Abstract
Chromosome deletions in the mouse have proven invaluable in the dissection of gene function. The brown deletion complex comprises >28 independent genome rearrangements, which have been used to identify several functional loci on chromosome 4 required for normal embryonic and postnatal development. We have constructed a 172-bacterial artificial chromosome contig that spans this 22-megabase (Mb) interval and have produced a contiguous, finished, and manually annotated sequence from these clones. The deletion complex is strikingly gene-poor, containing only 52 protein-coding genes (of which only 39 are supported by human homologues) and has several further notable genomic features, including several segments of >1 Mb, apparently devoid of a coding sequence. We have used sequence polymorphisms to finely map the deletion breakpoints and identify strong candidate genes for the known phenotypes that map to this region, including three lethal loci (l4Rn1, l4Rn2, and l4Rn3) and the fitness mutant brown-associated fitness (baf). We have also characterized misexpression of the basonuclin homologue, Bnc2, associated with the inversion-mediated coat color mutant white-based brown (Bw). This study provides a molecular insight into the basis of several characterized mouse mutants, which will allow further dissection of this region by targeted or chemical mutagenesis.
Keywords: brown locus, chromosome deletion, mouse genome sequence
The mouse brown (b) mutation is one of the oldest known loci and was one of the first to be cloned. The mutation is in the tyrosinase-related protein 1 (Tyrp1) gene, encoding a melanocyte enzyme required for the production of dark eumelanin (1). Homozygous loss of Tyrp1 results in a brown coat; because it was a simply scored phenotype, brown was used as part the mouse-specific locus test carried out at the Oak Ridge National Laboratories and elsewhere (reviewed in ref. 2). Wild-type mice were exposed to chemical or radiological mutagens and crossed with a tester stock homozygous for seven recessive visible mutations. The efficacy of the mutagenic treatments is assessed in the resulting F1 progeny. These experiments generated many chromosomal deletions that inactivated Tyrp1, particularly in the progeny of animals exposed to radiation or clastogenic chemical mutagens such as chlorambucil or melphalan.
Deletions around the seven specific loci have provided a unique opportunity to study gene function within these intervals. Individual deletions are hemizygous viable but when homozygous or in combination with others give lethal or visible phenotypes, which indicate the presence of essential genes neighboring the specific loci. Intercrossing deletions of different extents provided an avenue to map the resulting phenotypes. The deletions also provided the means to produce physical maps of genetic markers. Studies of this kind have been published for several loci, including albino (Tyr), piebald (Ednrb), pink-eyed dilution (p), and the brown deletion complex (2–6).
Studies of the brown deletions established genetic and physical maps of the interval by using a panel of 25 deletions spanning ≈8.5 cM of chromosome 4 (6–8). Complementation analyses defined phenotypic loci in the region, including three lethals and a phenotype, termed brown-associated fitness (baf), which results in postnatal runting and increased preweaning mortality. Previous studies have also described a chromosomal inversion, white-based brown (Bw), that was hypothesized to cause inappropriate expression of an unknown gene, resulting in melanocyte death and loss of hair pigmentation (9).
We have generated a finished and manually annotated sequence from a bacterial artificial chromosome (BAC) contig spanning the brown deletion complex. The region is very gene-poor and includes several large segments devoid of any protein-coding genes. We identified single-nucleotide polymorphisms that permit refinement of the deletion breakpoints that delineate known phenotypes. The sparse gene content means that we have identified one or a few candidate genes for each of the five phenotypes.
Results and Discussion
Sequence and Annotation of the Tyrp1 Deletion Complex.
