellular Ancestor (LUCA)]. Focusing meaningful evolutionary thinking on the translation apparatus led to the critical discovery that all cellular organisms belong to one of three cell lineages or domains, each characterized by a different type of ribosome (Archaea, Bacteria, and Eukarya) (1, 2). This natural division of the living world has now been confirmed by comparative genomics (3, 4). In each domain, the main informational processes (translation, transcription, and replication) display typical features, canonical patterns sensu Woese (5), that drastically distinguish each domain from the other two.
This unification of the cellular world has led to numerous attempts to draw a universal tree of life that would reflect the natural history of living organisms on our planet. Whereas the three-domains concept is now generally accepted, there is no consensus about how these domains originated and what are the evolutionary relationships among them. In the classical tree of life, two lineages diverged from LUCA, one leading to Bacteria and the other, to a common ancestor of Archaea and Eukarya (2). Alternatively, one of the two primordial lineages could have produced Eukarya, whereas the other led to a common ancestor of Archaea and Bacteria (6). In these scenarios, LUCA can be a progenote (7), a community of primitive organisms freely exchanging their genes (8), or a more sophisticated type of organism, already harboring some eukaryotic traits (6, 9). In all these cases, LUCA was a very different entity than its descendants. In contrast, Gupta and Cavalier-Smith (10, 11) have suggested that LUCA was a bacterium, and that Archaea originated from within Bacteria. Finally, several authors have proposed that Eukarya originated from the merging of an archaeal and a bacterial lineage (for review, see ref. 12).
All these models have drawbacks that are usually emphasized in turn by each of their proponents to repudiate the others. In my opinion, models involving the transformation of one domain into another, including chimera models, have little credibility, because they imply a dramatic and quite unrealistic change in the rate of protein evolution in only one particular bacterial or archaeal lineage. As stated by Carl Woese, “Modern cells are fully evolved entities which are sufficiently complex, integrated and individualized that further major change in their design does not appear possible” (13). The popular chimera models have additional pitfalls, because they do not explain the origin of eukaryotic specific proteins and complex apparatuses that appear to have no homologues (or even functional counterparts) in Archaea or Bacteria (14).
Models suggesting that Eukarya originated from an archaeal ancestor are also problematic, considering the stability of the prokaryotic cell type of organization (sensu Woese and Fox, ref. 7). It has been recognized for a long time that a deconstruction of the simple and efficient prokaryotic level of cellular organization to produce the complex organization of the eukaryotic cell is unlikely (6–8, 15, 16). To avoid this problem, Woese suggested that each domain originated independently from the progenote (7, 13). However, considering the high number of similarities between the informational apparatuses of Archaea and Eukarya (3), their common ancestor was probably an already sophisticated entity beyond the progenote stage. Another way to avoid the need for a transition in cell organization from a prokaryotic to a eukaryotic stage is to assume that LUCA already displayed some eukaryotic characters, including all features common to Archaea and Eukarya (6, 8). Nevertheless, one should now explain why most ancestral DNA replication proteins and many ribosomal proteins were replaced by functional analogues in the bacterial branch.
All models of early evolution previously discussed date back to the pregenomic era, and it was hoped at that time that revelations from genome sequencing would help to choose between them. This turned out to be wrong. Proponents of each model have stuck to their favorite one and have found in genome data arguments to support their case. This suggests that something critical may be missing from the complete picture.
Why Does Canonical Pattern Exist?
The three-domains concept was first based on the existence of three canonical ribosomal patterns (1). As recently stated by Woese, “Why canonical pattern exists is a major unanswered question” (17). At the individual protein level, the canonical patterns manifest as three “versions” (sensu Woese, ref. 5) of each ribosomal protein (and ribosomal RNA), one per domain. This means that, for example, one can recognize at first glance in a sequence alignment any archaeal ribosomal protein from its bacterial and eukaryal homologues. In addition to sequence divergence, the three versions always differ by the presence of several insertion/deletions (indels) and often by regions with very low or no similarity at all (5). How did these versions originate, and why can they still be recognized, despite subsequent divergence within each domain?
