Abstract
Wnt and Notch signaling have long been established as strongly oncogenic in the mouse mammary gland. Aberrant expression of several Wnts and other components of this pathway in human breast carcinomas has been reported, but evidence for a causative role in the human disease has been missing. Here we report that increased Wnt signaling, as achieved by ectopic expression of Wnt-1, triggers the DNA damage response (DDR) and an ensuing cascade of events resulting in tumorigenic conversion of primary human mammary epithelial cells. Wnt-1-transformed cells have high telomerase activity and compromised p53 and Rb function, grow as spheres in suspension, and in mice form tumors that closely resemble medullary carcinomas of the breast. Notch signaling is up-regulated through a mechanism involving increased expression of the Notch ligands Dll1, Dll3, and Dll4 and is required for expression of the tumorigenic phenotype. Increased Notch signaling in primary human mammary epithelial cells is sufficient to reproduce some aspects of Wnt-induced transformation. The relevance of these findings for human breast cancer is supported by the fact that expression of Wnt-1 and Wnt-4 and of established Wnt target genes, such as Axin-2 and Lef-1, as well as the Notch ligands, such as Dll3 and Dll4, is up-regulated in human breast carcinomas.
Keywords: breast cancer, DNA damage response, medullary carcinoma
Breast cancer is a complex malignancy comprising 18 distinct histopathological entities (1) that are thought to arise through a series of mutations (2). Although a large number of genes have been found mutated or misexpressed in breast carcinomas, it is unclear to what extent these changes contribute to tumorigenesis. The problem is linked to the fact that the earliest identifiable lesion, the carcinoma in situ, already contains most genetic changes and is difficult to study.
Transgenic mouse models are used extensively to model human breast cancer (3). Species-specific properties do not allow one to directly extrapolate findings in mice to humans; in fact, growth control pathways are wired differently in the two species (4). Moreover, genes frequently activated in mouse mammary carcinomas because of insertion of the mouse mammary tumor virus, such as Wnt-1, Notch1, and Notch4 (5), are distinct from the most common targets of mutation in human breast cancers (6). However, recent evidence suggests that Wnt or Notch signaling may also be deregulated in human breast cancer. Thus, expression of different Wnts is increased (7); an extracellular inhibitor of Wnt signaling, secreted Frizzled-related protein 1, is down-regulated in 80% of breast carcinomas (8, 9); and the positive regulator Disheveled is up-regulated (10). Similarly, a tumor suppressor function was suggested for the Notch inhibitor Numb (11).
Wnt signaling is initiated by the interaction of a Wnt ligand with a seven-transmembrane-domain Frizzled receptor and leads to stabilization of β̃-catenin, one of the central components of the pathway, by inhibiting glycogen-synthase kinase-3β. In the absence of Wnt signaling, β̃-catenin is targeted for degradation by phosphorylation through a complex of glycogen-synthase kinase-3β, adenomatous polyposis colon protein, protein phosphatase 2A, and Axin-1 (12). Stabilized β̃-catenin translocates to the nucleus, where it forms a bipartite complex with transcription factors of the T cell factor family and activates target gene expression (13). Notch signaling is triggered by the binding of one of five different membrane-bound ligands, Delta (Dll) 1, Dll3, Dll4, Jagged1, and Jagged2, to one of four Notch proteins on a neighboring cell. The interaction leads to two proteolytic cleavages of the receptor, resulting in the release of the Notch intracellular domain, which translocates to the nucleus, binds to a highly conserved DNA-binding transcription factor of the CSL family (RBP-Jk/CBF1 in mammals), and recruits coactivators to form a transcriptional activation complex (14).
Here we find that increased Wnt signaling in human mammary epithelial cells (HMECs), as achieved by ectopic Wnt-1 expression, elicits a DNA damage response (DDR), which has recently been shown to be an early event in human carcinogenesis (15, 16), followed by a cascade of events including Notch activation, resulting in transformation to a tumorigenic state. These findings suggest that deregulation of Wnt signaling may be an early event in mammary epithelial transformation.
Results
Effects of Wnt-1 Expression on Primary HMECs.
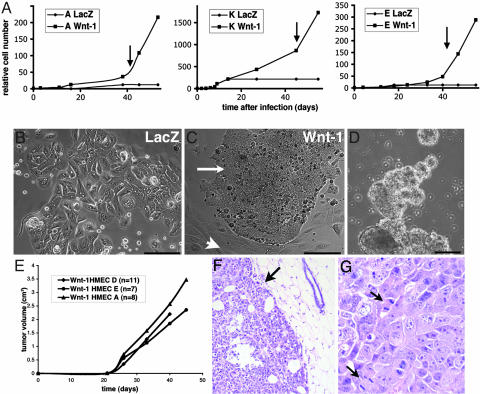
Wnt-1 was originally cloned as a frequent integration site for the mouse mammary tumor virus (17) and is a strong oncogene in the mouse mammary epithelium (18). It is functionally equivalent to other Wnts (19–21), some of which are overexpressed in human breast carcinomas (7). To assess potential effects of increased Wnt signaling in HMECs, we ectopically expressed Wnt-1 in primary HMECs. Specifically, we infected HMECs 10 days after they were derived from reduction mammoplasties, at passage 2 (P2) or P3, with high-titer retroviruses expressing full-length Wnt-1 cDNA or LacZ. Forty-eight hours later infection rates were determined by X-Gal staining to be ≈70%. To eliminate uninfected cells, G418 selection was applied. Ten days after infection, the Wnt-1-expressing cell populations showed increased proliferation compared with LacZ controls (Fig. 1A). Wnt-1 protein expression was readily detectable, and biological activity was ascertained in a reporter assay (data not shown). Thirty days after infection, LacZ-expressing cells and uninfected controls flattened out, showed vacuoles, and began to senesce, as expected (Fig. 1B). In contrast, in the Wnt-1-infected cultures, a subset of cells continued to grow, and some of the cells started to detach from the dish (Fig. 1C). The detached cells began to form spheroid structures around day 40, which continued to grow into more complex structures (Fig. 1D). Cells that were dissociated from these structures were capable of reforming them within 1–3 days, growing indefinitely thereafter (currently P36 after infection). The same observations were made on cells derived from four independent reduction mammoplasties (A, D, E, and K) (Fig. 1A); the cell strains obtained by ectopic Wnt-1 expression will be referred to below as Wnt-1-HMECs.
