Abstract
Single biomolecules in free solution have long been of interest for detailed study by optical methods, but Brownian motion prevents the observation of one single molecule for extended periods. We have used an anti-Brownian electrokinetic (ABEL) trap to trap individual protein molecules in free solution, under ambient conditions, without requiring any attachment to beads or surfaces. We also demonstrate trapping and manipulation of single virus particles, lipid vesicles, and fluorescent semiconductor nanocrystals.
Keywords: anti, Brownian electrokinetic trap, electrophoresis, feedback, single molecule, trapping
This year marks the 101st anniversary of Einstein’s explanation of Brownian motion. He showed that the jittering of small particles in water is the cumulative effect of countless collisions with thermally agitated water molecules (1). Brownian motion is a major transport process at the cellular and subcellular levels and thus is essential for life. Brownian motion also makes the task of studying subcellular structures in solution difficult: freely diffusing nano-objects do not hold still long enough for extended observation.
Laser tweezers have proved highly successful at manipulating objects in solution in the size range of 100 nm to 1 μm but require prohibitively large optical powers to trap objects much smaller than 100 nm (2). Surface-attachment chemistry often is used to immobilize individual molecules for extended study, but there remains a persistent doubt whether the immobilized molecules behave the same as their free-solution comrades (3). Recently, we described an anti-Brownian electrokinetic (ABEL) trap, which uses fluorescence microscopy and electrokinetic forces to overcome Brownian motion for 20- to 100-nm fluorescent polystyrene spheres (4, 5). Here, we show that the ABEL trap can suppress Brownian motion of individual protein molecules in free solution, under ambient conditions. We also demonstrate trapping and manipulation of single virus particles, lipid vesicles, and fluorescent semiconductor nanocrystals. To our knowledge, the trapping of individual biomolecules in free solution has not been previously described.
Enderlein (6) proposed to use feedback to translate either the laser focus or the microscope stage to keep a diffusing fluorescent molecule within the focal volume of a confocal microscope. Variants of this scheme have recently been implemented experimentally (7, 8) and there is continuing theoretical interest in developing optimal strategies for tracking diffusing molecules (9). Such approaches tend to be limited by the finite travel and slow response time of the mechanical feedback mechanisms.
The ABEL trap monitors the Brownian motion of a nanoparticle (by means of fluorescence microscopy) and then applies a feedback voltage to a microfluidic cell so that the resulting electrokinetic (i.e., electrophoretic and electroosmotic) forces produce a drift that exactly cancels the Brownian motion. The ABEL trap works on any object that can be imaged optically and is gentle enough to trap a variety of biological samples far smaller than can be trapped by other means. Trapping may be performed in most standard buffers or in distilled water.
A microfluidic cell is mounted in an inverted fluorescence microscope. Images are acquired on a high-sensitivity digital camera at frame rates of up to 300 Hz. A personal computer running custom software processes the images in real-time to extract the x,y-coordinates of a particle of interest and then applies a feedback voltage proportional to the offset between the measured position and a target position. The voltage induces a drift that pushes the particle toward the target position before the arrival of the next video image. The target position may be set to follow a predetermined two-dimensional trajectory or be controlled interactively by dragging with the computer mouse.
For optimal imaging of the fluorescent object, we built a microfluidic trapping cell made entirely of glass (Fig. 1). The fabrication process is described in detail in Supporting Text, Figs. 4 and 5, and Movies 1–4, which are published as supporting information on the PNAS web site. In a previous ABEL trap design, (4) this microfluidic cell was made of poly(dimethylsiloxane) (PDMS). PDMS is permeable to oxygen, and oxygen leads to rapid photobleaching of many of the dyes used on biomolecules. Efforts to remove oxygen from the solution were unsuccessful with the PDMS cell because the PDMS acted as a large reservoir of oxygen, continually replenishing the solution. With a glass cell, standard oxygen scavengers could be added to the trapping medium, extending the lifetime-to-photobleaching of individual molecules by a factor of ≈10.
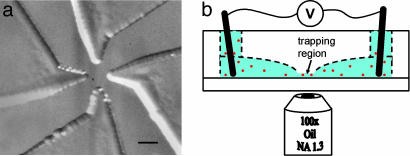
Fig. 1.