A minimal assumption to answer these questions is that three “dramatic evolutionary events” (6) or “major qualitative evolutionary changes” (13) occurred independently, at the origin of each domain, and produced a drastic modification in the rate of protein evolution (either reduction or acceleration). But what kind of events were these? Is it possible to be more specific? Putative answers were proposed by authors, who argue for the transmutation of one domain into another. Hence, Gupta suggested that Archaea originated from within Bacteria under selection pressure for antibiotic resistance (10), whereas Cavalier-Smith proposed that Archaea originated from within Bacteria under selection pressure for hyperthermophily (11). Both scenarios imply a drastic acceleration in the rate of protein evolution at the onset of the archaeal domain. In Gupta’s hypothesis, a particular lineage of Gram-positive Bacteria living in the soil among antibiotic producers replaced the antibiotic-sensitive bacterial versions of their informational proteins (and rRNA) by novel antibiotic-resistant ones (the archaeal versions). This is quite unlikely, because single point mutations are sufficient to produce drug-resistant bacterial versions of any antibiotic targets (not to mention the mobilization of plasmid genes conferring drug resistance). In Cavalier-Smith’s hypothesis, the transformation of Bacteria into Archaea was triggered by the “invention” of eukaryotic-like histones in some bacteria to protect their DNA against thermal denaturation (11). The modifications of the DNA environment induced by the presence of histones would have increased the rate of evolution of all informational proteins (a domino effect), leading to their archaeal versions. However, in direct contradiction to this idea, all DNA replication proteins of the archaeon Thermoplasma acidophilum are typically of the archaeal version, despite the replacement in this organism of the eukaryotic-like archaeal histone by the bacterial histone-like HU protein. This clearly indicates that domain-specific protein versions are stable in the course of evolution and cannot be dramatically modified by their interactions with new partners (there is no domino effect). As for the general proposal that transformation of some Bacteria into Archaea was triggered by adaptation to hyperthermophily, it is sufficient to say that regular hyperthermophilic bacteria also exist and have successfully adapted the “bacterial versions” of their proteins to function efficiently at high temperature.
The stability of protein domain-specific versions is well illustrated by the fate of proteins that have been displaced from one domain to another by horizontal gene transfer (HGT). These proteins have retained the typical signature of their original domain within the new setting, explaining why HGT can be detected via phylogenetic analyses. For instance, archaeal aminoacyl tRNA synthetases present in Bacteria, or bacterial DNA gyrase present in Archaea, cannot be distinguished from their homologues in their respective domain of origin (5, 18).
We can infer from these examples that the establishment of the three canonical versions of the informational proteins was probably due neither to any change in response to selection pressure of the environment nor to a sudden drastic modification in cellular organization. This also rules out an assumption of proponents of chimera models for the origin of Eukarya, suggesting that the merging of Archaea and Bacteria would have increased the evolutionary rate of the proteins of the archaeal parent, leading to their transformation into eukaryotic proteins.
In my opinion, the most convincing explanation for the origin of the three canonical versions of most informational proteins was proposed by Woese, who suggested that the rate of protein evolution was higher in the time frame between LUCA and the last common ancestor of each domain than it is today (1, 14). As a consequence, subsequent protein evolution occurring at a slower rate after the formation of the three domains was unable to erase the signatures of previous divergent evolution that occurred during the fast-track period.
The idea that proteins were fast evolving at the time of LUCA has its root in Woese’s conception of LUCA as a progenote (7), a primitive organism whose mechanisms for protein synthesis and genome replication were still error-prone. To explain the origin of the domains themselves, Woese suggested that the first cell lineages that diverged from LUCA were able to freely exchange their proteins by HGT, thus preventing the formation of coherent evolutionary units between organismal lineages and their proteins (14). At some point, the rate of HGT would have declined, and evolutionary coherent lineages would have appeared by “crystallization,” capturing a defined set of proteins and leading to modern evolution by speciation (the “Darwinian threshold”). The set of proteins captured at the origin of each domain then defined the canonical domain-specific versions of these proteins.
Although these ideas are appealing, there is clearly a missing link between the crystallization process leading to the origin of the three domains and the reduction in the evolutionary tempo of protein evolution. In particular, it is not clear to me how a reduction in the rate of HGT would have translated into a decline in protein evolutionary rate (or vice versa). As previously mentioned, the rate of protein evolution does not seem to be dramatically affected by HGT or by a domino effect triggered by the presence of new partners (or the absence of old ones). If the rate of protein evolution was reduced by a continuous increase in the number of protein–protein interactions and/or in protein optimization that occurred after the Darwinian threshold (increasing selection pressure against further modifications), it remains to be understood why this produced three discontinuities in the protein sequence space.