Fig. 1.
Effects of Wnt-1 on primary HMECs. (A) Growth curves of primary HMECs from three different individuals infected with Wnt-1- or LacZ-expressing retrovirus. Cell number relative to that at the time of infection is plotted over time. Wnt-1-HMEC strains continue to grow at 5 weeks after infection, whereas the LacZ-infected cells arrest. Arrows indicate when spheres start forming. (B and C) Micrographs of pLNCX-LacZ- and pLNCX-Wnt-1-infected HMECs 4 weeks after infection and selection (P5). pLNCX-LacZ-infected cells show hallmarks of senescence, whereas in pLNCX-Wnt-1 (P7)-infected cultures, small, rounded cells (arrows) appear next to flat, senescent cells (arrowheads). (Scale bars: 100 μm.) (D) More complex 3D structures formed by Wnt-1-HMECs 8 weeks after infections (P12). (Scale bar: 200 μm.) (E) Growth curves of tumors arising from three different Wnt-1-HMEC strains (P15–P20) injected into the mammary glands of RAG2−/− females. (F) Hematoxylin and eosin-stained sections of Wnt-1-HMEC tumors. Tumor cells grow as sheet and are surrounded by a pseudo capsule (arrow). (G) Higher magnification shows tumor cells with highly pleiomorphic nuclei and several mitotic figures (arrows).
Tumor Formation by Wnt-1-HMECs.
The prolonged lifespan and ability to grow independent of substrate of Wnt-1-HMECs suggested they might be transformed to a tumorigenic state. To test whether these cell populations were indeed tumorigenic, we injected them into mammary glands of 3-month-old immunocompromised RAG2−/− female mice (22). Twenty days after the injection, palpable tumors were detected, which grew to >1 cm in diameter within 6 weeks (Fig. 1E). The efficiency of tumor formation was high for all of the three cell strains tested, with tumors arising in 68% of the injected glands within 2 months (Table 1, which is published as supporting information on the PNAS web site). When the same number of control HMECs was injected, no tumors arose (data not shown).
The finding that increased Wnt expression is sufficient to induce a tumorigenic state contrasted with previous reports that expression of several oncogenes is required to transform HMECs (23). To address whether Wnt-1’s ability to transform HMECs is an artifact of our culture and/or infection conditions, we infected the primary HMECs from 10 different donors with retroviruses expressing either the oncogenic H-rasV12 or genomic SV40 large T (24). As expected, ectopic mutant Ras expression induced senescence (25). Large T expression transiently led to increased growth, but after several passages the cells flattened out and ceased to proliferate. None of these cells grew into spheres, and their injection into mouse mammary glands did not give rise to tumors (data not shown). Thus, increased Wnt-1 expression is unique in its ability to trigger, by itself, a cascade of events that result in oncogenic transformation of primary HMECs.
Histological analysis revealed that Wnt-1-HMEC tumors have round borders and are surrounded by a pseudo capsule (Fig. 1F, arrow). Tumor cells grow as sheets and with a syncytial pattern and do not form glandular structures or tubes. Cell nuclei were highly pleiomorphic, and several mitotic figures per high-powered field were found (Fig. 1G, arrows). Necrosis was frequent. All of these features are diagnostic criteria for typical medullary carcinoma of the breast, a histological subtype that represents ≈2% of breast carcinomas (26). To assess whether the Wnt-1-HMEC tumors resemble medullary carcinomas at the molecular level, we examined expression of characteristic markers by immunohistochemistry. Like typical medullary carcinomas, the Wnt-1-HMEC tumors were positive for cytokeratin 18 and negative for cytokeratin 14, estrogen receptors, and progesterone receptors (Fig. 5, which is published as supporting information on the PNAS web site). Strikingly, >70% of the tumor cell nuclei stained intensely for p53 (Fig. 5), as is characteristic of typical medullary carcinomas (27). A further hallmark of medullary carcinomas, a prominent lymphocytic infiltrate, was absent from Wnt-1-HMEC tumors, as was expected, because the RAG2−/− mice lack mature B and T cells (22). Thus, the Wnt-1-HMECs form tumors in mice that closely resemble a subtype of human breast tumors, medullary carcinomas, both morphologically and molecularly.
