Abstract
The sex pheromone biosynthetic pathways of lepidopterans require the concerted actions of multiple gene products. A number of pheromone gland (PG)-specific genes have been cloned in recent years and, whereas in vitro characterizations have indicated functions consistent with roles in pheromone production, there have been no clear demonstrations in vivo. Using an RNA interference-mediated loss-of-function approach, we injected newly formed Bombyx mori pupae with dsRNAs corresponding to genes of interest [i.e., PG fatty acyl reductase (pgFAR), B. mori PG Z11/Δ10,12 desaturase (Bmpgdesat1), PG acyl-CoA-binding protein (pgACBP), midgut ACBP, and pheromone biosynthesis activating neuropeptide receptor (PBANR)] to assess their specific roles during pheromonogenesis. In all cases, the introduced dsRNAs induced a dose-dependent reduction in sex pheromone production with the corresponding decrease in transcript levels. No effects on pupal development or adult emergence were observed. Disrupting the PBANR gene resulted in a loss of the lipase activity that liberates pheromone precursors, whereas knockout of the pgACBP gene prevented the daily accumulation and fluctuation of the triacylglycerols that function as the cellular deposits for the pheromone precursors. Taken together, our results provide unequivocal evidence that the pgACBP, Bmpgdesat1, pgFAR, and PBANR gene products are essential during pheromonogenesis and demonstrate the power of this methodology for dissecting the molecular interactions that comprise biosynthetic pathways.
Keywords: lipid droplet, sex pheromone biosynthesis, RNA interference
The remarkable abundance and diversity of the Lepidoptera (i.e., butterflies and moths) are mirrored by the vast assortment of low-molecular-weight compounds used in these insects as chemical cues regarding sexual maturation. Often used by females to lure conspecific males, most lepidopteran sex pheromones share a common progenitor that is de novo synthesized from acetyl-CoA via fatty acid synthesis in the pheromone gland (PG), a functionally differentiated organ located between the eighth and ninth abdominal segments. The species specificity of these multicomponent sex pheromones is based on the numerous structural and compositional variants present in the pheromone blends generated through enzymatic steps that alter chain length, the degree of unsaturation, and/or reductive modification of the carbonyl carbon (1–3).
(E,Z)-10,12-hexadecadien-1-ol (bombykol), the principal sex pheromone component in the silkmoth, Bombyx mori, is actively produced and released soon after adult emergence (4) from the single layer of epidermal cells located just below the endocuticle that comprise the pheromone-producing cells of the PG (5). In these cells, acetyl-CoA-derived palmitate (C16:Acyl) is stepwise converted to bombykol by the sequential actions of a bifunctional desaturase and a fatty acyl reductase; genes corresponding to each have recently been cloned and characterized as B. mori PG Z11/Δ10,12 desaturase (Bmpgdesat1) and PG fatty acyl reductase (pgFAR), respectively (6, 7). As with most lepidopteran sex pheromone biosynthetic pathways (2, 8), the bombykol biosynthetic pathway is regulated by a molecular interaction between the neurohormone pheromone biosynthesis-activating neuropeptide (PBAN) and its cognate G protein-coupled receptor (GPCR), a candidate of which has also been cloned (9).
Pheromonogenesis in the PG cells of B. mori is a dynamic period characterized by diverse intracellular events including the cytosolic accumulation of lipid droplets (5, 10), lipolysis of stored triacylglycerols (TG) from the lipid droplets (11), and the up-regulation of numerous PG-specific genes, such as PG acyl-CoA-binding protein (pgACBP), midgut ACBP (mgACBP), pgFAR, Bmpgdesat1, and PBAN receptor (PBANR) (6, 7, 9, 12, 13). Despite these advances, the precise molecular mechanisms underlying sex pheromone production are still not well understood. Furthermore, the functional roles of the genes currently linked to the biosynthetic pathways have not been demonstrated in vivo. Determination of gene function based on heterologous expression is always circumspect but is especially problematic for the PBANR given the crossreacting pheromonotropic activities of peptides containing the essential FXPRLamide sequence (2, 8). Indeed, similar EC50 values were reported for the Helicoverpa zea PBANR in calcium influx assays after stimulation with FXPRLamide peptides (14).
The gene knockdown effects of RNA interference (RNAi) can resolve these types of ambiguity, thereby allowing the unequivocal assignment of gene function. Although RNAi has been well documented in dipterans, reports detailing application of this methodology to lepidopterans are sparse (15–20). Here we provide evidence for the potential of RNAi to dissect the molecular interactions that constitute biosynthetic pathways and demonstrate the in vivo biological relevance of the pgACBP, mgACBP, Bmpgdesat1, pgFAR, and PBANR gene products as they relate to pheromonogenesis in B. mori. We also show that PBANR is indeed the GPCR that mediates initiation of the pathway and that pgACBP is critical for incorporation of the pheromone precursor fatty acyl groups into the TGs that comprise the cytoplasmic lipid droplets.