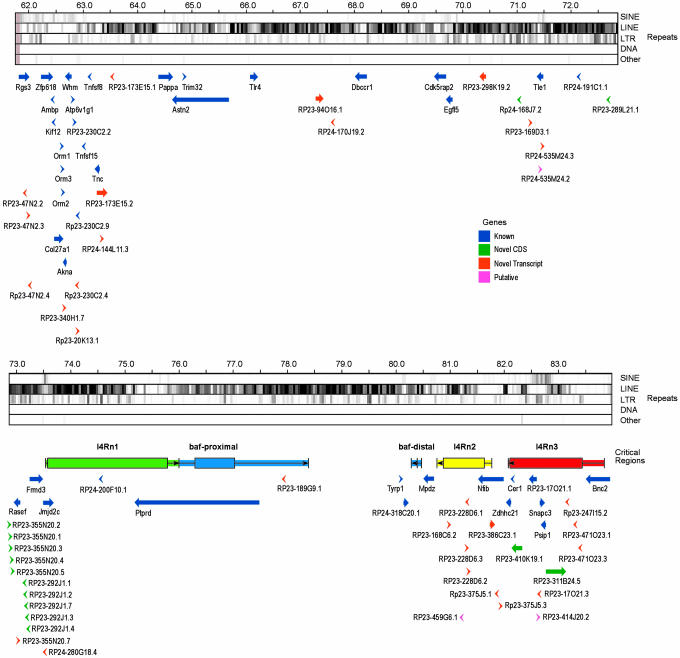
The finished sequence of each of 172 BACs from a clone contig encompassing the Tyrp1 deletion complex was individually annotated to identify genes, transcripts, and pseudogenes (Fig. 1; for a complete version of this figure, see Fig. 5, which is published as supporting information on the PNAS web site). The annotations can be viewed in the Vertebrate Genome Annotation (VEGA) browser (http://vega.sanger.ac.uk/index.html). The extent of the deletion complex was determined by locating the endpoints of those deletions extending furthest proximally (Tyrp111R30M) and distally (Tyrp18Pub). These deletions define an unusually gene-poor genomic segment of ≈21 megabases (Mb) from a point just distal to Rgs3 as far as, and including, Bnc2. This segment contains only 53 protein-coding genes (see Table 1, which is published as supporting information on the PNAS web site). We can be confident that 39 of these genes are true genes because they have orthologous protein-coding transcripts in other species. The remaining 14 genes are all identified by the existence of mouse cDNAs, which contain ORFs distributed across multiple exons and are predicted to encode peptides containing between 110- and 354-aa residues. One of them is derived from the 3′ end of the adjacent Frmd3 gene. None of the remaining thirteen, however, shows significant homology to transcripts from any other species nor do their predicted protein products have any recognizable motifs. Ten of these predicted genes arise from a 10-fold local amplification of a 15-kb genomic segment. Although there are EST matches to these amplified arrays, none of them are perfect, and it is not clear which of the repeated segments are transcribed. Furthermore, the human genome sequence contains a recognizable homologue neither to the transcript nor to the 15-kb amplified segment from which it derives. The rat genome does contain a single orthologous genomic segment, but this segment is located on rat chromosome 15 not on the rat homologue of mouse chromosome 4, which is chromosome 5.
Fig. 1.
Schematic representation of the annotation of the Tyrp1 deletion region. The deletion interval extends as far as, but not including, Rgs3 at the proximal end and includes Bnc2 at the distal end. Yellow blocks represent finished sequences, marked with accession numbers (available at http://vega.sanger.ac.uk/index.html), of individual BAC clones forming the tiling path across the region. A black block on the end of the clone represents an overlapping, redundant sequence. The five gray-black tracks show the distribution of different types of repeats, as defined by repeatmasker. The red line indicates a fraction of G and C nucleotides. Arrows show the position and orientation of annotated genes and pseudogenes, with the color of the arrows indicating the type as shown in the key. The five colored boxes are the extents of the critical regions for the deletion phenotypes as indicated. The wider box is the minimal critical region, the narrow box is the maximum, and the boxes are delineated by the deletions and markers listed in Tables 2 and 3. The ends of the boxes are the positions of the markers. The arrows in the boxes reflect the intact chromosomal ends, pointing into the deletion.
The deletion complex also gives rise to 28 spliced transcripts, identified by mouse cDNAs, which are derived from between two and five exons but contain neither any significant ORF nor show homology to transcripts from other species. In addition, there are 74 recognizable pseudogenes, almost all of them processed.
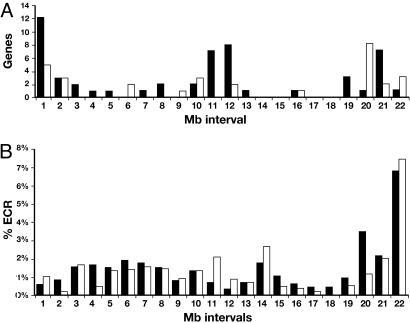
The overall gene density across this region is just >2 genes per Mb compared with the genome average of ≈10 genes per Mb. Furthermore, the genes are not evenly spaced (Fig. 2A). More than half are located in the megabases at each end of the complex and in a 2-Mb region in the center, leaving only 20 genes spaced across the remaining 18 Mb. There are seven stretches of >1 Mb, including one of 2.5 Mb, that are totally devoid of any protein-coding transcript. Another large segment of 1.9 Mb is solely occupied by 10 exons encoding isoforms of the 5′ UTR of the Ptprd gene. Furthermore, the 29 spliced noncoding transcripts are also not evenly distributed across the region; rather, they appear to be clustered in those regions that are enriched for genes (Fig. 2A).