The Multiple Versions and Erratic Distribution of DNA Informational Proteins Challenge All Extant Scenarios for Early Life Evolution
All scenarios produced during the last three decades to explain the evolutionary relationships among the three domains have been essentially based on the analysis of the translation and transcription apparatus. As a consequence, data gathered from the study of other informational systems, especially those dealing specifically with DNA, have usually been set aside. Indeed, the overall picture becomes more complex when proteins involved in DNA replication, recombination, or repair (DNA informational proteins) are taken into account. The reason is that many DNA informational proteins do not display the classical pattern, i.e., three homologous versions (one for each domain) (18–22). In particular, the major proteins involved in bacterial DNA replication (DNA polymerase, primase, and helicase) are not homologous to their archaeal/eukaryal counterparts (i.e., there is only one version of the DnaG primase, the bacterial one, and two versions of the archaeal/eukaryal primase). Generally speaking, cellular DNA informational proteins are often found in only one or two versions, instead of three. For instance, there are only two versions of cellular type II DNA topoisomerases of the A family, one in Bacteria and Archaea and another in Eukarya (18). Furthermore, many DNA informational proteins exist in different nonhomologous families (usually with several versions for one family). Hence, there are already six known nonhomologous families of cellular DNA polymerases (22). In the case of DNA polymerases of the B family, there is one version in Bacteria (only found in some proteobacteria), one in Archaea, and several in Eukarya (DNA polymerase, α, δ, ε, and ζ) (22). Furthermore, as seen in the few examples previously cited, the distribution of the different versions and families of cellular DNA informational proteins among domains is erratic most of the time and does not fit with any of the models proposed for the universal tree. A convincing scenario for the origin of the three domains thus should clearly explain why cellular DNA informational proteins do not follow the “one family, three versions” rule followed by universal ribosomal proteins. Interestingly, it is possible to explain this deviation by taking into account that living organisms are not all cellular and by integrating viruses into the overall picture.
Viruses as Major Players in Early Life Evolution
For a long time, viruses were not included in evolutionary scenarios, because they were considered only as nonliving entities, fragments of cellular genomes that escaped to become parasites. This prejudice was strengthened by the focus of evolutionary thinking on the translation apparatus: having no ribosome, viruses do not find their place in the universal tree of life. Recently, however, the importance of viruses and their potential role in early cellular evolution have been reevaluated (23, 24). In particular, the discovery of structural similarities among the capsid proteins and replicating enzymes of viruses infecting different domains has suggested that both RNA and DNA viruses may be more ancient than previously thought, possibly more ancient than LUCA itself (24). It was also realized from comparative genomic analyses that viruses can be the source of new proteins for cells (25). The most spectacular case occurred during the evolution of mitochondria from α-proteobacteria, because the original bacterial RNA polymerase, DNA polymerase, and helicase were replaced by viral proteins related to T3/T7 bacteriophages (26).
A critical point is that DNA informational proteins encoded by DNA viruses are usually not minor variations of host proteins but specific versions of a given viral lineage. Most of them indeed cluster in phylogenetic trees away from their cellular homologues (18, 22, 27). It is usually argued that this phenomenon is due the rapid rate of evolution of viral sequences. Although such explanation cannot be ruled out in some cases, it sounds like an ad hoc explanation to stick to the old prejudice that all viral genes originated from cellular ones. In other cases, however, the assumption that viral-specific versions of DNA informational proteins exist cannot be denied. For instance, the type II DNA topoisomerase encoded by the bacteriophage T4 cannot be an extremely derived version of a bacterial enzyme, because it contains an indel present in its eukaryotic but not in its bacterial homologues (28). Many viral DNA informational proteins are also clearly viral-specific simply because they have no cellular homologues, except for plasmid versions or viral remnants in cellular genomes (reviewed in ref. 28).
The extreme diversity of DNA informational proteins is not surprising considering the diversity of DNA viruses themselves and their probable antiquity (24). Different lineages of DNA viruses should have forged early on various types of DNA replication apparatuses, by recruiting independently different proteins originally involved in RNA transactions to perform the same function (20, 28). These RNA-informational proteins should have originated themselves in various lineages of RNA viruses (and cells) in the RNA world, at high evolutionary tempo. This could explain why many extant RNA and DNA informational proteins indeed exist in more than one family of nonhomologous proteins performing the same function and in many viral-specific versions of the same family (20, 28).
The existence of specific versions of viral DNA informational proteins immediately suggests an explanation for the erratic distribution of cellular DNA informational proteins among the different domains. It is likely that many DNA informational proteins encoded today in cellular genomes originated first in the viral world and were transferred later on randomly into the three cellular domains.