Mechanisms of Wnt-1-Induced Transformation.
These observations raised the question of how increased Wnt signaling triggers oncogenic transformation. Recent work indicates that a DDR similar to that caused by double-stranded breaks is activated at very early stages of tumorigenesis in the breast and other tissues (15, 16). Central mediator between insult and cellular response is the ATM (ataxia telangiectasia mutated) kinase (28). It autophosphorylates upon activation and proceeds to phosphorylate downstream effectors such as Chk2 (29) and histone H2AX (30). As a result, checkpoints are activated that ensure that a cell either repairs inflicted damage or undergoes apoptosis.
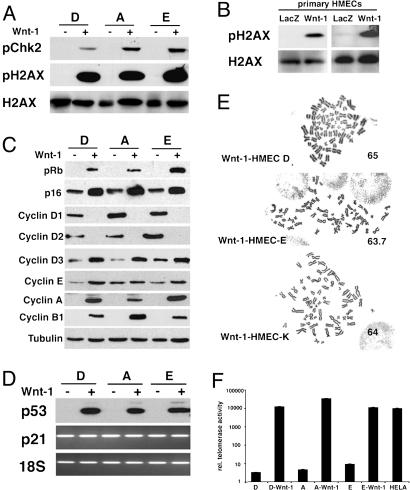
To assess whether Wnt-1 elicits a DDR, we compared levels of phospho Chk2 and phospho H2AX in Wnt-1-HMECs and parental control cells. Both proteins are phosphorylated in the Wnt-1-HMECs but not in the parental cells, whereas total H2AX levels are comparable, indicating that DDR is constitutively active in Wnt-1-transformed cells (Fig. 2A). We also assessed phospho H2AX levels in cells infected with Wnt-1 or LacZ control virus before the appearance of a transformed phenotype (P5). Whereas total H2AX protein levels were similar in the two cell populations, phospho H2AX was detectable in cells ectopically expressing Wnt-1 but not in control cells (Fig. 2B). Thus, the DDR triggered by increased Wnt signaling is an early event before overt signs of transformation.
Fig. 2.
Hallmarks of oncogenic transformation in Wnt-1-HMECs. (A) Expression of the phosphorylated form of Chk-2 and H2AX protein as well as total H2AX protein levels in three different Wnt-1-HMEC strains and the respective parental cells. (B) Expression of the phosphorylated form of H2AX protein as well as total H2AX protein levels in primary HMECs (P5) from two different donors that were infected with retroviruses expressing either LacZ or Wnt-1. (C) Expression of different cell-cycle proteins assessed by immunoblotting of lysates from three different Wnt-1-HMEC strains and respective parental cells. Cyclin D1 and D2 levels are down-modulated in Wnt-1-HMEC strains, whereas p16, phospho Rb, cyclin D3, cyclin A, and cyclin B1 levels are up-regulated. Tubulin was used as a loading control. (D) p53 protein levels assessed by immunoblotting are high in Wnt-1-transformed cells and below detection limit in parental controls. p21 mRNA levels are comparable. Real-time PCR quantification (data not shown) indicates that p21 mRNA levels normalized with 18S rRNA do not differ in a statistically significant manner. (E) Representative mitotic spreads of different Wnt-1-HMEC strains showing near triploid karyotype; average chromosome number for each cell strain is indicated (n = 10). (F) Telomerase activity in different Wnt-1-HMEC strains, their parental control cells, and HeLa cells assessed by real-time quantitative telomeric repeat amplification protocol.
As shown recently, the activation of a DDR at early stages of tumorigenesis provides selective pressure for inactivation of the G1/S-p16/Rb and the p53 checkpoints (15, 16). Consistent with cell-cycle checkpoint inactivation in Wnt-1-HMEC strains, hyperphosphorylated Rb protein was readily detected but below detection limit in the parental cells (Fig. 2C). To determine the mechanism that might account for the Rb hyperphosphorylation we analyzed the protein levels of different cyclins by immunoblotting. Cyclin D1 and D2 were present in control HMECs but below detection limits in Wnt-1-HMECs, whereas the expression levels of cyclin D3, cyclin A, and the mitotic cyclin B1 were increased in the Wnt-1 transformants (Fig. 2C). Wnt-1 transformation was accompanied by a substantial increase of p16 expression rather than down-regulation, as was previously reported for HMECs (31, 32). Thus, the Rb checkpoint is disrupted by a mechanism involving increased cyclin D3, cyclin A, and cyclin B1 expression and increased p16 levels.
Our finding that p53 protein levels are increased in the Wnt-1-HMEC tumors suggested that p53 is stabilized as a consequence of the DDR through mutations impairing its function (33). To test whether p53 protein levels are already changed during in vitro transformation or only later during in vivo growth, we determined p53 expression in cultured Wnt-1-HMECs and the parental control cells. p53 protein levels were found to be strongly up-regulated in all of the Wnt-1-HMEC strains compared with their parental cells (Fig. 2D). To assess whether p53 function is concomitantly disrupted, we measured mRNA expression levels of the p53 target gene p21 (34). In the presence of high p53 protein levels there was no up-regulation of p21 mRNA in the Wnt-1-HMECs compared with the parental controls (Fig. 2D); protein levels were actually decreased (data not shown). Together, these observations indicate that p53 protein levels are increased whereas its function is compromised during Wnt-1-induced transformation.