Fig. 2.
Distribution of genes and evolutionary-conserved regions (ECRs). (A) The number of protein-coding genes (black bars) and noncoding RNAs (white bars) in each 1-Mb interval across the region, beginning at the 3′ end of Rgs3. Where a gene or transcript spans more than one interval, it is placed in the one containing the 5′ end of the 5′ most transcript. (B) The percentage of ECR content in each 1-Mb interval, as in A. The fraction is calculated as the total length of sequence, after masking repeats and exons, that reaches the threshold. Black bars are mouse–human ECRs at 80% identity over >200 bp, and white bars are mouse–chicken ECRs at 70% identity over >100 bp.
The function, if any, of these noncoding RNA is unknown. Their localization near to established genes may indicate that the transcripts are hitherto unidentified parts of transcripts of the known protein-coding genes, or their expression may be a nonfunctional by-product of local transcriptional activity. Alternatively, it is possible that their transcription has a function in opening up the chromatin, or maintaining it in an open state, to facilitate expression of protein-coding genes.
Repeat content is correlated with gene density in the mammalian genome (10). Fig. 1 illustrates this correlation across the deletion region. Gene-poor segments are rich in LINE elements, whereas SINEs are more abundant in the gene-rich areas. The gene and transcript “deserts,” however, still contain substantial numbers of nontranscribed conserved sequences. Fig. 2B shows the distribution of nontrancribed segments that are conserved between the mouse sequence and the orthologous human and chicken sequences. Surprisingly, the locations of the conserved segments do not appear to correlate with transcript locations. The gene-rich first megabase of the region has a lower concentration of noncoding conserved segments, and the central part, which is also relatively enriched for genes, has a low level of noncoding conservation.
The function of most of the conserved segments in the genome is unknown, and they may indeed have a diversity of roles. The conserved sequences located close to genes are candidates for regulatory elements. Even those elements very distant from the 5′ end of a gene may still play a role; there is increasing evidence that sequences at several megabases distant can be essential for correct gene expression (11). Recently, Ovcharenko et al. (12) have examined gene deserts in the human, which they define as intergenic intervals of >640 kb, the longest 3% of all intergenic intervals. They find that the human–chicken sequence conservation in these deserts range from none to 12% and suggest that those with 2% or more conservation may be more likely to house long-range regulatory elements. Using the same criteria for conservation, we find that most intervals in this deletion region have <2% conservation.
A Site of Striking Evolutionary Rearrangement.
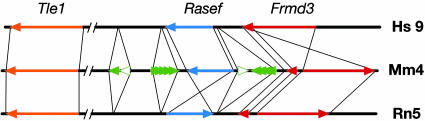
The entire region is homologous to human chromosome 9 but is not contiguous in humans. There is a striking correlation between gene density and human chromosome G-banding patterns. The deserts lie within dark G bands, and the gene-rich segments are in light bands (10). The proximal end of the region is homologous to 9q32-33, the distal end to 9p22-24, and an ≈3.5-Mb segment in the middle is homologous to 9q21.32. The distal end of this central segment has undergone considerable rearrangements over evolution. The protein-coding gene, RP23-289L21.1, is ≈600 kb from where the homology changes to 9p22-24. As mentioned, this gene is locally amplified ≈200 kb upstream, and in the opposite orientation are five tandemly arranged, almost identical, 15-kb segments that derive from the genomic segment encoding the gene. A further four copies of the segment are found another 100 kb further along the chromosome, between Rasef and Frmd3. Two pseudogenes derived from these genes are also presenting this region (Fig. 3). Characterization of this array of highly similar sequences required that the sequence produced for the region was high-quality and hand-finished. Earlier drafts of the region had only a single copy of the amplified array, which illustrates the value of hand-finished sequence.
Fig. 3.
