Abstract
Multiple mechanisms of tolerance induction limit autoimmunity, but their relative contribution for lymphocytes recognizing self-antigens of differing availability is incompletely understood. The mechanisms applied to arthritogenic B cells expressing antigen-specific B cell receptors (BCRs) with different affinities for glucose-6-phosphate isomerase (GPI) were examined in the corresponding Ig gene knock-in mice. This ubiquitously expressed and blood-borne enzyme is the target autoantigen in the K/BxN model of inflammatory arthritis and perhaps in some humans with arthritis. Negative selection of B cells expressing high-affinity anti-GPI specificities, whose surface receptors were occupied by GPI, operated mainly at the transitional B cell stages in the spleen, preventing their final differentiation and entry into follicular areas. Receptor editing contributed to the purging of cells displaying anti-GPI BCRs, and significant numbers of autoreactive cells escaped through expression of an additional Ig light (L) chain, accumulating gradually in lymphoid organs. In contrast, low-affinity anti-GPI B cells, whose surface receptors did not carry GPI, matured normally. The “escaped” dual-L-chain cells and the “ignored” low-affinity cells are the likely precursors of cells that produce pathogenic autoantibodies once T cell help is provided. These studies portray, in a single system, the range of tolerance mechanisms applied to potentially pathogenic B cells, and serve as a base for dissecting where T cell help intervenes and where therapeutic agents impinge.
Keywords: arthritis, autoimmunity, B cell tolerance
K/BxN mice spontaneously develop an autoimmune/inflammatory disease with many clinical and histopathological features in common with human arthritides (1). These animals carry a transgene encoding a self-reactive T cell receptor that recognizes cells presenting a peptide from glucose-6-phosphate isomerase (GPI), a glycolytic enzyme (2). Transgenic T cells are thereby activated, in turn providing help preferentially to B cells whose B cell receptor (BCR) recognizes GPI. K/BxN mice develop very high titers of antibodies (Abs) against GPI, predominantly of the IgG1 isotype (3, 4). Anti-GPI Abs directly mediate the joint pathology, and can induce arthritis in healthy recipients in the absence of T and B cells (3).
Given the central role of pathogenic Igs in K/BxN arthritis, B cells, and Abs from these mice have been characterized in some detail (4). A very high frequency of fusible Ab-producing cells with anti-GPI specificity was observed when hybridomas were generated with splenocytes from arthritic mice. All of these monoclonal antibodies (mAbs) had high affinity for GPI (Kd < 5 × 10−8 M), with V gene sequences suggesting that they had undergone extensive somatic hypermutation. Combinations of mAbs that recognized spaced epitopes on GPI were required for induction of arthritis.
GPI is normally cytoplasmic, but can be detected at low levels in serum (≈0.5–2 μg/ml) (5). It is not well understood how tolerance to a self-protein of this nature can be broken, either at the T or B cell levels. Multiple mechanisms of tolerance induction operate at the different stages of B cell differentiation. Clonal deletion, receptor editing, anergy, and helplessness have all been demonstrated, reflecting at least in part how and when particular B cells first encounter their antigens at different stages of differentiation, sites or concentrations, and in different forms (6–10). However, the relative contribution of these mechanisms, in particular which are used for a given autoantigen, is not well understood. Most relevant may be the hen egg lysozyme (HEL)/anti-HEL B cell tolerance model, where mice carrying a transgene encoding an Ab reactive with HEL are crossed to animals expressing a second transgene encoding HEL in a soluble form (6, 11).
Because GPI represents a true autoantigen with potential relevance to the pathogenesis of human arthritis, it seemed important to determine how B cell tolerance to GPI is achieved and maintained. Therefore, we generated knock-in mice in which the rearranged heavy (H) and L chain variable gene segments of an anti-GPI hybridoma were targeted into the corresponding germ-line J loci. The life histories of GPI-reactive cells expressing high affinity (H + L chain gene knock-ins) and low affinity (H chain gene only knock-in) were compared.
Results
Generation of Anti-GPI Ig Knock-In Mice.
From a panel of anti-GPI mAbs-derived from K/BxN mice (4), mAb 6.121 (hereafter called 121) was chosen for generation of Ig H- and L-chain knock-in mice. 121 is an IgG1 mAb that binds GPI with high affinity (Kd = 0.37 nM), and is arthritogenic when combined with other mAbs that recognize spaced epitopes on GPI (4). We first mutated the 121 H chain back to the putative germ-line sequence, but could no longer detect GPI binding by our assay when this H chain was combined with the 121 L chain (data not shown). Given this result and inherent uncertainties in guessing a corresponding germ-line sequence, we chose an alternative strategy for comparing arthritogenic BCRs of high and low affinity for GPI: the 121 H plus L chain combination versus the 121 H chain plus a diversity of endogenous L chains, some of which would permit GPI binding.