Inactivation of checkpoints results in genomic instability with abnormal karyotypes, a characteristic feature of most malignant tumors. Mitotic spreads from each of the four cell strains revealed two distinct cell populations: a larger one of near triploid cells (Fig. 2E) and a smaller one of diploid cells. At later stages of the transformation process telomerase activity is up-regulated to ensure continued adequate chromosome replication (2). Using a real-time quantitative telomeric repeat amplification protocol (35) to measure telomerase activity, we found telomerase activity close to detection limit in the parental control cells (P4 or P5). Wnt-1-HMECs showed a 1,000-fold increase in telomerase activity, comparable to that seen in HeLa cells (Fig. 2F).
Notch Signaling Activity in Wnt-1-HMECs.
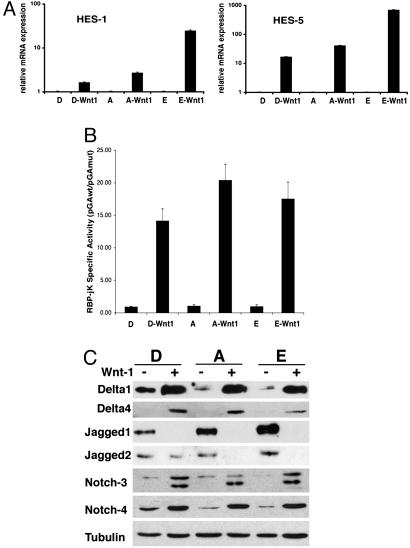
Activation of the DDR is a strong selective stimulus for genomic instability but is not sufficient to transform HMECs given that other oncogenes, such as SV40 large T, elicit a DDR (data not shown) yet fail to transform primary HMECs. To gain further insights into the molecular basis of Wnt-induced transformation we compared the gene expression profiles of Wnt-1-HMECs and parental cells. The Notch target HES-1 was among the most highly up-regulated genes. Quantification of the mRNA levels of the Notch target genes HES-1 and HES-5 (36, 37) by quantitative RT-PCR showed 10- and 250-fold increases, respectively, suggesting that the Notch signaling pathway might be involved in the transformation (Fig. 3A).
Fig. 3.
Notch signaling in Wnt-1-HMECs and in primary HMECs. (A) RT-PCR analysis of mRNA expression of the endogenous Notch target genes HES-1 and HES-5 in the parental HMECs and Wnt-1-HMECs. All real-time RT-PCR values are expressed as relative arbitrary units after internal normalization for 18S rRNA. (B) Notch signaling activity in different Wnt-1-HMECs (P15–P18) and parental HMECs (P4 and P5) derived from different reduction mammoplasties (D, A, and E) assayed with reporter plasmids containing an artificial Notch/RBP-Jk-responsive promoter (pGAwt) or a control reporter (pGAmut) without RBP-Jk-binding sites, plotted after internal normalization. (C) Immunoblot of Notch signaling components, Dll1, Dll4, Jagged1, and Jagged2, as well as Notch3 and Notch4 in parental HMECs (P4 and P5) and Wnt-1-HMECs (P15–P18) derived from different reduction mammoplasties (D, A, and E).
The best characterized mechanism by which Notch activation controls transcription is by converting the DNA-binding protein RBP-Jk/CBF-1 from a transcriptional repressor into an activator (14). Activity of an artificial promoter with concatemerized RBP-Jk-binding sites (pGAwt) provides a useful measure of Notch-RBP-Jk-dependent transcription and endogenous Notch signaling (38). The specific activity of this promoter, as determined after normalization with a negative control reporter plasmid lacking RBP-Jk-binding sites (pGAmut), was on average 17-fold higher in three different Wnt-1-HMECs than in the controls (Fig. 3B).
In parallel with these findings, expression of Notch ligands of the Delta family (Dll1, Dll3, and Dll4) as well as Notch3 and Notch4 receptors was found to be significantly increased in Wnt-1-HMECs at both protein and mRNA levels (Fig. 3C; see also Fig. 6, which is published as supporting information on the PNAS web site). Notch2 expression did not change, and Notch1 was below detection limit. Interestingly, expression of the ligands of the Jagged family was reduced, probably reflecting differences in amplified cell populations as discussed below.
The Role of Notch in Mediating Wnt-1-Induced Transformation and Tumorigenicity.
To assess the functional significance of increased Notch signaling in Wnt-induced transformation, we made use of the secreted form of Dll (sDll), which inhibits Notch signaling (39). We cocultured Wnt-1-HMECs with NIH 3T3 cells expressing sDll. Dissociated Wnt-1-HMECs that were plated onto control-infected NIH 3T3 cells formed spheres, whereas sphere formation of the Wnt-1 transformants was completely suppressed when the cocultured 3T3 cells expressed sDll (Fig. 7A, which is published as supporting information on the PNAS web site).
To determine whether Notch signaling is not only required but sufficient for substrate-independent growth, we infected primary HMECs with a retrovirus expressing the constitutively active Notch1 intracellular domain (40). Strikingly, 4 days after infection, cells already began to detach from the plates and to form spheres (Fig. 7B) similar to those arising from the Wnt-1 cultures with a much longer delay, but more compact. However, unlike the Wnt-1-expressing cells, cells in the Notch intracellular domain-induced spheres failed to proliferate and did not give rise to tumors when injected into mouse mammary glands (data not shown). Thus, Notch signaling appears to be both required and sufficient for detachment and formation of 3D structures, whereas additional events are required for Wnt-1-induced proliferation.