Schematic representation of a region of striking evolutionary rearrangement. The orthologous regions of human chromosome 9 (Hs9), mouse chromosome 4 (Mm4), and rat chromosome 5 (Rn5) are shown. The orientations of the genes are indicated by the following colored arrows: Tle1 in orange, Rasef in blue, and Frmd3 in red. Lines connect the 3′ and 5′ ends of each. The second, short red arrow in mouse and rat genome represents the RP23-292J1.4 transcript, which apparently derives from a 3′ end duplication and inversion of Frmd3. The 10 green arrowheads in the mouse line represent the 9-fold amplification encoding the transcripts related to RP23-289L21.1, which are absent from the rat and human lines. Open green arrowheads are the two pseudogenes of the same gene.
The chromosomal orthology breakpoint is immediately adjacent to the 3′ end of Frmd3, and this gene also seems to have undergone considerable evolutionary rearrangement. In mice, Rasef and Frmd3 are divergently transcribed from opposite strands, whereas the human orthologues are transcribed from the same strand (Fig. 3). The 3′ portions of both human and mouse Frmd3 are transcribed from within an intron by using an alternative, conserved exon. In mouse, a homologue of this shorter transcript is present in a second copy (RP23-292J1.4), in reverse orientation (the same orientation as the human transcript). There appears to have been a duplication accompanying the inversion (Fig. 3), and the human gene orientation is ancestral. Examination of the rat genome sequence reveals that Frmd3 in this species has the same orientation as in mouse and also has the duplicated 3′ transcript, indicating that the rearrangement occurred before the mouse and rat lineages diverged. In rat, however, the Rasef gene is reversed compared with the mouse, apparently a result of another gene inversion in the rat lineage (Fig. 3). The genomic arrangement in zebrafish is different yet again; in this case, Rasef and Frmd3 are transcribed from opposite strands, as in mouse, but are divergent not convergent (data not shown).
Alignment of the entire 22 Mb Tyrp1 deletion region reveals at least two other small inversions in the mouse genome with respect to the human genome of 80 and 100 kb, but nether of these inversions include any genes or transcripts.
Identification of Deletion Phenotypes.
To accurately identify candidate genes for loci previously mapped to the deletion complex, we mapped the ends of deletion breakpoints using SNPs between the strains of origin of the deletion chromosomes and Mus spretus in deletion/spretus heterozygotes. We focused on those deletions that had been used to define mutant loci. The minimal and maximal extents of key deletion intervals are illustrated in Fig. 1. Tables 2 and 3, which are published as supporting information on the PNAS web site, list the deletions and markers used to define the endpoints. The relevant markers from Table 3 map on Fig. 1 as the maximum and minimum extents of each deletion.
Embryonic Lethal Phenotypes.
Combinations of deletions identify three embryonic or neonatal lethal phenotypes (7, 8, 13), and their endpoints define the locations of essential genes deleted or mutated by them.
l4Rn1 is a deletion phenotype resulting in in utero death before E14.5 (13). The physical map of Bell et al. (8) indicates that it lies between the proximal end points of the Tyrp18Pub and Tyrp111Pu deletions. Tyrp18Pub breaks within the Ptprd gene and is the most proximal viable deletion, whereas Tyrp111Pu breaks within the Jmjd2c gene and is the most distal lethal deletion. l4Rn1, therefore, lies in the 2.45-Mb interval defined by these endpoints (Fig. 1). Despite the length of this segment, only three genes are within the interval. We can exclude Ptprd as a candidate because Tyrp18Pub disrupts Ptprd yet does not delete l4Rn1. The remaining two candidates are Jmjd2c and an uncharacterized gene, RP24-200F10.1, defined by the Institute of Physical and Chemical Research (Japan) (RIKEN) clone no. 3110001D03, which is the mouse orthologue of human C9orf123. Jmjd2c is the orthologue of a characterized human gene, GASC1, which is amplified in esophageal cancer cell lines (14). The encoded protein contains two jumonji domains, jmjC and jmjD, which are found together in numerous mouse proteins and may indicate function as a transcription factor. Any potential function of the product of the C9Orf123 orthologue is difficult to predict; the protein is only 111 aa in length but has two potential transmembrane domains that are conserved in humans.