The 121 VHDHJH and VkJk gene segments were cloned from hybridoma cells, and used to generate targeting constructs for recombination into the JH or Jk locus (see Fig. 6, which is published as supporting information on the PNAS web site) of an embryonic stem cell line (PC3, 129/Sv background). The Neor gene, used to select homologous recombinant clones, was deleted during spermatogenesis in male chimera mice because of the protamine-promoter-driven Cre transgene in PC3 (12). The deletion of Neor in progeny was confirmed by PCR analysis. The targeted Ig H and L chain alleles were maintained in heterozygous state (designated H121/+ and L121/+, respectively, where 121 denotes the knock-in allele, and + is the wild-type allele). Both targeted alleles were expressed quite efficiently: 72–86% of the B cells in the bone marrow of H121/+L121/+ mice expressed the IgMa allotype of the knock-in H chain (not shown), and 82–95% of the IgMa+ immature B cells in the bone marrow of H121/+L121/+ mice expressed the knock-in L chain allele (see below).
Anti-GPI B Cells in 121 Knock-In Mice and Their Receptor Occupancy.
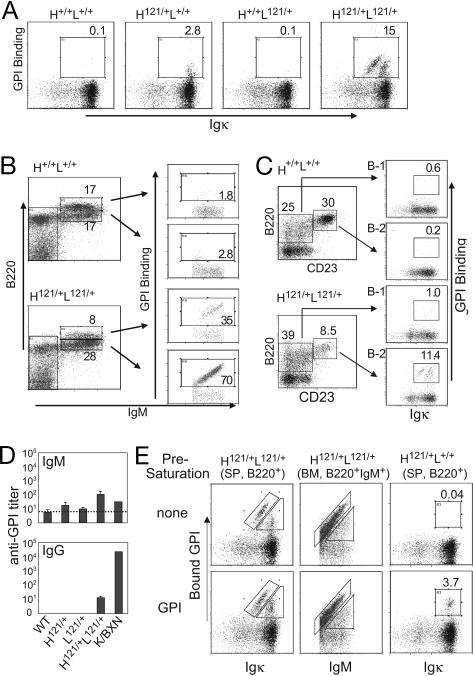
The expression of GPI-reactive receptors on B cells was detected by using biotinylated recombinant GPI protein (referred to as GPI-binding) or with a secondary anti-GPI Ab compatible with 121 binding (referred to as bound GPI) (4). In 8- to 12-week-old H121/+L121/+ mice, 15–25% of B cells were GPI-reactive (Fig. 1A Right). The GPI-binding cells formed two distinct populations based on expression of κ L chain and on GPI staining. One population showed a proportional intensity of κ display and GPI staining (i.e., fell on a diagonal), whereas the other had lower GPI staining (0.5-fold on average) and higher κ expression (2.8-fold); these populations are explored in detail below. In H121/+L+/+ mice, 1 to 3% of B cells bound GPI at low intensity (Fig. 1A Middle Left). H+/+L121/+ and H+/+L+/+ animals had only background numbers of GPI-stainable B cells. The bone marrow of H121/+L121/+ mice showed an increased proportion of immature (B220lo) B cells and a decreased fraction of mature (B220hi) B cells vis-a-vis H+/+L+/+ controls (Fig. 1B). The vast majority of the immature B cells in H121/+L121/+ mice were GPI-reactive, but this proportion dropped in the more mature B cells (which mainly represent recirculating peripheral B cells) (Fig. 1B Bottom Right). Several studies have demonstrated that self-reactive B cells can accumulate preferentially in the peritoneal cavity as B-1 cells (13). This was not the case here, as hardly any of the B220loCD23− cells bound GPI (Fig. 1C).
Fig. 1.
Anti-GPI B cells in 121 knock-in mice. (A) B220+ splenocytes from adult knock-in mice (H121/+L121/+) and control littermates stained with anti-Igκ and GPI-biotin. (B) Bone marrow B cells of adult knock-in (H121/+L121/+) and wild-type (H+/+L+/+) mice, stained with anti-B220, anti-IgM, and GPI-biotin, and gated on lymphocyte population. (C) Igκ and GPI profiles for B-1 (B220loCD23−) and B-2 (B220hiCD23+) lineages of the peritoneal cavity lavage cells. (D) ELISA titers of anti-GPI IgM or IgG antibodies in the serum of H121/+L121/+ and control littermates (n = 3–5). Titer is defined here as the serum dilution that gave an optical density of 2× background. A pooled serum from K/BxN arthritic mice was included as a reference. (E) The extent of occupancy by self-antigen of anti-GPI surface receptors was analyzed by staining cells directly (Upper), or after preincubation with saturating amounts of GPI (100 μg/ml; Lower). Cytometry profiles representative of 3–10 independent mice.
H121/+L121/+ animals had only 3–10 times higher anti-GPI IgM titers than did single-chain littermates, whose titers were similar to those of wild-type littermates (Fig. 1D Upper). With respect to IgG isotypes (Fig. 1D Lower), only H121/+L121/+ mice had detectable anti-GPI titers, at levels considerably lower than those of K/BxN transgenics. These low titers suggest that tolerance was operating and that anti-GPI B cells were not activated by cognate autoantigen.