To address whether Notch signaling is also required for in vivo tumor formation, we mixed sDll-expressing or control NIH 3T3 cells, after a mitomycin C treatment, with Wnt-1-HMECs and injected the two combinations of cells into contralateral mouse mammary glands. The presence of control NIH 3T3 cells had no effect on tumor growth, whereas the sDll-expressing cells either completely suppressed it, in the case of two of the tested strains (Wnt-1-HMEC-A and Wnt-1-HMEC-E), or reduced it to a significant extent, in the case of the third strain tested (Wnt-1-HMEC-D) (Fig. 7C). Thus, Notch signaling is required for Wnt-1-induced transformation of primary HMECs both in vitro and in vivo.
Wnt Signaling Activity in Human Breast Cancer.
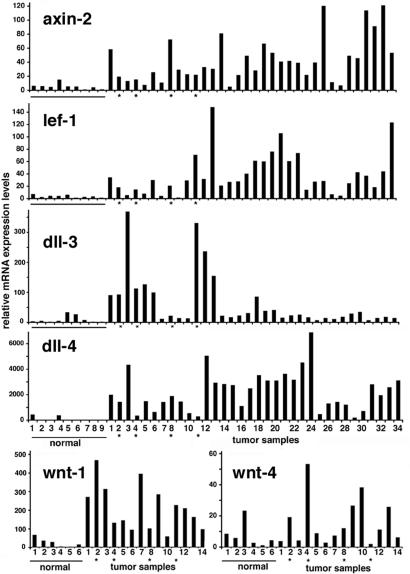
To test whether there is an increase in Wnt signaling in clinically occurring tumors that may be linked to up-regulation of Notch Dll ligand expression, we initially assessed levels of Wnt signaling activity in a panel of 34 human breast carcinomas by determining mRNA levels of the direct and specific Wnt target genes Axin-2 (41–43) and Lef-1 (44–46). Expression of these genes, as determined by real-time RT-PCR, was consistently higher in breast carcinomas than in normal breast tissue samples (on average 8- and 12-fold) (Fig. 4). Concomitantly, expression of the Notch ligand Dll4 was remarkably increased in >90% of the tumors (on average 25-fold), with expression of Dll3 being also augmented in a third of them. Of the existing 16 human Wnt family members, Wnt1, 2, 4, 5a, 5b, 7a, 8b, 9a, 9b, 10b, and 11 are expressed in normal human breast samples (our unpublished observations). Of these, in parallel with the increased Wnt signaling, Wnt-1 and Wnt-4 were overexpressed in tumors, whereas the other family members were not (Fig. 4).
Fig. 4.
Axin-2, Lef-1, Dll-3, Dll-4, Wnt-1, and Wnt-4 mRNA expression levels in normal human breast tissue (n = 9 or 6) and breast carcinoma samples (n = 34 or 14) quantified by real-time RT-PCR. All samples were run in triplicate and normalized to 18S rRNA. To account for tumor-to-tumor variability in gene expression levels, each bar represents a single tumor. Asterisks indicate medullary carcinomas.
Discussion
Taken together, our results show that increased Wnt signaling is sufficient to cause transformation of primary HMECs, with early activation of the DDR followed by a cascade of events resulting in the tumorigenic phenotype. Expression of Notch ligands Dll1, Dll3, and Dll4 is increased, and Notch activation is required for Wnt-induced transformation both in vitro and in vivo. Finally, analysis of a substantial number of human breast carcinomas indicates that these findings are likely to be relevant to the clinical situation.
The transforming effects of increased Wnt signaling are unique, in that neither expression of SV40 small and large T antigens nor high levels of H-ras-V12 or hTERT were, by themselves, able to induce transformation of HMECs (23). Recent evidence points to the DDR being a very early step in the genesis of breast cancer and other malignancies (15, 16). Consistent with this possibility, we have found that increased Wnt signaling in HMECs leads to a DDR before any detectable signs of deranged growth and transformation. Wnt may elicit a DDR by the mechanism proposed for other oncogenic insults, that is, through inappropriate growth stimulation (16). Alternatively, increased Wnt signaling may trigger a DDR through a more direct mechanism, as suggested by recent reports that the established Wnt signaling components adenomatous polyposis colon protein and Dvl affect microtubule stability (47, 48). A scenario seems plausible whereby Wnt signaling may affect the spindle checkpoint, leading to genomic instability and a subsequent DDR.
The DDR activates checkpoints that ensure cells cease to proliferate and redirect their activity toward repair of DNA damage. In the context of an oncogenic stimulus, the DDR provides a setting in which further growth relies on strong selective pressure for the inactivation of these checkpoints. In agreement with this view, we have found functional inactivation of p53 in Wnt-1-transformed HMECs, with increased levels of the p53 protein but no increase in p21 expression. Along the same lines, cell-cycle control is deregulated with loss of the G1/S checkpoint and high levels of hyperphosphorylated Rb protein.