The Possible Viral Origin of Cellular DNA Genomes
To explain why DNA replication proteins and DNA replication mechanisms first originated in the viral world, I have proposed that DNA itself appeared in ancestral viral lineages (20, 29). In that hypothesis, DNA originated as a modified form of RNA resistant to the host cellular defenses mechanisms targeted against viral RNA genomes. This would have provided an immediate benefit for the virus (Darwinian selection). This scenario is supported by the fact that many modern viruses encode viral-specific versions of ribonucleotide reductases and thymidylate synthases (the two enzymatic activities required to produce DNA precursors). To explain how DNA was later transferred to cells, one can imagine that a DNA virus living in a carrier state in an RNA cell (persistent infection, ref. 30) lost the genes encoding for capsid proteins and lytic functions and became established as DNA extrachromosomal elements (linear or circular DNA plasmids) in an RNA cell (31). These plasmids could then have enlarged by picking up RNA genes from the cellular chromosome via retrotranscription. If the DNA plasmid was replicated more efficiently and/or was more stable than the RNA chromosome, it might have become advantageous for the cell to have essential genes carried on the invader DNA plasmid rather than on the original RNA genome. This would have ended up in the complete replacement of the cellular RNA genome by the former DNA plasmid, now a cellular DNA chromosome (31). In such a scenario, the RNA cell was transformed, from within, into a DNA cell.
Originally, two independent transfers of DNA from viruses to cells were suggested to explain the existence of two nonhomologous DNA replication machineries (one in Bacteria, the other in Archaea and Eukarya) (29). Recently, as an extension of this proposal, I suggested that the DNA replication machineries of each domain could have also originated from three different viruses (31). Interestingly, in addition to explaining the origin and idiosyncrasies of DNA replication machineries, this could also explain why there are three canonical versions of ribosomal proteins (see below). This new model for the origin of the cellular domains is therefore presented in more detail here, as a possible solution to the many puzzling questions surrounding the topology of the universal tree of life.
The Independent Transformation of Three Lineages of RNA Cells into DNA Cells
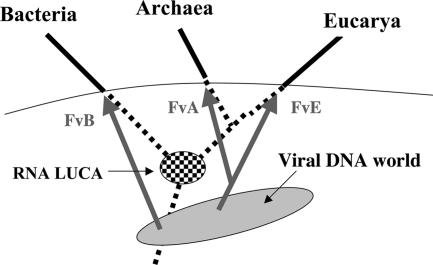
In the present theory (hereafter called the three viruses, three domains theory), each cellular domain originated independently from the fusion of an RNA cell and a large DNA virus (Fig. 1). These independent RNA to DNA transitions at the cellular level thus correspond to the “major qualitative evolutionary changes” or “dramatic evolutionary events” previously postulated at the origin of Archaea, Bacteria, and Eukarya (6, 14). The three nascent DNA cells and their descendants would have rapidly outcompeted all contemporary lineages of RNA cells, because they were able to accumulate more useful genes in larger genomes. An important consequence of the complete removal of ancestral RNA cells from the biosphere is that spreading of DNA cells eliminated by the same token the possibility for the origin of additional domains. In contrast, the model in which Archaea originated from Bacteria or in which Eukarya originated from the merging of Archaea and Bacteria did not explain why new domains are not continuously produced by the same mechanisms that could a priori still operate today.
Fig. 1.
The three viruses, three domains theory. Dotted lines correspond to RNA cell lineages, and bold lines correspond to DNA cell lineages. FvA, FvB, and FvE are founder viruses for Archaea, Bacteria, and Eukarya, respectively. The cellular tree is rooted in the “bacterial branch” (1), but other rootings are possible (discussed in ref. 6). The arrows between FvA and FvE are connected to symbolize specific evolutionary relationships between the two viruses that provide their DNA replication machineries to the archaeal and the eukaryal domains.
Because DNA genomes can be replicated more faithfully than RNA genomes (32), the viral-induced transformation of an RNA cell into a DNA cell would have been immediately followed by a drastic drop in the evolutionary tempo of protein evolution for all proteins that were previously encoded by RNA genes. The present theory thus readily explains the formation of the three canonical patterns for ribosomal proteins, one for each domain. As soon as Carl Woese and others suggested that LUCA had an RNA genome (1, 33), it was tempting to propose that the RNA–DNA transition was responsible for the reduction in the rate of protein evolution that led to the existence of the three canonical patterns of informational proteins. However, there was a difficulty with this idea. Indeed, because the major proteins involved in archaeal and eukaryotic DNA replication (DNA replicase, helicase, and primase) are homologous (3), it was a priori evident that these two domains had originated from an ancestor with a DNA genome. If this ancestor was cellular in nature, we were left with only two RNA–DNA transitions for three canonical versions. In the theory proposed here, this contradiction is solved, because the Archaea and Eukarya did indeed share an ancestor with a DNA genome, but this ancestor was a DNA virus (Fig. 1).