Glass microfluidic cell for the ABEL trap. (a) Trapping region, showing the patterned glass cell. Molecules are trapped in the center. Four channels ≈17 μm deep (the regions shaped like a bird’s beak) extend to the edge of the image and terminate in macroscopic electrodes. (Scale bar, 100 μm.) (b) The microfluidic cell sits above the oil-immersion objective of an inverted optical microscope capable of observing single molecules. The lower part of the cell is formed by a glass or fused silica slide. The top and bottom of the cell can be separated for cleaning or surface-treatments. The aqueous solution (blue) containing fluorescent biomolecules (red dots) sits above the coverslip and is confined by the cell material (shown transparent in this image). Molecules in the trapping region are confined to a thin fluid layer several hundred nanometers thick, preventing diffusion out of the focal plane of the microscope. The voltages applied across the electrodes provide the electrokinetic forces to counteract Brownian motion.
Objects within the disk-shaped trapping region are confined to the focal plane of the microscope but are free to diffuse within this plane. Four fluidic channels convey voltages from macroscopic control electrodes to the corners of the trapping region. Applying a voltage V = (Vx, Vy) to the control electrodes leads to a force F ∝ V on objects in the trapping region. The force arises through two distinct mechanisms: (i) charged particles are acted on directly by the electric field, resulting in an electrophoretic drift; and (ii) the electric field leads to an electroosmotic flow in the trapping region, which imparts a hydrodynamic force on all particles. As in capillary electrophoresis, the relative contributions of these two mechanisms may be adjusted by tailoring the composition of the solution and the surface chemistry of the microfluidic channels (10).
In contrast to optical forces, electrokinetic forces are so strong that they cease to limit the trapping strength of the ABEL trap. Rather, the trapping strength is limited by the latency of the feedback loop: the particle undergoes position fluctuations along each axis with mean-square amplitude σ2 ≈ 2Dtr, where D is the diffusion coefficient of the particle and tr is the response time of the feedback loop. The effective spring constant of the trap is given by keff = kBT/σ2. Because D ∝ 1/ηa, where η is the viscosity of the trapping medium and a is the radius of the particle, we have keff ∝ ηa. In contrast, the effective spring constant of laser tweezers scales as keff ∝ a3. Thus, the ABEL trap scales more favorably than laser tweezers for trapping small particles, and furthermore the trapping strength of the ABEL trap may be increased by increasing the viscosity of the trapping medium.
Increasing the field of view (FOV) increases the distance the particle has to go before it escapes from the trap. This increase could allow trapping of smaller particles at a given feedback latency but with increased spatial fluctuations. In addition, if an increase in the FOV is achieved by reading out a larger number of pixels from the charge-coupled device (CCD) (we normally only read out a small fraction of the CCD), then the frame rate of the camera slows down, and the image-processing software runs more slowly; these effects combine to counteract the gain from the increased FOV. The other way to increase the FOV is to use a lower-magnification objective or hardware binning of the pixels. However, once the width of the point-spread-function of the microscope becomes smaller than the dimensions of one pixel, the signal-to-noise ratio decreases. It becomes increasingly difficult to locate the particle amidst the fuzz from pixel noise and background autofluorescence. As cameras, computers, and image-processing software improve, undoubtedly we will be able to trap smaller objects.
Results and Discussion
We trapped individual particles of fluorescently labeled tobacco mosaic virus (TMV; ≈300 nm long × 15 nm in diameter) (see Movie 1). From the record of the position, r(tn), at each time step tn and the feedback voltage, V(tn), it is useful to extract a “pseudofree trajectory” (i.e., a trajectory similar to the one the particle would have followed had it not been trapped) as follows. The electrokinetic mobility was calculated from a linear regression of the displacements r(tn+1) − r(tn) between successive iterations of the feedback against the applied voltages V(tn). From the mobility and the record of the applied voltages, we calculated the field-induced displacement for each time step. This field-induced displacement was subtracted from the measured displacements. The resulting residual displacements (which are uncorrelated with the applied voltage) are due to Brownian motion. Summing these residuals leads to a pseudofree trajectory, constrained to start and end at the origin. Fig. 2 illustrates the measured and pseudofree trajectories for 13 trapped particles of TMV.
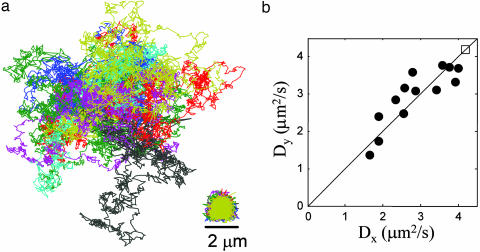
Fig. 2.