Besides the DDR and ensuing biochemical events, Wnt-1-induced transformation of HMECs is associated with increased Notch signaling through a mechanism that is likely to involve up-regulation of Notch ligands of the Dll family. Our findings that dll3 and 4 are overexpressed in human breast cancers suggest that this observation is likely to be clinically relevant. In the case of jagged ligands, a recent report shows that high jagged1 expression in human breast carcinomas correlates with poor prognosis (49). The tumors that we obtained by Wnt-1 transformation have low levels of jagged1 and resemble typical medullary carcinomas of the breast, which are characterized by a good prognosis despite their high-grade characteristics (nuclear morphology and high mitotic index). Thus, they fall into the categories of breast tumors that are expected to show relatively low jagged expression. Additionally, in the normal human breast, at least jagged1 expression is localized specifically to myoepithelial rather than luminal cells (49). In Wnt-1-HMEC cultures, immunostaining revealed expression of the luminal marker cytokeratin 18, whereas in the control cultures 30 days after infection <5% of the cells express this marker, with the remaining population expressing cytokeratin 14, a marker of myoepithelial cells (S. Gass and C.B., unpublished observations). Thus, the loss of myoepithelial cells during Wnt-1-induced transformation may also account for the low Jagged1 levels, which contrast with the up-regulation of Dll expression and associated increase in Notch signaling activity.
Irrespective of the specific ligands involved, Notch activation is likely to play an important role in breast carcinogenesis. In fact, we have shown that increased Notch activity is required for Wnt-induced transformation and is by itself sufficient to reproduce significant aspects of this process (formation of 3D structures). Both Wnt and Notch signaling pathways are important in maintaining and amplifying progenitor cells in different tissues (50–52), including the breast (53–55). Thus, in concomitance with the biochemical events described above, a further factor to be considered is the existence of subpopulations of HMECs with different susceptibility to malignant transformation that may be selectively amplified by activation of Wnt and/or Notch signaling pathways. The crosstalk between these pathways may impinge especially on early steps of breast carcinogenesis, with potential impact on novel treatments and/or prevention of breast cancer.
Methods
Cell Culture and Retroviral Infection.
Normal human breast tissue was obtained from women undergoing reduction mammoplasties, with no previous history of breast cancer, who gave their informed consent. All samples were confirmed by histopathological examination to be free of malignancy. Primary HMECs were derived from these specimens as described (56). At P2 or P3, cells were spin-infected with high-titer amphotropic retroviruses as described (57) and selected with G418 (200 μg/ml). Notch intracellular domain cDNA was subcloned into MSCV2.2 (57) with a neomycin resistance gene. NIH 3T3 cells stably expressing sDll or Jagged (39) were grown in DMEM/10% calf serum and treated for 2 h with 5 μg/ml mitomycin before mixing with Wnt-1-HMECs.
Quantitative Real-Time PCR.
Total RNA (1 μg) was reverse-transcribed by using reverse transcriptase (Invitrogen) and random hexamers (Roche). The resulting cDNAs were used for quantitative PCR analysis by using the iCycler apparatus (Bio-Rad) and the SYBR Green PCR Core Reagents system (Qiagen). Results were evaluated with icycler iq real-time detection system software (Bio-Rad). Samples were run in triplicate, and all quantifications were normalized to endogenous control 18S rRNA. For primer sequences, see Supporting Methods, which is published as supporting information on the PNAS web site.
Transient Luciferase Assays.
Transient transfections were performed by using FuGENE (Roche) according to the manufacturer’s instructions. A total of 104 cells were cotransfected with 25 ng of luciferase reporter for RBP-Jk activity, either pGAwt or pGAmut, and 25 ng of phRL-TK (Promega) for internal normalization. After 24 h, the cells were assayed for luciferase and Renilla activity by using the dual-luciferase reporter assay system (Promega, E1910).
Western Blot Analysis.
For details, see Supporting Methods.
Mice.
RAG2−/− (129SV/C57BL/6) mice were purchased from Taconic Farms and bred and housed under conventional conditions with a 12-h cycle of light and dark in filter-top cages. They were supplied ad libitum with irradiated feed and water. Tumor size was measured every 3–4 days.
Tumor Assay.
A total of 106 Wnt-1-HMECs were resuspended on their own or mixed with 3 × 106 mitomycin-treated NIH 3T3 cells in 100 μl of PBS and injected into inguinal mammary glands of 3-month-old RAG2−/− females.
RNA Isolation.
Frozen tumor samples and reduction mammoplasty tissue, enriched for epithelial components, were used for total RNA extraction with TRIzol (GIBCO). After RNeasy (Qiagen) treatment, RNA was dissolved in RNase-free water.
Histopathology.
Samples were fixed in 10% buffered formalin and embedded in paraffin. Six-micrometer sections were stained with hematoxylin and eosin according to standard protocols. Antibodies against cytokeratin 18 and 14, estrogen receptors, progesterone receptors (Neomarkers, MS-142, RB-9020, RB-9101, and RB-1492), and p53 (Novocastra NCL-p53-D01) were diluted as indicated by the supplier and applied overnight at 4°C. Biotinylated secondary antibodies were detected with a Vectastain Elite Kit (Vector Laboratories).
Real-Time Quantitative Telomeric Repeat Amplification Protocol.
Real-time quantitative telomeric repeat amplification protocol was performed as described in ref. 35 by using 0.25 μg of protein extract per 25-μl reaction, with the SYBR Green PCR Core reagents system (Qiagen).