The existence of three distinct DNA viruses (the founder viruses) at the origin of DNA cells readily explains the erratic distribution of cellular DNA informational proteins versions and families in Archaea, Bacteria, and Eukarya, because each domain obtained its DNA informational proteins from a different virus. In particular, in addition to accounting for the difference between the archaeal and bacterial DNA replication apparatus, the present theory also readily explains those (often neglected) between the archaeal and eukaryotic DNA replication machineries, such as the existence of domain-specific DNA polymerases (family D in Archaea, DNA polymerase α in Eukarya) and DNA topoisomerases (family IIB in Archaea and IB in Eukarya).
A major problem in early cellular evolution is why eukaryotes, supposed to be a sister group of Archaea, have “bacterial-like” lipids in all their membranes (cytoplasmic, reticulum, and nuclear). Archaeal and bacterial (eukaryal) lipids have opposite chirality, because the formation of the covalent linkages between the glycerol and the acyl chains are formed by nonhomologous enzymes with different stereochemistry (34). These enzymes were recruited from two different dehydrogenase superfamilies that were likely both present in LUCA (34). Although the nature of the lipids present in LUCA cannot be inferred from comparative genomic analysis, it is reasonable to suggest that during the diversification of RNA cells from LUCA, some cellular lineages ended up by using specifically one of the two types of lipids. These RNA cells were most likely infected by different families of DNA viruses, independently of their lipid composition, because viruses recognize only the protein component of their host membrane, their receptors. In other words, two viruses with related DNA replication machineries could have infected RNA cells with different types of lipids to produce Archaea and Eukarya, respectively. In contrast, viruses of the same family tend to infect cells with related mechanisms for protein synthesis, because they have usually coevolved with them to take advantage of the protein machinery of their hosts. It thus makes sense that the two viruses that provide DNA for Archaea and Eukarya infected two RNA cells that had similar translation apparatus, explaining why all informational processes are finally similar between Archaea and Eukarya.
Corollaries
The Nature of LUCA.
The three viruses, three domains theory implies that LUCA had an RNA genome, in agreement with earlier proposals (1, 7, 33) (Fig. 1). The idea that LUCA was still a member of the RNA world was previously difficult to reconcile with the presence of several DNA informational proteins in the list of the universal proteins encoded in all completely sequenced genomes: the RecA-like recombinases, the Rad50/Mre11 proteins, the DNA topoisomerase I of the A family, and the DNA polymerase processivity factors (clamp and clamp loader) (23). However, in the present theory, one has simply to postulate that the three founder DNA viruses shared this small set of homologous DNA informational proteins (specific viral versions of these proteins are indeed encoded by modern viruses and/or plasmids).
Some readers will be skeptical regarding the present theory, because they consider RNA cells too primitive to be at the origin of individual domains. Many biologists used to think that RNA genomes could not be repaired or replicated faithfully at all. However, it has now been experimentally established that molecular mechanisms for RNA repair exist in modern cells, as well as mechanisms to increase the fidelity of RNA synthesis. As stated by Poole and Logan in a recent review, “This lends credibility to the proposal that the LUCA had an RNA genome” (35).
Why More than One Domain? Why only Three?
Thirty years after the discovery of Archaea and despite the explosion of environmental microbial ecology and the extensive search for new organisms using universal primers for PCR, all cellular organisms can still be grouped into “only” three domains. It is possible that the viral-induced transformation of an RNA cell into a DNA cell was a rare event that occurred indeed only three times (Fig. 1). Alternatively, one can also imagine that more than three lineages of DNA cells initially originated, but that only three of them survived the competition among nascent DNA cells. On the other hand, one can also wonder why the first lineage of DNA cells failed to take over the whole planet by rapidly preventing the possibility of further DNA cell formation, leading to the existence of a single domain. Possibly the three transfers occurred in different locations, leaving enough time for large populations of three different DNA cells to evolve separately before encountering each other. Considering that the last common ancestor of Archaea was probably a hyperthermophile (36), it is tempting to suggest that ancestral DNA cells at the origin of the Archaea were initially outcompeted by Bacteria and survived only by successfully invading high-temperature environments that were still free from their competitors. One can also imagine that the RNA cell at the origin of Eukarya was already a large predatory organism, whose lifestyle did not compete with the small rapidly dividing RNA cells at the origin of the bacterial and archaeal domains (k versus r selection) (37).
The Origin of Plasmids and “Prokaryotic” Chromosomes.