Results
In B. mori, PBAN interacts with its cognate receptor to stimulate an influx of extracellular calcium that activates an enzymatic cascade, which in turn regulates the terminal reductive step in bombykol production. In recent years, a number of genes have been identified that are thought to play important roles in the bombykol biosynthetic pathway, including pgFAR, Bmpgdesat1, mgACBP, pgACBP, and PBANR. Although molecular characterization of these genes has indicated functionalities consistent with a role in the bombykol biosynthetic pathway, there has been no demonstration of the functional relevance of these gene products in vivo. Previous studies have shown that mRNA transcripts corresponding to these genes accumulate 1 day before adult emergence. Given this transcriptional timeframe, we reasoned that if these gene products are indeed crucial components of the bombykol biosynthetic pathway, then injecting pupae with the corresponding dsRNAs would result in reduced bombykol production. To test this, pupae from different developmental stages were injected with 5 μg of dsRNA corresponding to pgACBP, mgACBP, pgFAR, or Bmpgdesat1 and maintained under normal conditions until adult emergence. We assessed bombykol production from the RNAi-treated females after decapitation, overnight clearance of endogenous PBAN, and injection of 5 pmol of synthetic PBAN and compared bombykol production with control pupae injected with diethyl pyrocarbonate (DEPC)-treated H2O. Disruption of the targeted genes had no effect on pupal development or on adult emergence but did affect bombykol production, with the most pronounced effects observed when pupae were injected 1 day after the larval–pupal molt (see Fig. 8, which is published as supporting information on the PNAS web site).
Dose-Dependent Reduction in Bombykol Production.
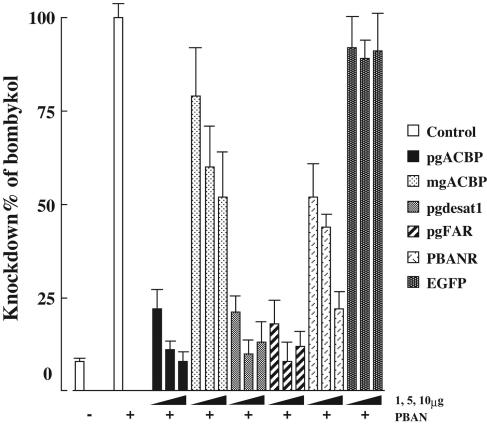
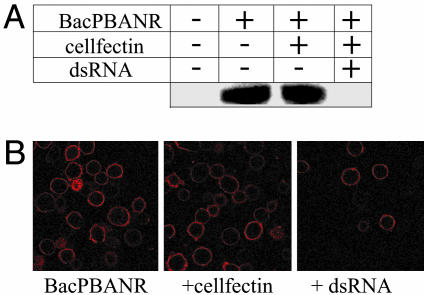
We expanded on the above findings by injecting 1-day-old pupae with varying concentrations (1, 5, and 10 μg) of dsRNAs corresponding to the full length pgFAR mRNA; the ORFs of Bmpgdesat1, mgACBP, and pgACBP; and nucleotides 379–796 of the PBANR ORF. For controls, pupae were injected with either DEPC-treated H2O or dsRNA corresponding to enhanced GFP. In all cases, disruption of the targeted gene resulted in a significant dose-dependent reduction in bombykol production, albeit with varying levels of efficacy (Fig. 1). Injection of dsRNAs corresponding to pgACBP, Bmpgdesat1, and pgFAR elicited the largest reduction in bombykol, from 77% reduction with 1-μg injections to >90% reduction with 10-μg injections. Although the gene silencing effects of dsRNAs corresponding to mgACBP and PBANR were not as pronounced, significant reduction was achieved with 10-μg dsRNA injections, 50% reduction for mgACBP, and 80% reduction for PBANR. No decrease in bombykol production was observed in the GFP dsRNA controls, indicating that disruption of bombykol production was specific to the dsRNA sequence. The variances observed in the efficacy of the injected dsRNAs, in particular the mgACBP and PBANR dsRNAs, could be an indication that the sequences used to generate them are not as suitable for promoting gene silencing (21–23). To investigate the quality of the PBANR dsRNA, we examined its gene silencing ability in vitro by using BmN cells infected with baculovirus containing a modified PBANR gene with an N-terminal His-tag (BacPBANR). Western blot analysis of infected cells harvested 48 h postinfection (h.p.i.) showed that overnight incubation with 50 nM PBANR dsRNA effectively abolished PBANR expression (Fig. 2A). These results were confirmed in binding experiments by using a rhodamine red-conjugated analog of PBAN, because infected cells incubated with PBANR dsRNA showed a marked reduction in binding (Fig. 2B), and suggest that, in the case of PBANR, the variance in bombykol production is likely attributable to poor dsRNA uptake.