To measure the extent of BCR occupancy by GPI, we used a secondary anti-GPI mAb (or polyclonal Abs) known not to interfere with binding of the 6.121 mAb, with or without preincubating the cells with saturating amounts of soluble recombinant GPI (Fig. 1E). Splenic B cells from H121/+L121/+ mice (Fig. 1E Left) displayed essentially the same intensity of GPI staining with or without presaturation of the surface BCRs by GPI, suggesting that most, if not all, surface Igs were occupied by endogenous GPI. This observation was true of GPI-binding cells in either of the two GPI-positive populations. By the same criteria, immature B cells in the bone marrow (Fig. 1E Center) also showed high receptor occupancy, confirming that anti-GPI B cells first encountered cognate antigen in the bone marrow. In contrast, detection of anti-GPI B cells in H-chain-only H121/+L+/+ mice was completely dependent on preincubation with GPI, indicating that the H121-bearing receptors were most likely empty in these animals (Fig. 1E Right). We estimated by Scatchard analysis that the average Kd was on the order of 150 nM for GPI-reactive B cells of the H121/+L+/+ mice (data not shown), which is much less than the subnanomolar affinity of the full 121 BCR.
Allelic Inclusion and Receptor Editing in Anti-GPI B Cells.
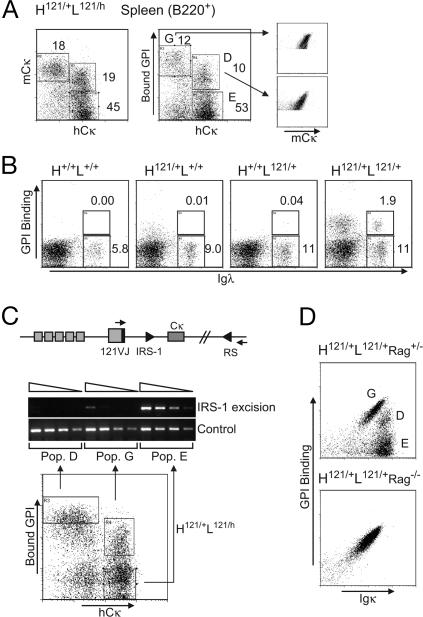
Considering the two distinct populations of GPI-binding B cells in H121/+L121/+ splenocytes (Fig. 1A), we hypothesized that the one showing lower binding to GPI relative to its Cκ levels might have resulted from the expression of additional Ig L chains, originating from rearranged endogenous Ig loci. To test for such events at the L chain locus, we crossed H121/+L121/+ animals to a mouse line in which the Ig L chain gene Cκ segment (mCκ) was replaced by its human ortholog (hCκ) (14), thereby generating H121/+L121/h mice. Rearrangement and expression from the 121 and hCκ loci were assessed by staining cells with Abs specific for mCκ or hCκ. Two populations of GPI-binding B cells were found in H121/+L121/h mice (Fig. 2A). One stained most brightly with GPI and anti-mCκ, and did not express hCκ (referred to as population “G” hereafter). The second population bound GPI less intensely, and was positive for hCκ, indicating that the cells were dual-expressors (population “D”), pointing to allelic inclusion for L chains in 121 knock-in mice. This conclusion was confirmed by testing for expression of the Igλ locus in H121/+L121/+ (Fig. 2B) or H121/+L121/h mice (data not shown). A distinct population of B cells bound GPI and expressed the λ chain (λ1, λ2, λ3); such cells were absent in all other mice. Thus, the allelic inclusion associated with the combination of 121 knock-ins can involve both the κ and λ loci. The lower frequency of λ/κ, compared with κ/κ, dual-expressors is consistent with the lower efficiency of λ, compared with κ, rearrangement. By extension, we interpret similarly the two GPI-binding populations shown in Fig. 1A Right: the off-diagonal population with stronger mCk but duller GPI staining should be composed of cells that use both the knock-in and the endogenous wild-type κ loci.
Fig. 2.
Receptor editing at the light chain locus. (A) Allelic inclusion revealed by the expression of the hCκ chain in B220+ splenocytes from H121/+L121/h mice; populations “G,” “D,” and “E” (abbreviations stand for “GPI only,” “Dual,” and “Eliminated”) are defined in relation to their GPI-binding capacity (Middle). (B) Isotypic inclusion and the usage of λ L chains in B220+ splenocytes of adult H121/+L121/+ mice and control littermates. (C) RS-IRS-1 excision (a 1.1-kb band) was determined with 3-fold serial dilutions of genomic DNA from sorted cells of the populations “G,” “D,” and “E” (starting with amount equivalent to 5,000 cells) using flanking PCR primers (small arrowheads). (D) B220+ splenocytes of H121/+L121/+Rag+/− and H121/+L121/+Rag−/− mice. Representative of three to five mice.
In addition, the B cell compartment of H121/+L121/h mice contained a third population, which displayed only hCκ and did not bind GPI (population “E” in Fig. 2A), likely equivalent to the GPI-negative B cells of Fig. 1A Right. These cells may represent those in which the 121 L chain was never expressed, or was expressed but secondarily altered by receptor editing. In the 121 L chain gene knock-in construct, there were no additional J segments downstream of the rearranged VJ region, thus preventing a secondary rearrangement between the upstream V and downstream J segments. Inactivation of 121VJ by recombining sequence (RS) recombination (15) was assessed by semiquantitative PCR of genomic DNA prepared from the sorted populations “G,” “D,” and “E”. As shown in Fig. 2C, RS-IRS-1 excision was easily detectable in population “E” but not in “G” and only weakly in “D,” confirming that population “E” resulted from the inactivation of the L121 knock-in locus by RS recombination.