Trapping of individual particles of TMV. (a) Measured (Right) and pseudofree (Left) trajectories of 13 trapped particles of TMV. Each particle was trapped for 6.8 s (2,000 video frames at 3.4 ms per frame). The pseudofree trajectories are offset for clarity. (b) Diffusion coefficients along the x and y axes for the 13 particles trapped in a. If there were no statistical errors in the measurements, the data points would fall along the line. The rms deviation (rmsd) of Dx from Dy, 〈(2(Dx − Dy)/(Dx + Dy))2〉1/2, is 9%, whereas the rmsds of Dx and Dy from their ensemble-averaged values are 28% and 26%, respectively, indicating heterogeneity in the ensemble above the noise level of the measurement. The square indicates the value from dynamic light scattering.
These pseudofree trajectories show significant heterogeneity in the diffusion coefficient of the trapped particles: heterogeneity that would not have been apparent from a bulk experiment. The extracted mobility had an average value of 5 × 10−4 cm2/V·s, consistent with the literature value of 5.2 × 10−4 cm2/V·s (11). Fig. 2b shows the diffusion coefficients along the x- and y-axes. From the strong correlation between Dx and the independently measured Dy, we conclude that the heterogeneity reflects an intrinsic property of the trapped particles rather than a statistical error due to the stochastic nature of the measurement. No particles showed a diffusion coefficient larger than the literature value of 4.19 μm2/s (12), from which we conclude that the heterogeneity is due to aggregation of the TMV virions. TMV has a rotational relaxation rate of 318 s−1, which guarantees that the rotational anisotropy averages out after a few video frames, whereas the diffusion coefficients are calculated by averaging the displacements over 6.8 s. Thus, anisotropy of the TMV does not contribute to the measured heterogeneity of D. Because of the many different ways in which multiple TMV particles can bind together, we do not expect to see clustering of the diffusion coefficients corresponding to distinct numbers of particles in an aggregate. In contrast to the experiments on TMV, trapped 200-nm fluorescent polystyrene nanospheres showed considerably less heterogeneity in D.
In one of the first papers on optical trapping, Ashkin and Dziedzic (13) demonstrated trapping of TMV particles in a concentrated solution. In that experiment, it was not possible to control the number of TMV virions in the trap because each virion experienced essentially the same attractive optical potential. In contrast, the ABEL trap always traps exactly one particle: the Brownian motion of distinct particles is uncorrelated, so the force used to cancel the motion of one particle on average augments the motion of all of the others. Movie 1 of a trapped TMV particle shows that when two particles enter the trapping region, the mutual center of brightness is trapped, until one of the particles exits the trapping region, at which point the remaining particle is trapped.
To identify the dimmest fluorescent object that could be trapped, we formed ≈100-nm-diameter vesicles of egg-phosphatidylcholine (Egg-PC) doped with one or two fluorescent lipid molecules per vesicle. Despite their low fluorescence intensity, we were able to trap these vesicles and observe photobleaching in discrete steps (Fig. 3a). Vesicles containing individual fluorophores were stably held in the trap until photobleaching rendered the vesicles undetectable.
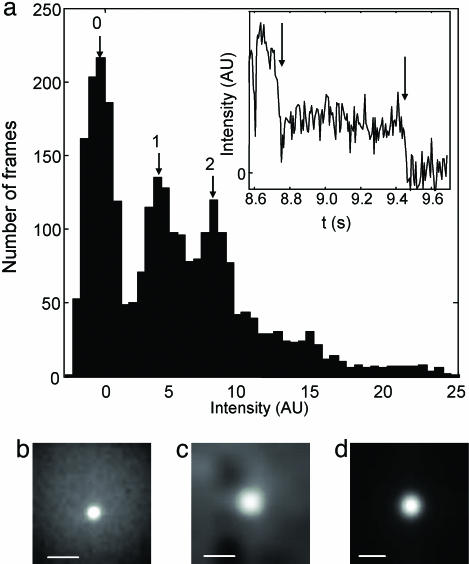
Fig. 3.
Trapping of individual fluorescent objects. (a) Histogram of intensities of individual trapped fluorescently labeled vesicles. The vesicles had an average diameter of 100 nm and were composed of egg-phosphatidylcholine with 1 part in 105 of the fluorescent lipid N-(6-tetramethylrhodaminethiocarbamoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine. A total of 26 vesicles were trapped, for a cumulative 2,690 video frames (at 6.5 ms per frame). Intensities were computed from a sliding average with a 26-ms window. Peaks indicate vesicles containing zero, one, or two fluorophores, as well as an unresolved contribution from more highly labeled vesicles. Some intensity values are negative because the background subtraction was set to yield zero mean intensity when the trap was empty. (Inset) A typical trajectory showing two-step photobleaching (arrows) of a vesicle containing two fluorophores. The vesicle was trapped with only one active fluorophore for ≈700 ms (between arrows). (b–d) Time-averaged images of trapped single molecules and CdSe nanocrystals. (b) A single molecule of B-phycoerythrin (average of 500 images taken over 2.2 s). (c) GroEL labeled with Cy3 (average of 10,000 images taken over 45 s). During this interval, several single molecules were sequentially trapped, eventually photobleached, and released from the trap. (d) A single CdSe fluorescent nanocrystal (average of 20,000 images taken over 90 s). (Scale bar in b–d, 2 μm.)