Mice homozygous for l4Rn2 deletions die neonatally, and the locus is defined by the distal end of deletion Tyrp111R30M and the distal end of deletion Tyrp19R75VH (6). Our deletion mapping indicates that this interval contains only part of only one gene, nuclear factor I-B (Nfib), encoding a transcription factor expressed at high levels in the embryonic lung (15) (Fig. 1). Nfib mutant pups die shortly after birth as a result of severe pulmonary hypoplasia (16), and this phenotype is entirely consistent with that of mice bearing homozygous deletions of l4Rn2. We suggest deletion of Nfib is responsible for the l4Rn2 phenotype.
The final early embryonic lethal deletion locus, l4Rn3, lies between the distal ends of the Tyrp146UThc deletion and Tyrp18Pub (Fig. 1). This interval contains six genes: RP23-410K19.1, which is the Frem1 gene (17); the PC4 and SFRS1 interacting protein 1, Psip1; the small nuclear activating complex protein 3 (Snapc3); basonuclin 2 (Bnc2); and two uncharacterized genes, RP23-17O21.2, encoding the cDNA 1810054D07Rik, and RP23-311B24.5, encoding A330015D16Rik, which are the mouse orthologues of human C9Orf52 and C9Orf93, respectively. We can exclude Frem1 as a candidate as loss-of-function mutations of this gene result in the head blebs (heb) phenotype (17). The SNAPC3 protein forms part of the small nuclear RNA-activating protein complex and reduction of protein in vitro leads to inhibition of RNA polymerase II- and III-mediated small nuclear RNA gene transcription (18). PSIP is a nuclear protein important for cellular protection against stress-induced apoptosis in which activity is regulated by caspase-mediated cleavage (19, 20). Neither protein has been knocked out but, given their cellular functions, either would make a good candidate for l4Rn3. Of the two unknown function transcripts, only the C9Orf93 orthologue has any recognizable protein domain, including coiled coils and tropomyosin motifs. Very few of the brown deletions were mapped relative to l4Rn3; we were unable to further reduce this locus, and all five genes remain candidates.
Brown-Associated Fitness (baf).
Previous studies of the deletion complex showed that appropriate combinations of chromosomal deletions complement the embryonic lethal phenotypes. All complementing deletions reported result in mice with poor growth rates, alterations in behavior, and compromised survival (6, 7). This phenotype, termed brown-associated fitness (baf), must result from the loss of a gene or genes very close to Tyrp1. However, certain deletion combinations, namely Tyrp11OZ with Tyrp137DTD and Tyrp147UTHc with Tyrp137DTD, Tyrp133G, or Tyrp11THO-IV result in phenotypically normal mice, indicating that the gene(s) in which deletion results in baf must lie outside these deletions (E. M. Rinchik, unpublished data). Paradoxically, each of these deletions, in combination with others, can produce baf mice. We must conclude that there are two genetic elements, one on each side of Tyrp1, the deletion of either of which gives rise to the baf phenotype (Fig. 1).
The maximum deletion interval on the proximal side that defines baf contains a part of only one gene, Ptprd. This gene has been mutated by gene targeting and results in mice with neurological defects that fail to thrive without ground feeding (21). This phenotype is consistent with baf, and we conclude that the phenotype caused by deletions proximal to Tyrp1 is due to loss of Ptprd. It is worth noting that these deletions remove a 2.5-Mb gene desert in addition to Ptprd, yet the mice have a phenotype no more severe than a targeted mutation of Ptprd alone. The cause of the distal baf phenotype is less clear. Seventy kilobases distal of Tyrp1 is an uncharacterized gene RP24-318C20.1, the orthologue of human C9Orf150. However, deletion of all or part of this gene, from Tyrp11THO-IV and Tyrp137DTD, does not result in the baf phenotype. Rather, the distal baf is defined by the endpoints of Tyrp11THO-IV Tyrp1173G (Fig. 1), which encompass a 180-kb segment between the C9Orf150 orthologue and Mpdz, containing no predicted genes or transcripts. Comparison of this sequence with other mammalian species reveals numerous segments with a high level of sequence identity consistent with evolutionary conservation of regulatory element(s). It is possible that these are long-range control element(s) necessary for the expression of Ptprd, for which the 5′ end is some 3 Mb away, and this finding would account for the consistency of phenotype between distal and proximal baf deletions.
Characterization of White-Based Brown (Tyrp1B-w).