To solidify these conclusions, we crossed the RAG1 knock-out mutation into H121/+L121/+ line, preventing any alternative or secondary rearrangements. As shown in Fig. 2D, H121/+L121/+Rag−/− mice contained only population “G,” and all B cells bound GPI in proportion to the amount of surface Ig they displayed.
The analyses of the H chain locus, by staining for the IgMa and IgMb allotypes carried by the knock-in and wild-type alleles, showed that H chain allelic inclusion did not contribute significantly to the generation of GPIdull cells (Fig. 7, which is published as supporting information on the PNAS web site).
Evolution of GPI-Binding Populations over Time.
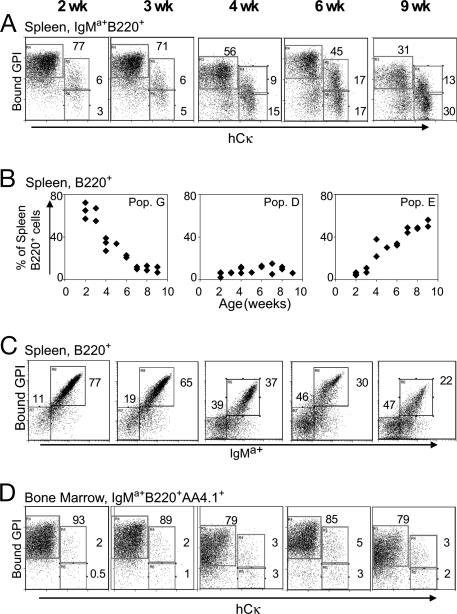
The different abundance of GPI-binding B cells in the bone marrow versus spleen of H121/+L121/+ mice, as well as the editing events at the L chain loci, suggested that GPI-binding cells were under negative pressure in the secondary lymphoid organs. To determine when this negative selection took place, we examined H121/+L121/h mice of different ages (Fig. 3). Over time, population “G” decreased, whereas “E” increased, and “D” increased only slightly (Fig. 3A). This change was most dramatic when gated on total B220+ cells (Fig. 3B), due to a progressive increase in IgMa-negative cells in the spleen (Fig. 3C). In contrast, the distribution among immature B cells in the bone marrow remained about the same through this age window, most cells keeping a high GPI-binding ability (Fig. 3D). These results suggest that the edited populations accumulated preferentially in the peripheral organs of H121/+L121/h mice, outcompeting the cells with high affinity for GPI. However, one cannot tell whether the actual editing occurred in the bone marrow or in the peripheral organs. No age-dependent changes in numbers of the low-affinity B cells of H121/+L+/+ mice were observed (data not shown).
Fig. 3.
Age-dependent shift in anti-GPI populations of H121/+L121/h mice. L chain allelic inclusion in relation to GPI-binding capacity in B220+ IgMa+ splenocytes (A) and immature B cells in bone marrow expressing the knock-in H chain (gated as IgMa+B220+AA4.1+) (D). (B) The proportion of populations “G,” “D,” and “E” among total B220+ splenocytes. Each diamond represents an individual mouse. (C) H chain usage in total B220+ splenocytes.
High-Affinity GPI-Binding B Cells Arrested in Maturation in the Peripheral Lymphoid Organs.
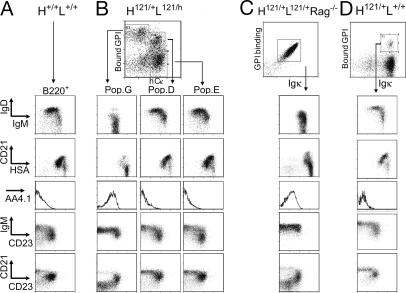
Since the time-period during which the changes of different populations were greatest corresponded to that when B cells undergo full maturation in normal mice (16), the maturation state of the three B cell populations in spleens of H121/+L121/h mice was evaluated by immunophenotyping with a panel of differentiation and activation markers. For comparison, the homogenous GPI-binding B cells of H121/+L121/+Rag−/− mice and the low-affinity GPI-binding cells in the H121/+L+/+ mice were also analyzed (Fig. 4). The cells of population “G” had a profile characteristic of immature B cells: little IgD but a high level of IgM, albeit subtly lower than on immature B cells from the wild-type H+/+L+/+ mice. The same down-regulation was true for Igκ staining (Fig. 1A). They displayed little or no CD21 or CD62L (data not shown), showed high levels of heat shock antigen (HSA) and were AA4.1+ (Fig. 4B), all characteristic of immature B cells of the T1 subpopulation as defined in ref. 17, or of the T1/T2 subpopulation defined in ref. 18. CD21/CD23 plots also distinguished population “G” cells from conventional follicular and marginal zone B cells (typically CD21loCD23+ and CD21hiCD23−, respectively) confirming that they are immature cells. This immature profile was also characteristic of the homogenous GPI-binding cells in H121/+L121/+Rag−/− mice (Fig. 4C). “G” cells also had slightly lower expression of MHC class II molecules and CD19, but normal levels of CD44, CD25, CD69, CD80, and CD86 (data not shown). In contrast, populations “D” and “E” had very different phenotypes, comparable in all respects with that of mature B cells in wild-type mice (Fig. 4 A and B). Therefore, the expression of an endogenous L chain somehow attenuated the impact of the autoreactive BCR and rescued anti-GPI B cells from their maturation arrest, as did the complete elimination of the autoreactive receptor. “D” cells were found both in the marginal zones and follicles by CD21/CD23 staining (Fig. 4B). In the 3H9 system, κ/λ dual-expresssors preferentially localized to the marginal zones (19). This difference might be due to the nature of the antigen involved.