Fig. 1.
Dose-dependent effects of RNAi treatment on in vivo bombykol production. One-day-old pupae were injected with 1, 5, and 10 μg of dsRNAs corresponding to pgACBP, mgACBP, Bmpgdesat1, pgFAR, PBANR, or EGFP. Results are expressed as percent reduction in bombykol levels compared with non-RNAi-treated females after intraabdominal injections of 5 pmol of PBAN. Bars represent mean values ± SD (n = >9).
Fig. 2.
Silencing of recombinant PBANR. (A) Western blot analysis. BacPBANR-infected BmN cells were transfected at 3 h.p.i. with cellfectin ± 50 nM PBANR dsRNA overnight. At 48 h.p.i., cell lysates were immunoblotted and probed with an anti-His antibody. Uniformity of protein loading was confirmed by Coomassie stain. (B) Binding of a fluorescent PBAN analog. BmN cells treated as before were incubated with 50 nM rhodamine red-labeled PBAN for 1 h at 4°C. Cells were then fixed and imaged with a confocal fluorescence microscope.
Altered Transcript Levels.
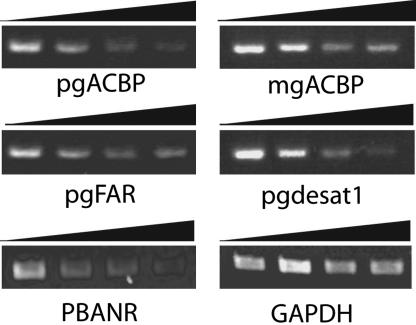
To confirm that the dsRNA injections had affected the transcript abundance of the targeted genes, RT-PCR analyses were performed by using ≈500 ng of total RNA prepared from newly emerged adults ± RNAi treatment (1, 5, and 10 μg). In all cases, the gene in question exhibited a dose-dependent reduction in transcript levels compared to non-RNAi-treated controls (Fig. 3). The reduction in transcript levels was limited to the genes targeted, because levels of a low expression control gene, GAPDH, remained uniform despite the introduction of dsRNAs. Transcript levels were not affected in PGs treated with GFP dsRNA, even at 10 μg per injection (data not shown), further supporting that the phenotypes observed in the RNAi-treated PGs were specific to the targeted genes.
Fig. 3.
Dose-dependent RNAi-induced suppression of targeted gene transcripts. Transcript analysis was performed by using cDNAs prepared from the total RNA of pupae injected with 0, 1, 5, or 10 μg of dsRNAs corresponding to pgACBP, mgACBP, pgFAR, Bmpgdesat1, or PBANR. Bar over images indicates increasing amounts of dsRNA injected. Transcript levels of the low expressing gene GAPDH are shown to demonstrate specificity of the RNAi-induced suppression.
Fluctuation and Accumulation of Cytoplasmic Lipid Droplets.
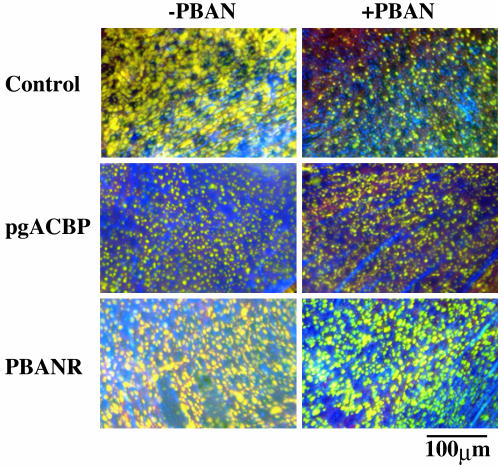
Previous reports have shown that accumulation of the cytoplasmic lipid droplets that permeate the pheromone-producing cells fluctuates in accordance with the daily bombykol titer (5, 10), and that these droplets are predominantly composed of bombykol precursors (11). When stained with the fluorescent lipid marker, Nile red, numerous large droplets that decreased in size and number after PBAN injection were observed in the cytoplasm of PGs prepared from non-RNAi-treated pupae (Fig. 4). In contrast, PBAN had no effect on lipid droplet dynamics in glands prepared from pupae treated with PBANR dsRNA (Fig. 4), indicating that PBANR is indeed the GPCR-mediating bombykol biosynthesis as well as confirming earlier studies (5, 10) that suggested PBAN mediated liberation of the bombykol precursors from the lipid droplets. Lipid droplets in PGs prepared from pupae treated with pgACBP dsRNA were noticeably smaller than those observed in control glands (Fig. 4), supporting the role proposed previously that pgACBP donates the acyl-CoAs required for expansion of the lipid droplets (13). In addition, there was no PBAN-induced depletion of the lipid droplets, an indication that the substrate hydrolyzed by the PBAN-activated lipase is not present in the lipid droplets. Taken together, these results demonstrate the essential role that pgACBP plays in bombykol precursor storage.