If the present theory is correct, the profound structural differences between eukaryotic and prokaryotic cell types could be explained by differences in both the type of the founder viruses and the type of RNA cells at the origin of the different domains. The similar and simple structure of archaeal and bacterial genomes (usually a single circular chromosome) suggests that Archaea and Bacteria both originated from a single transfer involving a DNA virus with a large circular genome. These viruses probably had the capacity to replicate their DNA in the cytoplasm and thus to couple the transcription of their DNA with the translation of their mRNA into the cytoplasm of the RNA cell, a hallmark of the “prokaryotic” cell type of architecture. Interestingly, the process that initially transformed a viral chromosome into a cellular DNA plasmid in the ancestral RNA cell could have continued later on in nascent DNA cells, in such a way that DNA cells from the emerging archaeal and bacterial lineages rapidly accumulated new plasmids by incorporating the genomes of other infecting viruses. Therefore, this theory can explain the origin of plasmids and their ubiquity in Bacteria and Archaea.
In my opinion, the present evolutionary connection between viruses, plasmids, and chromosomes in Archaea and Bacteria (30) is a strong argument for the present theory. It should be obvious to everyone that plasmids originated from viruses (for instance, rolling-circle plasmids from rolling-circle viruses with homologous Rep proteins) and not the reverse (otherwise, one has to explain how a plasmid can invent a capsid in the absence of preexisting viruses). It is also clear that there is an evolutionary link between plasmids and chromosomes (28, 31). For instance, it has been shown that the second chromosome of Vibrio cholerae utilizes a plasmid-like replication origin (38). Because bacteriophages can have very large DNA genomes (www.giantvirus.org/top.html), it is tempting to suggest that bacterial megaplasmids (or second chromosomes) are ancient viral genomes that are now permanently established in their bacterial hosts. In this case, why not imagine that the same occurred with the bacterial chromosomes itself, i.e., it is a descendant of the viral genome at the origin of the bacterial domain!
The Origin of the Eukarya and of the Cell Nucleus.
The paucity of plasmids in Eukarya and the completely different structure of their chromosomes suggest a different and more complex type of DNA transfer at the origin of the eukaryotic cells. The virus that donated its DNA to the RNA cell at the origin of Eukarya probably had a linear DNA genome (with possibly multiple chromosomes). One can also postulate that the RNA cell at the origin of Eukarya was more complex in terms of its molecular biology (especially RNA processing) than the RNA cells at the origin of Archaea and Bacteria. This elaborate RNA cell possibly already had a cytoskeleton and internal membrane system. Considering the complexity of the molecular biology of extant eukaryotic cells, it might be that this domain emerged in its present state only after the integration of more than one large DNA virus. This could explain the existence of three RNA polymerases (if they are of viral origin) and of several DNA polymerases of the B family (α, δ, and ε) in Eukarya. It is usually assumed that these are paralogous proteins that originated by duplication in the early evolution of Eukarya. However, in reconstructed phylogenies, these various groups of DNA and RNA polymerases are not monophyletic but are usually interspersed with homologous archaeal polymerases and different groups of viral enzymes (22, 39, 40). Accordingly, one cannot exclude that the different versions of RNA and DNA polymerases found in Eukarya are not paralogues but originated from different founder viruses that contribute to the complete design of these astonishing types of cell.
The founder virus at the origin of Eukarya (or at least one of them) could have been a complex enveloped virus of the nucleocytoplasmic large DNA viruses (NCLDV) superfamily. Some of these viruses (e.g., Poxviruses) replicate in the cytoplasm, form small nuclei, and produce their envelope by recruiting their membrane from the endoplasmic reticulum (all features common to the eukaryotic nucleus itself). Such viral nuclei could have evolved to produce the modern eukaryotic nucleus, in agreement with the viral eukaryogenesis hypothesis (41, 42). The recent discovery of the Mimivirus, an NCLDV with a genome of 1.2 Mb (40), gives credibility to the idea that such giant viruses could have provided the nascent DNA cell at the origin of Eukarya with all enzymes required for the replication and transcription of its genome. The capsid proteins of the Mimivirus and other NCLDVs are homologous to those of the Adenovirus and of several bacterial and archaeal viruses (43), suggesting that these viruses indeed preexisted the formation of the eukaryotic lineage.
How to Test the Theory?