Fig. 4.
Maturation status of different GPI-binding populations. B220+ splenocytes from control H+/+L+/+ mice (A); H121/+L121/h mice, split into populations G, D, and E as defined in Fig. 2 (B); H121/+L121/+Rag−/− (C) or H121/+L+/+ (D) mice. Representative of five independent experiments.
Interestingly, the GPI-binding B cells in H121/+L+/+ mice were not blocked at the immature stage, and had a mature phenotype (Fig. 4D). Most likely, the reduced affinity of their BCR for GPI, and the resulting low occupancy, allowed them to bypass the block in differentiation.
To understand the population kinetics of the different B cells of the H121/+L121/h mice, we compared their turnover rate by BrdUrd labeling in vivo. Continuous labeling for different periods via BrdUrd in the drinking water revealed that population “G” had a much faster turnover rate than populations “D” and “E” (Fig. 8, which is published as supporting information on the PNAS web site). Analyses of the turnover rate of the homogeneous immature B cells in H121/+L121/+Rag−/− mice (and TCRα−/− mice, which are also devoid of T cells, used as controls) showed again a high turnover rate (Fig. 8D). The faster turnover rate of population “G” was thus intrinsic to the immature phenotype of the cells and/or their fully occupied receptors, and does not result from competition with mature cells. This conclusion contrasts with that made in the HEL system (20).
GPI-Binding B Cells Are Excluded from B Cell Follicles.
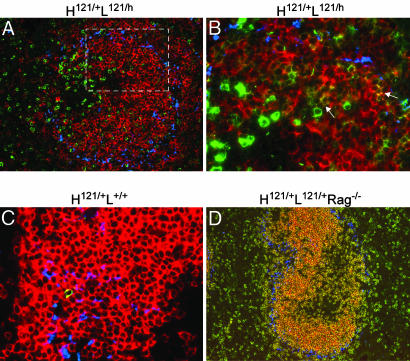
To examine the anatomical localization of anti-GPI B cells, we carried out immunofluorescence analysis of spleen sections of various mice; the B cell populations defined above were identified by staining with labeled GPI and anti-hCκ mAb: population “G” stained with GPI alone, “D” was identified by the colocalization of GPI and hCκ staining, and “E” was identified by hCκ staining alone. Most GPIbright cells of population “G” were found to be excluded from B cell follicles, residing in the T cell zone of the splenic white pulp, yet concentrated on the T–B cell border area (Fig. 5A). The GPI-negative cells of population “E” were essentially all found in the follicles, as were the dual-labeled cells of population “D” (Fig. 5B), consistent with their phenotypes. The rare GPI+ B cells of H121/+L+/+ mice were also found in B cell follicles (Fig. 5C). In H121/+L121/+ Rag−/− mice, in contrast, most GPIbright B cells did localize to the B cell follicles (Fig. 5D). This finding indicates that exclusion of immature GPI-binding B cells from the follicles does represent a competitive event, as had been described in the HEL system (20).
Fig. 5.
Localization of GPI-binding B cells in spleen. Spleen sections from various mice were stained with FITC-GPI (shown in green) and Moma-1 antibody (shown in blue to mark the boundary of the marginal zone). (A) Spleen sections costained with hCκ (shown in red) (×16 objective). (B) Magnification of the area marked by the white rectangle in A (×40 objective). GPI-binding cells appear in green, and allelicly included cells coexpressing the alternate hCκ chain appear in yellow (arrows). (C and D) Spleen sections costained with mCκ (shown in red). Representative of sections from three or more animals at 8–12 weeks of age.
Response Capacity of GPI-Binding B Cells.
To determine whether anti-GPI B cells were anergic, we tested their proliferative responses to different stimuli in vitro. Because sorting populations “G,” “D,” and “E” would have required engaging their BCRs and attempts to distinguish proliferating cells by label dilution also proved unsuccessful, we focused on the homogeneous cells from H121/+L121/+Rag−/− mice that have the same immature phenotype as the GPI-binding B cells of population “G” in H121/+L121/h animals. Responses to BCR and LPS were reduced only 3- to 5-fold for GPI-binding B cells versus controls (Fig. 9 A and B, which is published as supporting information on the PNAS web site). On the other hand, stimulation by anti-CD40 mAb was more profoundly affected (10-fold on average in several experiments) (Fig. 9C), but this unresponsiveness was partially relieved in the presence of IL-4 (Fig. 9D).