Attempts to study transmembrane proteins in lipid vesicles at the single-molecule level have been hampered by the diffusion-limited observation time of free vesicles (14). Furthermore, transmembrane proteins in planar-supported lipid bilayers are often immobile (15), although adding an extra bilayer ameliorates this problem (16). We expect that the ABEL trap will open avenues for single-molecule studies of transmembrane proteins in a close-to-native environment.
The response time of the ABEL trap was insufficient to trap objects smaller than ≈20 nm diameter in buffer (D > 22 μm2/s), although this goal will be possible in the future with faster imaging and feedback. To trap smaller objects, we increased the viscosity of the trapping medium by adding glycerol or sucrose. In solutions of 50% glycerol (η ∼ 6 ηH2O), we were able to trap single molecules of large proteins (GroEL and B-phycoerythrin) and single fluorescent CdSe nanocrystals (Fig. 3 b–d and Movies 2–4). GroEL was labeled with, on average, six fluorophores per tetradecamer, and stepwise photobleaching was often observed in the trapped molecules. B-phycoerythrin is intrinsically fluorescent and was trapped without any artificial labeling. The molecules of B-phycoerythrin showed single-step photobleaching, consistent with the understanding that the many fluorophores in the molecule are strongly coupled and form a single excitonic system (17). Trapped nanocrystals showed fluorescence blinking and were typically lost from the trap whenever they experienced an off-time greater than a few milliseconds.
With the ability to overcome the effects of Brownian motion, we expect that the ABEL trap will enable new biophysical measurements. It is clear that other spectroscopic techniques can be brought to bear on trapped molecules, such as fluorescence polarization/anisotropy, fluorescence lifetimes, binding events, and Förster resonant energy transfer (FRET). The ABEL trap could provide the analysis station for a device analogous to the well known FACS (fluorescence-activated cell sorter) but now for single biomolecules. The ability to trap single viruses and trigger the release of their genetic material at a defined location is a tantalizing prospect (18). By trapping lipid vesicles containing transmembrane proteins, one may study the action of these proteins at the single-molecule level in a near-native environment; these proteins are notoriously sensitive to departures from the cellular milieu. Enzymes that produce a fluorescent substrate are particularly attractive candidates for study because they are not subject to photobleaching (19). The ABEL trap would provide a nonperturbative way to study the folding of single proteins in solution (20). In addition to aiding single-molecule studies, the ABEL trap might allow one to assemble nanostructures one molecule at a time.
Materials and Methods
Trapping of TMV.
Particles of TMV (American Type Culture Collection) were suspended at a concentration of 50 nM in a buffer of 0.1 M NaHCO3 (pH 8.0). They were incubated with 1 mM Cy3-succinimidyl ester (Molecular Probes) at 4°C for 48 h for labeling of exposed amines. Unreacted dye was removed by gel filtration followed by dialysis against distilled water. The trapping was performed in distilled water at a TMV concentration of 20 pM. The viruses were excited at 532 nm.
Trapping of Lipid Vesicles.
Lipid vesicles of egg-phosphatidylcholine were formed according to the procedure provided by Avanti Polar Lipids. Egg-phosphatidylcholine doped with 1 part in 105 of the fluorescent lipid TRITC-DHPE [N-(6-tetramethylrhodaminethiocarbamoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine; Avanti Polar Lipids] was dissolved in chloroform and then dried under vacuum. Lipids were hydrated in buffer and then homogenized by repeated extrusion through a polycarbonate membrane with 100-nm pores. Vesicles then were diluted in distilled water to a concentration of ≈20 pM. The vesicles were excited at 532 nm.
Trapping of GroEL.
GroEL was fluorescently labeled at exposed amines with an average of six molecules of Cy3-succinimidyl ester (Molecular Probes) per tetradecamer of GroEL. A solution of 20 pM GroEL was dissolved in a buffer of 1 mM DTT, 50 mM Tris·HCl, 50 mM KCl, and 5 mM MgCl2 (pH 7.4). An equal volume of glycerol was added to increase the viscosity. The molecules were excited at 532 nm.