The best test of the present theory would be to transform an RNA cell into a DNA cell using a DNA virus. Unfortunately, there is no longer an RNA cell wandering around (something predicted by the theory!). Alternatively, it could be possible to transform cells containing reverse transcriptase with in vitro engineered RNA plasmids to check the possibility of gene transfer in vivo from RNA to DNA genomes. Another realistic but difficult line of experimental research could be to play with modern DNA cells and viruses to create new “domains” of life (at least cell lineages with novel DNA replication and transcription apparatus). This could help us understand the barriers that have prevented nonorthologous displacements of DNA informational proteins in real life by viral ones, once the three canonical DNA replication apparatuses have been established (44). Interestingly, these barriers have indeed been eliminated in the evolution of Bacteria into modern mitochondria, because the ancestral DNA replication mechanism has been fully replaced by a viral one (26).
If the theory is correct, extensive screening for new viruses and plasmids in all kingdoms of the three domains could lead to the discovery of modern DNA viruses that still have close evolutionary affinities with DNA founder viruses. For instance, it should be fascinating to find new archaeal and/or bacterial viruses encoding specific viral versions of most cellular DNA replication proteins from one of the two prokaryotic domains. The recent discovery of a bacterial prophage encoding a homologue of the archaeal replicative helicase minichromosome maintenance protein (MCM) is a first step in that direction (45). The problem again will be to demonstrate that these proteins have not been stolen from their host by the viruses (something difficult to imagine in the last example). Hopefully, more sound phylogenetic analyses (for instance, the use of an indel, as in the case of type II DNA topoisomerases; ref. 32) will help us to polarize with more confidence the direction of transfers of DNA informational proteins among viruses and cells. A systematic polarization of ancient transfers in the direction from viruses to cells would be a further step in the validation of the theory.
Conclusion
As with most evolutionary scenarios, the three domains, three viruses theory cannot be easily falsified. However, it has great explanatory power, because it explains the formation of the canonical protein patterns characteristic of each domain of life, the formation of a discrete numbers of domains, or else the puzzling distribution of DNA informational proteins among the three cellular domains. The theory is compatible with an RNA-based LUCA and at the same time with the existence of a few homologous DNA informational proteins in the three domains. Finally, it takes into account the whole biosphere (cells, viruses, and plasmids). The unification of cellular life, a major achievement of the last century, has left aside viruses as nonliving entities. In the present theory, both viruses and plasmids find their place in the history of life as critical players in the origin of DNA genomes and modern cells.
Acknowledgments
This paper is dedicated to the memory of Wolfram Zillig, who recognized early on the antiquity of viruses. I am grateful to Carl Woese, who encouraged me to write this manuscript. For advice and editing, I also thank Dave Musgrave, David Prangishvili, Jonathan Berthon, Chlöe Terras, Simonetta Gribaldo, and Celine Brochier. I thank two anonymous referees for helpful comments, criticisms, and suggestions. The work in my laboratory in Orsay on DNA informational proteins is supported by grants from the Human Frontier Science Program and Association pour la Recherche sur le Cancer.
Abbreviations
- LUCA
Last Universal Cellular Ancestor
- HGT
horizontal gene transfer.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Woese C. R. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woese C. R., Kandler O., Wheelis M. L. Proc. Natl. Acad. Sci. USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olsen G. J., Woese C. R. Cell. 1997;89:991–994. doi: 10.1016/s0092-8674(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 4.Makarova K. S., Koonin E. V. Curr. Opin. Microbiol. 2005;8:586–594. doi: 10.1016/j.mib.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Woese C. R., Olsen G. J., Ibba M., Soll D. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forterre P., Philippe H. BioEssays. 1999;21:871–879. doi: 10.1002/(SICI)1521-1878(199910)21:10<871::AID-BIES10>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 7.Woese C. R., Fox G. E. J. Mol. Evol. 1977;10:1–6. doi: 10.1007/BF01796132. [DOI] [PubMed] [Google Scholar]