Supplementary Material
Acknowledgments
We thank S. Mallepell and R. Rajaram for technical assistance; A. Kispert (Institute for Molecular Biology, Medizinische Hochschule, Hannover, Germany) and R. A. Weinberg (Whitehead Institute, Cambridge, MA) for providing the pLNCX-mWnt-1, pLNCX-LacZ, and pBabe-genomic large T and the pBabe-H-rasV12 retroviral vectors, respectively; and J. Storre and J. Ligner for the critical reading of the manuscript. This work was supported by funds from the National Center of Competence in Research in Molecular Oncology and Oncosuisse. G.C. was supported by a Swiss National Science Foundation grant.
Abbreviations
- HMEC
human mammary epithelial cell
- DDR
DNA damage response
- Pn
passage n.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Ellis I. O., Schnitt S. J., Sastre-Garau X., Bussolati G., Tarassoli F. A., Eusebi V., Peterse J. L., Mukai K., Tabár L., Jacquemeir J. In: Pathology and Genetics of Tumours of the Breast and Female Genital Organs. Tavassoli F., Devilee P., editors. Vol. 5. Lyon, France: International Agency for Research on Cancer; 2003. pp. 13–59. [Google Scholar]
- 2.Hanahan D., Weinberg R. A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Cardiff R. D., Bern H. A., Faulkin L. J., Daniel C. W., Smith G. H., Young L. J., Medina D., Gardner M. B., Wellings S. R., Shyamala G., et al. Comp. Med. 2002;52:12–31. [PubMed] [Google Scholar]
- 4.Rangarajan A., Weinberg R. A. Nat. Rev. Cancer. 2003;3:952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- 5.Gallahan D., Callahan R. J. Virol. 1987;61:66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchholz T. A., Wazer D. E. Semin. Radiat. Oncol. 2002;12:285–295. doi: 10.1053/srao.2002.35248. [DOI] [PubMed] [Google Scholar]
- 7.Howe L. R., Brown A. M. Cancer Biol. Ther. 2004;3:36–41. doi: 10.4161/cbt.3.1.561. [DOI] [PubMed] [Google Scholar]
- 8.Ugolini F., Adelaide J., Charafe-Jauffret E., Nguyen C., Jacquemier J., Jordan B., Birnbaum D., Pebusque M. J. Oncogene. 1999;18:1903–1910. doi: 10.1038/sj.onc.1202739. [DOI] [PubMed] [Google Scholar]
- 9.Ugolini F., Charafe-Jauffret E., Bardou V. J., Geneix J., Adelaide J., Labat-Moleur F., Penault-Llorca F., Longy M., Jacquemier J., Birnbaum D., Pebusque M. J. Oncogene. 2001;20:5810–5817. doi: 10.1038/sj.onc.1204706. [DOI] [PubMed] [Google Scholar]
- 10.Nagahata T., Shimada T., Harada A., Nagai H., Onda M., Yokoyama S., Shiba T., Jin E., Kawanami O., Emi M. Cancer Sci. 2003;94:515–518. doi: 10.1111/j.1349-7006.2003.tb01475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pece S., Serresi M., Santolini E., Capra M., Hulleman E., Galimberti V., Zurrida S., Maisonneuve P., Viale G., Di Fiore P. P. J. Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polakis P. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 13.Behrens J., von Kries J. P., Kuhl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 14.Artavanis-Tsakonas S., Rand M. D., Lake R. J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 15.Bartkova J., Horejsi Z., Koed K., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J. M., Lukas C., et al. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 16.Gorgoulis V. G., Vassiliou L. V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R. A., Jr., Kastrinakis N. G., Levy B., et al. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 17.Nusse R., Varmus H. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 18.Tsukamoto A. S., Grosschedl R., Guzman R. C., Parslow T., Varmus H. E. Cell. 1988;55:619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 19.Herzlinger D., Qiao J., Cohen D., Ramakrishna N., Brown A. M. Dev. Biol. 1994;166:815–818. doi: 10.1006/dbio.1994.1360. [DOI] [PubMed] [Google Scholar]
- 20.Kispert A., Vainio S., McMahon A. P. Development (Cambridge, U.K.) 1998;125:4225–4234. doi: 10.1242/dev.125.21.4225. [DOI] [PubMed] [Google Scholar]
- 21.Shimizu H., Julius M. A., Giarre M., Zheng Z., Brown A. M., Kitajewski J. Cell Growth Differ. 1997;8:1349–1358. [PubMed] [Google Scholar]
- 22.Shinkai Y., Rathbun G., Lam K. P., Oltz E. M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A. M., et al. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 23.Elenbaas B., Spirio L., Koerner F., Fleming M. D., Zimonjic D. B., Donaher J. L., Popescu N. C., Hahn W. C., Weinberg R. A. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn W. C., Counter C. M., Lundberg A. S., Beijersbergen R. L., Brooks M. W., Weinberg R. A. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 25.Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 26.Roses D. F. Breast Cancer. Philadelphia: Churchill Livingstone; 1999. [Google Scholar]