Trapping of B-Phycoerythrin.
B-phycoerythrin (Molecular Probes) was dialyzed against a buffer of 100 mM phosphate/100 mM NaCl (pH 7.4). Just before trapping, the solution was mixed with an equal volume of glycerol, and 1 mg/ml BSA was added to prevent adsorption. The molecules were excited at 532 nm.
Trapping of CdSe Nanocrystals.
Streptavidin-coated CdSe nanocrystals (QD565; Quantum Dot, Hayward, CA) were dissolved to a concentration of 20 pM in a solution of 47% distilled water, 48% glycerol, 4% 2-mercaptoethanol, and 1% antiadsorption polymer (Applied Biosystems). The nanocrystals were pumped at 488 nm.
Supplementary Material
Acknowledgments
We thank Stefanie Nishimura, So Yeon Kim, and Lawrence Klein for valuable assistance in sample preparation, Willy Wiyatno (Applied Biosystems) for providing a polymer to suppress electroosmotic flow, and Mary Tang for help with microfabrication. A.E.C. was supported by a Hertz Foundation Graduate Fellowship. This work was supported in part by U.S. Department of Energy Grant DE-FG02-04ER63777, National Science Foundation Grant CHE-0554681, and by the Stanford Nanofabrication Facility (a member of the National Nanotechnology Infrastructure Network), which is supported by National Science Foundation Grant ECS-9731293.
Abbreviations
- ABEL
anti-Brownian electrokinetic
- TMV
tobacco mosaic virus.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Einstein A. Annalen Physik. 1905;17:549–560. [Google Scholar]
- 2.Ashkin A., Dziedzic J. M., Bjorkholm J. E., Chu S. Opt. Lett. 1986;11:288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 3.Moerner W. E. J. Phys. Chem. B. 2002;106:910–927. [Google Scholar]
- 4.Cohen A. E., Moerner W. E. Appl. Phys. Lett. 2005;86:093109. [Google Scholar]
- 5.Cohen A. E. Phys. Rev. Lett. 2005;94:118102. doi: 10.1103/PhysRevLett.94.118102. [DOI] [PubMed] [Google Scholar]
- 6.Enderlein J. Appl. Phys. B. 2000;71:773–777. [Google Scholar]
- 7.Berglund A. J., Mabuchi H. Opt. Express. 2005;13:8069–8082. doi: 10.1364/opex.13.008069. [DOI] [PubMed] [Google Scholar]
- 8.Levi V., Ruan Q., Gratton E. Biophys. J. 2005;88:2919–2928. doi: 10.1529/biophysj.104.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson S. B. Appl. Phys. B. 2005;80:809–816. [Google Scholar]
- 10.Khaledi M. G., editor. High Performance Capillary Electrophoresis: Theory, Techniques, and Applications. New York: Wiley; 1998. [Google Scholar]
- 11.Grossman P. D., Soane D. S. Anal. Chem. 1990;62:1592–1596. doi: 10.1021/ac00214a011. [DOI] [PubMed] [Google Scholar]
- 12.Wilcoxon J., Schurr J. M. Biopolymers. 1983;22:849–867. doi: 10.1002/bip.360221011. [DOI] [PubMed] [Google Scholar]
- 13.Ashkin A., Dziedzic J. M. Science. 1987;235:1517–1520. doi: 10.1126/science.3547653. [DOI] [PubMed] [Google Scholar]
- 14.Zarrabi N., Zimmermann B., Diez M., Graeber P., Wrachtrup J., Boersch M. Proc. SPIE; 2005. pp. 175–188. [Google Scholar]
- 15.Groves J. T., Wulfing C., Boxer S. G. Biophys. J. 1996;71:2716–2723. doi: 10.1016/S0006-3495(96)79462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parthasarathy R., Groves J. T. Proc. Natl. Acad. Sci. USA. 2004;101:12798–12803. doi: 10.1073/pnas.0402168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goulian M., Simon S. M. Biophys. J. 2000;79:2188–2198. doi: 10.1016/S0006-3495(00)76467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evilevitch A., Lavelle L., Knobler C. M., Raspaud E., Gelbart W. M. Proc. Natl. Acad. Sci. USA. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rondelez Y., Tresset G., Tabata K. V., Arata H., Fujita H., Takeuchi S., Noji H. Nat. Biotechnol. 2005;23:361–365. doi: 10.1038/nbt1072. [DOI] [PubMed] [Google Scholar]
- 20.Lipman E. A., Schuler B., Bakajin O., Eaton W. A. Science. 2003;301:1233–1235. doi: 10.1126/science.1085399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.