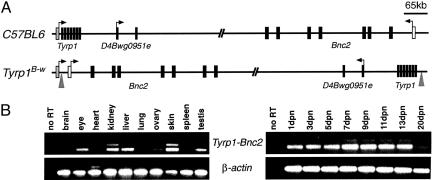
White-based brown (Tyrp1B-w) is a mutation of the brown locus that arose spontaneously during a radiation mutagenesis experiment (22). The Tyrp1B-w chromosome carries a dominant mutation that results in the absence or reduction in pigment at the base of the hair follicle but which is also a recessive loss-of-function allele of Tyrp1. This curious phenotype has been partially explained by preliminary molecular characterization showing that it results from an inversion that inactivates Tyrp1 by interrupting the gene in the first intron (9, 23). It has been proposed that the dominant pigmentation loss in the mice results from ectopic expression of a second gene from the Tyrp1 promoter, which either abrogates pigment production or leads to melanocyte death during the latter stages of the hair growth cycle. Short sequences bordering the inversion breakpoints have been cloned (9), and the genome sequence of the interval allowed us to determine the exact nature of the rearrangement.
The proximal end of the inversion is 105 bp from the exon-2 splice acceptor site in Tyrp1 intron 1 (Fig. 4A). The other breakpoint lies distal of the brown deletion complex, ≈15-kb upstream of the recently characterized Basonuclin 2 (Bnc2) gene (24). RT-PCR analysis demonstrated the expression of a Tyrp1-Bnc2 fusion transcript in several Tyrp1B-w tissues (Fig. 4B). Normal Tyrp1 expression is restricted to melanocytes; however, we detect expression of the fusion transcript in several other tissues such as the kidney, liver, ovary, and testis. This finding indicates that enhancer elements associated with Bnc2 are driving ectopic expression of Tyrp1 in these tissues or that Tyrp1 expression is normally repressed by elements subsequently separated from the locus by the inversion. To determine whether melanocyte death occurred during the hair follicle cycle, we examined expression of the fusion transcript during the first hair cycle. Tyrp1 is normally activated shortly after birth when melanocytes begin to produce pigment during anagen, and this expression stops by 13 days postnatal. However, we observed persistent expression of the fusion transcript in the skin both at birth and after 13 days postnatal, presumably as a consequence of the ectopic expression.
Fig. 4.
White-based brown inversion. (A) Schematics of the C57BL/6J and Tyrp1B-w chromosomes showing the regions around the Tyrp1 and Bnc2 genes, and the intervening D4Bwg0951e gene for orientation. Arrows indicate transcriptional start sites. The gray arrowheads mark the breakpoints of the inversion. (B) RT-PCR showing transcription of the Tyrp1-Bnc2 fusion transcript in a range of adult tissues and in the skin of mice between 1 and 20 days postnatal (dpn).
We sequenced multiple RT-PCR products of the Tyrp1-Bnc2 fusion transcript. Normal splicing of Tyrp1 exons 1 and 2 uses three different donor sites (25), which were all detected in the fusion transcripts spliced to Bnc2. We identified promiscuous splicing of Tyrp1 to splice acceptor sites in Bnc2 exons 1a and 2 (Fig. 4A) of which products of splicing to 1a would produce normal protein products. Because expression of the coding region of Tyrp1 at the proximal end of the inversion is not detected (23), we propose that overexpression of Bnc2 underlies the dominant loss of pigment in Tyrp1B-w animals.
Other Deletion Complexes.
The surprisingly low gene density of this deletion complex may account for the viability of large deletions encompassing Tyrp1. Certain other specific locus regions, such as Tyr and Ednrb, also have large, viable deletions and appear to be gene-poor. By contrast, the agouti region is relatively gene rich, but relatively few deletions have been recovered from the region. Once other regions of the mouse genome are annotated at high quality, it will be interesting to correlate gene content with the presence or absence of large deletions that have been identified at many other chromosomal sites following radiation mutagenesis.
Materials and Methods
BAC Contig Construction, Sequencing, and Annotation.