- 27.de Cremoux P., Salomon A. V., Liva S., Dendale R., Bouchind’homme B., Martin E., Sastre-Garau X., Magdelenat H., Fourquet A., Soussi T. J. Natl. Cancer Inst. 1999;91:641–643. doi: 10.1093/jnci/91.7.641. [DOI] [PubMed] [Google Scholar]
- 28.Shiloh Y. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 29.Matsuoka S., Rotman G., Ogawa A., Shiloh Y., Tamai K., Elledge S. J. Proc. Natl. Acad. Sci. USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 31.Huschtscha L. I., Noble J. R., Neumann A. A., Moy E. L., Barry P., Melki J. R., Clark S. J., Reddel R. R. Cancer Res. 1998;58:3508–3512. [PubMed] [Google Scholar]
- 32.Romanov S. R., Kozakiewicz B. K., Holst C. R., Stampfer M. R., Haupt L. M., Tlsty T. D. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- 33.Borresen-Dale A. L. Hum. Mutat. 2003;21:292–300. doi: 10.1002/humu.10174. [DOI] [PubMed] [Google Scholar]
- 34.el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 35.Hou M., Xu D., Bjorkholm M., Gruber A. Clin. Chem. 2001;47:519–524. [PubMed] [Google Scholar]
- 36.Kuroda K., Tani S., Tamura K., Minoguchi S., Kurooka H., Honjo T. J. Biol. Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 37.de la Pompa J. L., Wakeham A., Correia K. M., Samper E., Brown S., Aguilera R. J., Nakano T., Honjo T., Mak T. W., Rossant J., Conlon R. A. Development (Cambridge, U.K.) 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 38.Kato H., Taniguchi Y., Kurooka H., Minoguchi S., Sakai T., Nomura-Okazaki S., Tamura K., Honjo T. Development (Cambridge, U.K.) 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 39.Trifonova R., Small D., Kacer D., Kovalenko D., Kolev V., Mandinova A., Soldi R., Liaw L., Prudovsky I., Maciag T. J. Biol. Chem. 2004;279:13285–13288. doi: 10.1074/jbc.C300564200. [DOI] [PubMed] [Google Scholar]
- 40.Capobianco A. J., Zagouras P., Blaumueller C. M., Artavanis-Tsakonas S., Bishop J. M. Mol. Cell. Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan D., Wiesmann M., Rohan M., Chan V., Jefferson A. B., Guo L., Sakamoto D., Caothien R. H., Fuller J. H., Reinhard C., et al. Proc. Natl. Acad. Sci. USA. 2001;98:14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lustig B., Jerchow B., Sachs M., Weiler S., Pietsch T., Karsten U., van de Wetering M., Clevers H., Schlag P. M., Birchmeier W., Behrens J. Mol. Cell. Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jho E. H., Zhang T., Domon C., Joo C. K., Freund J. N., Costantini F. Mol. Cell. Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kengaku M., Capdevila J., Rodriguez-Esteban C., De La Pena J., Johnson R. L., Belmonte J. C., Tabin C. J. Science. 1998;280:1274–1277. doi: 10.1126/science.280.5367.1274. [DOI] [PubMed] [Google Scholar]
- 45.Hovanes K., Li T. W., Munguia J. E., Truong T., Milovanovic T., Lawrence Marsh J., Holcombe R. F., Waterman M. L. Nat. Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 46.Filali M., Cheng N., Abbott D., Leontiev V., Engelhardt J. F. J. Biol. Chem. 2002;277:33398–33410. doi: 10.1074/jbc.M107977200. [DOI] [PubMed] [Google Scholar]
- 47.Ciani L., Krylova O., Smalley M. J., Dale T. C., Salinas P. C. J. Cell Biol. 2004;164:243–253. doi: 10.1083/jcb.200309096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zumbrunn J., Kinoshita K., Hyman A. A., Nathke I. S. Curr. Biol. 2001;11:44–49. doi: 10.1016/s0960-9822(01)00002-1. [DOI] [PubMed] [Google Scholar]
- 49.Reedijk M., Odorcic S., Chang L., Zhang H., Miller N., McCready D. R., Lockwood G., Egan S. E. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 50.Korinek V., Barker N., Moerer P., van Donselaar E., Huls G., Peters P. J., Clevers H. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 51.Oishi K., Kamakura S., Isazawa Y., Yoshimatsu T., Kuida K., Nakafuku M., Masuyama N., Gotoh Y. Dev. Biol. 2004;276:172–184. doi: 10.1016/j.ydbio.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 52.Duncan A. W., Rattis F. M., DiMascio L. N., Congdon K. L., Pazianos G., Zhao C., Yoon K., Cook J. M., Willert K., Gaiano N., Reya T. Nat. Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 53.Li Y., Welm B., Podsypanina K., Huang S., Chamorro M., Zhang X., Rowlands T., Egeblad M., Cowin P., Werb Z., et al. Proc. Natl. Acad. Sci. USA. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dontu G., Jackson K. W., McNicholas E., Kawamura M. J., Abdallah W. M., Wicha M. S. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu B. Y., McDermott S. P., Khwaja S. S., Alexander C. M. Proc. Natl. Acad. Sci. USA. 2004;101:4158–4163. doi: 10.1073/pnas.0400699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stampfer M. R., Bartley J. C. Proc. Natl. Acad. Sci. USA. 1985;82:2394–2398. doi: 10.1073/pnas.82.8.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hawley R. G., Lieu F. H., Fong A. Z., Hawley T. S. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.