Discussion
A full understanding of the pathogenesis of arthritis needs to encompass both the local inflammatory events in the joint, largely mediated by players of the innate immune system, and the break in self-tolerance during the adaptive autoimmune phase that lies upstream. There has been a recent resurgence of interest in interpretations of rheumatoid arthritis (RA) pathogenesis that portray pathogenic B cells and autoantibodies as playing a central role in generating joint-specific inflammation. Arguments in favor of this notion have come from mechanistic insights from animal models (3, 21), from the prevalence and specificity of anti-citrullin autoantibodies in human RA along with the demonstration that anti-citrullin Abs can be pathogenic (22), and from the impact of therapies directed at B lymphocytes (23). Thus, it is important to understand how arthritogenic autoantibodies are normally controlled, but are unleashed in arthritic mice or humans.
We tackled this question by generating Ig knock-in mice expressing the rearranged Ig H and L chain genes of an arthritogenic mAb derived from K/BxN mice. This approach allowed us to follow the life history of cells expressing two types of receptors: a BCR composed of both chains from the 121 mAb, with high affinity for the soluble autoantigen, and a high degree of BCR occupancy by antigen from the earliest stages in the bone marrow; or lower affinity BCRs, generated by pairing of endogenously encoded L chains with the knock-in H chain, receptors that remain largely unoccupied by GPI.
Cells displaying the high-affinity BCR did emerge in the repertoire of splenic B cells, but a variety of roadblocks prevented them from reaching the fully mature B cell pool. These blocks entailed several mechanisms observed in other models of B cell tolerance (6, 8–11, 24–26). The gene encoding the high-affinity anti-GPI BCR could be edited out by RS recombination, edited down by allelic inclusion, sequestered in blocked immature B cells where it is poorly responsive, or excluded from normal follicular areas. Ultimately, the amount of IgG made by the potentially monoclonal B cell repertoire was orders of magnitude lower than that elicited by transgenic KRN T cells in association with polyclonal B cells.
Although anergy is an oft-invoked form of B cell tolerance to soluble antigen (6, 24–26), our results suggest that receptor editing also plays an important role, as was recently shown for HEL L-chain knock-in mice (27). RS recombination was used to inactivate the 121 light chain. In 121 mice, the generation of cells that did not express the 121 L chain or the anti-GPI cells that expressed two L chains probably depended on the relative efficiencies of RS recombination of the knock-in allele and the productive secondary rearrangements of the endogenous allele. We have not formally proven that the secondary rearrangements observed were dependent on GPI (a practical impossibility because the absence of GPI is lethal in very early embryogenesis; ref. 28), or that expression of the endogenous allele occurred simply because of the inefficiency of the knock-in L chain gene to exclude rearrangement on the other allele. Yet, the demonstration of RS excision in population “E” by PCR of genomic DNA suggests that it is an active process. The increased percentage of λ+ cells and the generation of λ/κ cells (Fig. 2B) also support this view. The receptor occupancy analysis confirmed that immature GPI-binding B cells first encountered antigen in the bone marrow. Although we did not formally establish that no editing took place in the peripheral lymphoid organs, the kinetics of the system were consistent with a regular output of edited B cells from the bone marrow, which accumulated in the secondary lymphoid organs because they had a competitive advantage in maturation and survival.
An obvious question is how the expression of an additional L or H chain in the dual-expressors masks their autoreactive nature, enabling them to bypass the maturational arrest. The Weigert laboratory (8, 19, 29) demonstrated that autoreactive B cells in the 3H9 anti-DNA models could escape tolerance induction by expressing two L chains. In those contexts, the expressed second L chain could decrease the density of the original receptor by competing for the potentially autoreactive H chain and/or by covering the cell surface with a non-autoreactive BCR. Comparison of mean fluorescence intensities of anti-BCR staining on the B cells of H121/+L121/+ mice showed that cells of population “D” had 3-fold higher total Igκ but half the labeled GPI staining (Fig. 1A), suggesting that GPI-reactive receptors constituted only 1/6 of the total surface receptors on average. The degree of BCR cross-linking by antigen may be further reduced by the formation of chimeric BCR molecules (one arm anti-GPI and the other not, turning the divalent Ig into a monovalent one in terms of GPI binding). An alternative interpretation is that a positive signal provided by the non-autoreactive BCR can override the negative signal from the autoreactive BCR.
It is important to understand the effect of autoreactive receptor affinity on the mechanisms of tolerance induction. It has been suggested that affinity does not play an important role for deletion or receptor editing when membrane-bound autoantigens are concerned (30, 31). However, our results indicate that induction of tolerance to a soluble autoantigen can be highly dependent on receptor affinity. The low-affinity GPI-binding cells in H121/+ mice appeared to mature and colonize lymphoid organs quite normally. This behavior is probably linked to their low level of receptor occupancy. It is worth keeping in mind that when the antigen concentration was low, even the high-affinity anti-HEL B cells showed ignorance (32). By analogy, one might propose that it is not the intrinsic affinity of the repeated engagements of the BCR by autoantigen that matters, but rather the overall level of persistent BCR occupancy, as shown recently (33).