A BAC contig spanning the entire brown deletion complex was constructed by using clones from RCPI-23 and -24 C57BL/6J strain mouse libraries (26). Clones containing genes known to map to the interval were identified by hybridization to gridded libraries, and contigs were extended by using probes produced by overlapping oligonucleotides (overgos) designed to the ends of these BACs. The contig was also extended by using BAC fingerprint data (27). Contiguous BAC clones were sequenced by the Rosalind Franklin Centre for Genome Research and the Sanger Institute, as described in ref. 28. Manual annotation was performed by using established criteria and methodologies (www.sanger.ac.uk/HGP/havana/havana.shtml). Detailed annotation of the complete brown deletion sequence is available at the Vega web site (http://vega.sanger.ac.uk/index.html). Details of phenotypes defined in the region are in the Mouse Genome Database (www.informatics.jax.org).
Deletion Mapping.
Deletion endpoints were mapped by using a panel of deletion/Spretus DNAs (7). Primer pairs were predominantly designed to amplify nonrepetitive BAC end sequences, although in some cases were designed specifically to map genes against deletions. Primers used for mapping key endpoints are listed in Table 3. Additional primer sequences and their amplification conditions are available on request.
Analysis of the Bw Inversion.
The proximal end of the Bw inversion was identified by using the short sequence known to border the inversion (12). The genomic structure and sequence of mouse Basonuclin 2 (Bnc2) was determined by assembling ESTs and comparing these with both the Basonuclin 1 (Bnc1) sequence and expanding the gene structure in the human, fugu, and zebrafish genome assemblies by using genewise (29). RT-PCR analysis of splicing between Tyrp1 and Bnc2 was performed by using a forward primer in Tyrp1 exon 1 (TCA GGG GAA AAG CAG ACA TC) and a reverse primer in exon 3 of Bnc2 (GGT GCA GTT TAC CAA TGT GC). RNA was extracted from homogenized mouse tissues by using the Total RNA Isolation System kit (Promega) and cDNA and PCR were undertaken by using the Access RT-PCR Kit (Promega) following the manufacturers protocols.
Supplementary Material
Acknowledgments
The following people contributed at the Wellcome Trust Sanger Institute by finishing the sequence of between one and five BAC clones: Lucy Matthews, Harminder Sehra, Matthew Ellwood, Karen Oliver, Matthew Dunn, Neil Sycamore, Alan Tracey, Jeff Almeida, Jamieson Lovell, Ben Hopkins, Mike Kay, Rebekah Hall, Anthony Tromans, Sarah McGuire, Elliot Harrison, Michelle Smith, Debbie Wright, Phil Howden, Nicholas Matthews, Heena Lad, Siobhan Whitehead, David Lloyd, Paul Heath, Giselle Kerry, Joanna Chapman, Andrew Ellington, and Sophie Williams. This work was supported by the Medical Research Council (MRC) U.K. Mouse Genome Sequencing Program, the Wellcome Trust, and the U.S. Department of Energy. I.M.S. was a Wellcome Trust Fellow. Collaboration between the MRC Human Genetics Unit and the Oak Ridge National Laboratories has been aided by North Atlantic Treaty Organization Collaborative Research Grant CRG910945.
Abbreviations
- BAC
bacterial artificial chromosome
- Mb
megabase.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequences reported in this paper have been deposited in the EMBL/GenBank database. For a complete list of accession nos., see Table 4, which is published as supporting information on the PNAS web site.
References
- 1.Bennett D. C., Huszar D., Laipis P. J., Jaenisch R., Jackson I. J. Development (Cambridge, U.K.) 1990;110:471–475. doi: 10.1242/dev.110.2.471. [DOI] [PubMed] [Google Scholar]
- 2.Davis A. P., Justice M. J. Genetics. 1998;148:7–12. doi: 10.1093/genetics/148.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelsey G., Schutz G. Curr. Opin. Genet. Dev. 1993;3:259–264. doi: 10.1016/0959-437x(93)90032-k. [DOI] [PubMed] [Google Scholar]
- 4.Peterson K. A., King B. L., Hagge-Greenberg A., Roix J. J., Bult C. J., O’Brien T. P. Genomics. 2002;80:172–184. doi: 10.1006/geno.2002.6818. [DOI] [PubMed] [Google Scholar]