- 8.Woese C. R. Proc. Natl. Acad. Sci. USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penny D., Poole A. Curr. Opin. Genet. Dev. 1999;9:672–677. doi: 10.1016/s0959-437x(99)00020-9. [DOI] [PubMed] [Google Scholar]
- 10.Gupta R. S. Rev. Microbiol. 2000;26:111–131. doi: 10.1080/10408410091154219. [DOI] [PubMed] [Google Scholar]
- 11.Cavalier-Smith T. Int. J. Syst. Evol. Microbiol. 2002;52:7–76. doi: 10.1099/00207713-52-1-7. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Garcia P., Moreira D. Trends Biochem. Sci. 1999;24:88–93. doi: 10.1016/s0968-0004(98)01342-5. [DOI] [PubMed] [Google Scholar]
- 13.Woese C. R. Proc. Natl. Acad. Sci. USA. 2000;97:8392–8396. doi: 10.1073/pnas.97.15.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartman H., Fedorov A. Proc. Natl. Acad. Sci. USA. 2002;99:1420–1425. doi: 10.1073/pnas.032658599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reanney D. C. J. Theor. Biol. 1974;48:243–251. doi: 10.1016/0022-5193(74)90194-5. [DOI] [PubMed] [Google Scholar]
- 16.Poole A., Jeffares D., Penny D. BioEssays. 1999;21:880–889. doi: 10.1002/(SICI)1521-1878(199910)21:10<880::AID-BIES11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 17.Woese C. R. Proc. Natl. Acad. Sci. USA. 2002;99:8742–8747. doi: 10.1073/pnas.132266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gadelle D., Filée J., Buhler C., Forterre P. BioEssays. 2003;25:232–242. doi: 10.1002/bies.10245. [DOI] [PubMed] [Google Scholar]
- 19.Leipe D. D., Aravind L., Koonin E. V. Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forterre P., Filée J., Myllykallio H. In: The Genetic Code and the Origin of Life. Ribas de Pouplana L., editor. New York: Springer; 2004. pp. 145–168. [Google Scholar]
- 21.Myllykallio H., Lipowski G., Leduc D., Filée J., Forterre P., Liebl U. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 22.Filée J., Forterre P., Sen-Lin T., Laurent J. J. Mol. Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- 23.Forterre P. Res. Microbiol. 2003;154:4–6. doi: 10.1016/s0923-2508(03)00111-6. [DOI] [PubMed] [Google Scholar]
- 24.Bamford D. H. Res. Microbiol. 2003;154:231–236. doi: 10.1016/S0923-2508(03)00065-2. [DOI] [PubMed] [Google Scholar]
- 25.Daubin V., Ochman H. Curr. Opin. Genet. Dev. 2004;14:616–619. doi: 10.1016/j.gde.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Filée J., Forterre P. Trends Microbiol. 2005;13:510–513. doi: 10.1016/j.tim.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Miller W., Kutter E., Mosig G., Arisaka F., Kunisawa T., Ruger W. Microbiol. Mol. Biol. Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forterre P. Virus Res. 2006. in press. [DOI] [PubMed] [Google Scholar]
- 29.Forterre P. Curr. Opin. Microbiol. 2002;5:525–532. doi: 10.1016/s1369-5274(02)00360-0. [DOI] [PubMed] [Google Scholar]
- 30.Villarreal L. P. In: Viruses and the Evolution of Life. Villarreal L. P., editor. Washington, DC: Am. Soc. Microbiol. Press; 2005. [Google Scholar]
- 31.Forterre P. Biochimie. 2005;87:793–803. doi: 10.1016/j.biochi.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 32.Lazcano A., Guerrero R., Margulis L., Oro J. J. Mol. Evol. 1988;27:283–290. doi: 10.1007/BF02101189. [DOI] [PubMed] [Google Scholar]
- 33.Mushegian A. R., Koonin E. V. Proc. Natl. Acad. Sci. USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereto J., Lopez-Garcia P., Moreira D. Trends Biochem. Sci. 2004;29:469–477. doi: 10.1016/j.tibs.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Poole A. M., Logan D. T. Mol. Biol. Evol. 2005;22:1444–1455. doi: 10.1093/molbev/msi132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forterre P., Brochier C., Philippe H. Theor. Popul. Biol. 2002;61:409–422. doi: 10.1006/tpbi.2002.1592. [DOI] [PubMed] [Google Scholar]
- 37.Carlile M. Trends Biochem. Sci. 1982;7:128–130. [Google Scholar]
- 38.Egan E. S., Waldor M. K. Cell. 2000;114:321–330. [Google Scholar]
- 39.Villarreal L. P., DeFilippis V. R. J. Virol. 2000;74:7079–7084. doi: 10.1128/jvi.74.15.7079-7084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raoult D., Audic S., Robert C., Abergel C., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J. M. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 42.Bell P. J. J. Mol. Evol. 2001;53:251–256. doi: 10.1007/s002390010215. [DOI] [PubMed] [Google Scholar]
- 43.Takemura M. J. Mol. Evol. 2001;52:419–425. doi: 10.1007/s002390010171. [DOI] [PubMed] [Google Scholar]
- 44.Benson S. D., Bamford J. K., Bamford D. H., Burnett R. M. Mol. Cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 45.Iyer L. M., Leipe D. D., Koonin E. V., Aravind L. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 46.McGeoch A. T., Bell S. D. Cell. 2005;120:167–168. doi: 10.1016/j.cell.2004.11.031. [DOI] [PubMed] [Google Scholar]