The multiple roadblocks to the differentiation of a fully effective autoreactive B cell still permitted the appearance of potentially dangerous cells. There are potentially three scenarios to explain how T cell help can elicit autoantibody production from these cells. First, the monoreceptor high-affinity cells might become activated. Although immature and less responsive to BCR or toll-like receptor triggers, this population responded robustly to coupled stimulation via CD40 and the IL-4 receptor. Second, the dual expressor cells might be activated. These cells mature normally and should be reactive to stimulation, either through GPI or through their alternative receptor. Finally, low-affinity B cells, as exemplified by the GPI-binding cells of H121/+L+/+ single knock-in mice, are also candidate precursors of fully pathogenic B cells. The combinatorial arrangement of the 121 H with endogenous L chains mainly generates low-affinity BCRs on cells that mature unhampered; these receptors would form an excellent starting point for affinity maturation. Which of these explanations proves right, and how T cells eventually subvert the state of partial tolerance in GPI-reactive B cells, are essential issues to resolve.
Materials and Methods
Mice.
The targeting constructs containing 6.121 VDJ and VJ were transfected into the 129/Sv-derived ES cell line PC3. Verified homologous recombinant clones were injected into C57BL/6 (B6)-derived blastocysts, and the resulting chimeric mice were bred to B6 mice for germline transmission (see Supporting Text, which is published as supporting information on the PNAS web site, for details). Experimental mice are on a mixed B6/129 background or partially backcrossed to B6.
Expression and Purification of Recombinant GPI.
The coding region of the mouse GPI cDNA (2) was cloned into a pET21a vector (Novagen) and the resulting GPI-histidine fusion protein was expressed and purified from Escherichia coli BL21 cells by a nickel column (Qiagen) following the manufacturer’s instructions.
Immunohistochemistry.
For detection of GPI-specific B cells, cryosections of spleen were stained with FITC-labeled GPI, and the signal was amplified sequentially with an Alexa Fluor 488 rabbit anti-fluorescein and an Alexa Fluor 488 goat anti-rabbit IgG reagent (Molecular Probes/Invitrogen).
Flow Cytometric Analyses.
Biotin-labeled GPI was used at 5–10 μg/ml to stain anti-GPI B cells. See Supporting Text for details.
In Vitro Proliferation of Cultured B Cells.
Cells were cultured in 96-well plates (3 × 105 cell per well) with or without various stimuli. F(ab′)2 fragment of goat anti-mouse IgM (Jackson ImmunoResearch), LPS (Sigma), anti-CD40 (clone 1C10; eBioscience), and IL-4 (recombinant; Becton Dickinson) were used at the indicated concentrations. After culturing for 48–60 h, 1 μCi of [3H]thymidine was added to culture (1 Ci = 37 GBq). Cells were harvested 12–18 h later for scintillation counting.
BrdUrd Labeling and Detection.
See Supporting Text for details.
Supplementary Material
Acknowledgments
We thank Drs. K. Rajewsky (CBR Institute for Biomedical Research, Boston), D. Allman (University of Pennsylvania, Philadelphia), and M. C. Nussenzweig (The Rockefeller University, New York) for different reagents; Q. M. Pham and V. Tran for help with mice; and Drs. K. Rajewsky, M. C. Carroll, T. I. Novobrantseva, and M. Weigert for advice and critical reading of the manuscript. This work was supported by National Institutes of Health Grants R01 NIH-AR046580-06 (to D.M. and C.B.) and R01 AI14782 (to J.F.K.), as well as by the Joslin National Institute of Diabetes and Digestive and Kidney Diseases-supported Diabetes and Endocrinology Research Center cores. H.H. was supported by Damon Runyon Cancer Research Foundation Grant DRG-1616.