- 5.Rinchik E. M., Bultman S. J., Horsthemke B., Lee S. T., Strunk K. M., Spritz R. A., Avidano K. M., Jong M. T., Nicholls R. D. Nature. 1993;361:72–76. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- 6.Rinchik E. M., Bell J. A., Hunsicker P. R., Friedman J. M., Jackson I. J., Russell L. B. Genetics. 1994;137:845–854. doi: 10.1093/genetics/137.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson E. H., Suffolk R., Bell J. A., Jordan S. A., Johnson D. K., Hunsicker P. R., Weber J. S., Justice M. J., Jackson I. J. Mamm. Genome. 2000;11:58–63. doi: 10.1007/s003350010011. [DOI] [PubMed] [Google Scholar]
- 8.Bell J. A., Rinchik E. M., Raymond S., Suffolk R., Jackson I. J. Mamm. Genome. 1995;6:389–395. doi: 10.1007/BF00355638. [DOI] [PubMed] [Google Scholar]
- 9.Javerzat S., Jackson I. J. Mamm. Genome. 1998;9:469–471. doi: 10.1007/s003359900798. [DOI] [PubMed] [Google Scholar]
- 10.Craig J. M., Bickmore W. A. Bioessays. 1993;15:349–354. doi: 10.1002/bies.950150510. [DOI] [PubMed] [Google Scholar]
- 11.Kleinjan D. A., van Heyningen V. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ovcharenko I., Loots G. G., Nobrega M. A., Hardison R. C., Miller W., Stubbs L. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinchik E. M. Genetics. 1994;137:855–865. doi: 10.1093/genetics/137.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z. Q., Imoto I., Fukuda Y., Pimkhaokham A., Shimada Y., Imamura M., Sugano S., Nakamura Y., Inazawa J. Cancer Res. 2000;60:4735–4739. [PubMed] [Google Scholar]
- 15.Chaudhry A. Z., Lyons G. E., Gronostajski R. M. Dev. Dyn. 1997;208:313–325. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 16.Grunder A., Ebel T. T., Mallo M., Schwarzkopf G., Shimizu T., Sippel A. E., Schrewe H. Mech. Dev. 2002;112:69–77. doi: 10.1016/s0925-4773(01)00640-2. [DOI] [PubMed] [Google Scholar]
- 17.Smyth I. M., Du X., Taylor M. S., Justice M. J., Beutler B., Jackson I. J. Proc. Natl. Acad. Sci. USA. 2004;101:13560–13565. doi: 10.1073/pnas.0402760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry R. W., Ma B., Sadowski C. L., Kobayashi R., Hernandez N. EMBO J. 1996;15:7129–7136. [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma P., Singh D. P., Fatma N., Chylack L. T., Jr., Shinohara T. Biochem. Biophys. Res. Commun. 2000;276:1320–1324. doi: 10.1006/bbrc.2000.3606. [DOI] [PubMed] [Google Scholar]
- 20.Wu X., Daniels T., Molinaro C., Lilly M. B., Casiano C. A. Cell Death Differ. 2002;9:915–925. doi: 10.1038/sj.cdd.4401063. [DOI] [PubMed] [Google Scholar]
- 21.Uetani N., Kato K., Ogura H., Mizuno K., Kawano K., Mikoshiba K., Yakura H., Asano M., Iwakura Y. EMBO J. 2000;19:2775–2785. doi: 10.1093/emboj/19.12.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunsicker P. R. Mouse News Lett. 1969;40:41. [Google Scholar]
- 23.Jackson I. J., Chambers D., Rinchik E. M., Bennett D. C. Genetics. 1990;126:451–459. doi: 10.1093/genetics/126.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanhoutteghem A., Djian P. Proc. Natl. Acad. Sci. USA. 2004;101:3468–3473. doi: 10.1073/pnas.0400268101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson I. J., Chambers D. M., Budd P. S., Johnson R. Nucleic Acids Res. 1991;19:3799–3804. doi: 10.1093/nar/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osoegawa K., Tateno M., Woon P. Y., Frengen E., Mammoser A. G., Catanese J. J., Hayashizaki Y., de Jong P. J. Genome Res. 2000;10:116–128. [PMC free article] [PubMed] [Google Scholar]
- 27.Marra M. A., Kucaba T. A., Dietrich N. L., Green E. D., Brownstein B., Wilson R. K., McDonald K. M., Hillier L. W., McPherson J. D., Waterston R. H. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallon A. M., Wilming L., Weekes J., Gilbert J. G., Ashurst J., Peyrefitte S., Matthews L., Cadman M., McKeone R., Sellick C. A., et al. Genome Res. 2004;14:1888–18901. doi: 10.1101/gr.2478604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birney E., Durbin R. Genome Res. 2000;10:547–548. doi: 10.1101/gr.10.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.