Abbreviations
- GPI
glucose-6-phosphate isomerase
- BCR
B cell receptor
- HEL
hen egg lysozyme
- H
heavy
- L
light.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Kouskoff V., Korganow A.-S., Duchatelle V., Degott C., Benoist C., Mathis D. Cell. 1996;87:811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- 2.Matsumoto I., Staub A., Benoist C., Mathis D. Science. 1999;286:1732–1735. doi: 10.1126/science.286.5445.1732. [DOI] [PubMed] [Google Scholar]
- 3.Korganow A.-S., Ji H., Mangialaio S., Duchatelle V., Pelanda R., Martin T., Degott C., Kikutani H., Rajewsky K., Pasquali J.-L., et al. Immunity. 1999;10:451–461. doi: 10.1016/s1074-7613(00)80045-x. [DOI] [PubMed] [Google Scholar]
- 4.Maccioni M., Zeder-Lutz G., Huang H., Ebel C., Gerber P., Hergueux J., Marchal P., Duchatelle V., Degott C., van Regenmortel M., et al. J. Exp. Med. 2002;195:1071–1077. doi: 10.1084/jem.20011941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsumoto I., Maccioni M., Lee D. M., Maurice M., Simmons B., Brenner M., Mathis D., Benoist C. Nat. Immunol. 2002;3:360–365. doi: 10.1038/ni772. [DOI] [PubMed] [Google Scholar]
- 6.Goodnow C. C., Crosbie J., Adelstein S., Lavoie T. B., Smith-Gill S. J., Brink R. A., Pritchard-Briscoe H., Wotherspoon J. S., Loblay R. H., Raphael K., et al. Nature. 1988;334:676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 7.Nemazee D. A., Burki K. Nature. 1989;337:562–566. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- 8.Gay D., Saunders T., Camper S., Weigert M. J. Exp. Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radic M. Z., Erikson J., Litwin S., Weigert M. J. Exp. Med. 1993;177:1165–1173. doi: 10.1084/jem.177.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiegs S. L., Russell D. M., Nemazee D. J. Exp. Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phan T. G., Amesbury M., Gardam S., Crosbie J., Hasbold J., Hodgkin P. D., Basten A., Brink R. J. Exp. Med. 2003;197:845–860. doi: 10.1084/jem.20022144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Gorman S., Dagenais N. A., Qian M., Marchuk Y. Proc. Natl. Acad. Sci. USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H., Clarke S. H. Curr. Opin. Immunol. 2004;16:246–250. doi: 10.1016/j.coi.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Casellas R., Shih T. A., Kleinewietfeld M., Rakonjac J., Nemazee D., Rajewsky K., Nussenzweig M. C. Science. 2001;291:1541–1544. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]
- 15.Moore M. W., Durdik J., Persiani D. M., Selsing E. Proc. Natl. Acad. Sci. USA. 1985;82:6211–6215. doi: 10.1073/pnas.82.18.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allman D. M., Ferguson S. E., Lentz V. M., Cancro M. P. J. Immunol. 1993;151:4431–4444. [PubMed] [Google Scholar]
- 17.Loder F., Mutschler B., Ray R. J., Paige C. J., Sideras P., Torres R., Lamers M. C., Carsetti R. J. Exp. Med. 1999;190:75–89. doi: 10.1084/jem.190.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allman D., Lindsley R. C., DeMuth W., Rudd K., Shinton S. A., Hardy R. R. J. Immunol. 2001;167:6834–6840. doi: 10.4049/jimmunol.167.12.6834. [DOI] [PubMed] [Google Scholar]
- 19.Li Y., Li H., Weigert M. J. Exp. Med. 2002;195:181–188. doi: 10.1084/jem.20011453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cyster J. G., Goodnow C. C. Immunity. 1995;3:691–701. doi: 10.1016/1074-7613(95)90059-4. [DOI] [PubMed] [Google Scholar]
- 21.Stuart J. M., Dixon F. J. J. Exp. Med. 1983;158:378–392. doi: 10.1084/jem.158.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill J., Cairns E., Bell D. A. J. Rheumatol. 2004;31:1471–1473. [PubMed] [Google Scholar]
- 23.Edwards J. C., Leandro M. J., Cambridge G. Biochem. Soc. Trans. 2002;30:824–828. doi: 10.1042/bst0300824. [DOI] [PubMed] [Google Scholar]
- 24.Mandik-Nayak L., Bui A., Noorchashm H., Eaton A., Erikson J. J. Exp. Med. 1997;186:1257–1267. doi: 10.1084/jem.186.8.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santulli-Marotto S., Retter M. W., Gee R., Mamula M. J., Clarke S. H. Immunity. 1998;8:209–219. doi: 10.1016/s1074-7613(00)80473-2. [DOI] [PubMed] [Google Scholar]
- 26.Rojas M., Hulbert C., Thomas J. W. J. Immunol. 2001;166:3194–3200. doi: 10.4049/jimmunol.166.5.3194. [DOI] [PubMed] [Google Scholar]
- 27.Hippen K. L., Schram B. R., Tze L. E., Pape K. A., Jenkins M. K., Behrens T. W. J. Immunol. 2005;175:909–916. doi: 10.4049/jimmunol.175.2.909. [DOI] [PubMed] [Google Scholar]
- 28.West J. D., Flockhart J. H., Peters J., Ball S. T. Genet. Res. 1990;56:223–236. doi: 10.1017/s0016672300035321. [DOI] [PubMed] [Google Scholar]
- 29.Prak E. L., Weigert M. J. Exp. Med. 1995;182:541–548. doi: 10.1084/jem.182.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartley S. B., Goodnow C. C. Int. Immunol. 1994;6:1417–1425. doi: 10.1093/intimm/6.9.1417. [DOI] [PubMed] [Google Scholar]
- 31.Lang J., Jackson M., Teyton L., Brunmark A., Kane K., Nemazee D. J. Exp. Med. 1996;184:1685–1697. doi: 10.1084/jem.184.5.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adelstein S., Pritchard-Briscoe H., Anderson T. A., Crosbie J., Gammon G., Loblay R. H., Basten A., Goodnow C. C. Science. 1991;251:1223–1225. doi: 10.1126/science.1900950. [DOI] [PubMed] [Google Scholar]
- 33.Gauld S. B., Benschop R. J., Merrell K. T., Cambier J. C. Nat. Immunol. 2005;6:1160–1167. doi: 10.1038/ni1256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.