Fig. 4.
Accumulation and fluctuation of cytoplasmic lipid droplets. Images are pheromone-producing cells in the PG of 1-day-old decapitated females treated as pupae with DEPC-H2O (control), 5 μg of pgACBP dsRNA, or 10 μg of PBANR dsRNA. Images indicate ± PBAN treatment. Lipid droplets were stained with the fluorescent lipid marker, Nile red.
TG Content of Lipid Droplets.
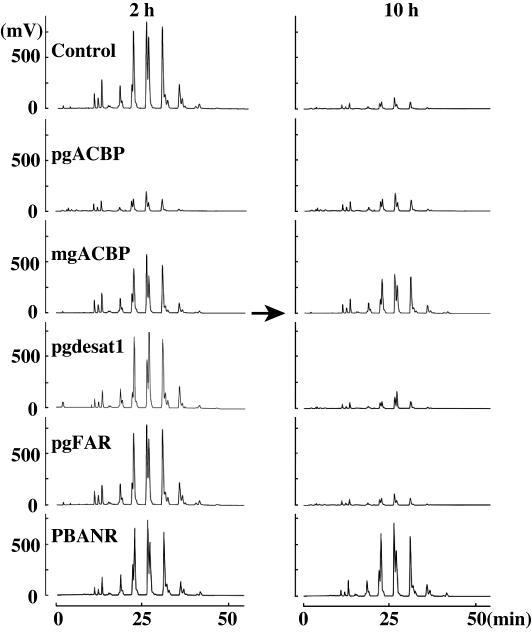
Previously, Matsumoto et al. (11) reported that the bombykol precursors are stored as fatty acid esters in the TGs that comprise the cytoplasmic lipid droplets. Consequently, we sought to determine the effects of RNAi treatment on the accumulation and fluctuation of these TGs. PGs from RNAi-treated pupae were dissected at 2 and 10 h postemergence, and the lipid components were analyzed by HPLC on a PEGASIL-Silica column. In agreement with previous results (11), the lipid droplet TG component eluted collectively over a 5-min period between 13 and 18 min (see Fig. 9A, which is published as supporting information on the PNAS web site). RNAi-treated samples exhibited a TG elution profile similar to that of the control at 2 h postemergence; however, overall TG content was reduced ≈75% in the pgACBP dsRNA-treated samples (see Fig. 9B). This pgACBP dsRNA-mediated reduction supports our earlier observation regarding lipid droplet formation (Fig. 4) and is a clear indication that pgACBP is necessary for TG accumulation. By 10 h postemergence, the stored TGs had been depleted, presumably from PBAN-induced hydrolysis, because the TG levels in the PBANR dsRNA-treated samples remained high.
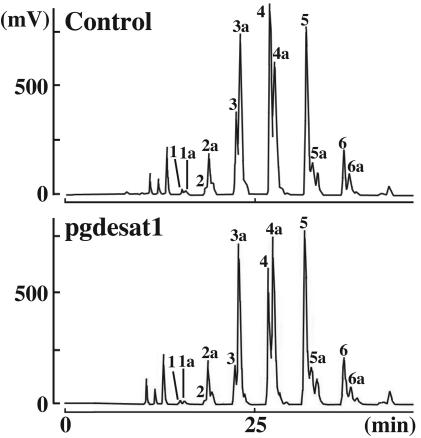
To assess the effects of gene silencing on the incorporation of individual fatty acyl components, the lipid droplet TG components from PGs dissected at 2 and 10 h postemergence were separated by using reversed-phase HPLC on a C22 docosil column. In agreement with the previous separation, the TG components from the RNAi-treated samples exhibited elution profiles similar to that of the non-RNAi-treated control (Fig. 5) and, as indicated by the reduction in peak size observed in the samples taken 10 h postemergence, underwent PBAN-induced depletion. Tandem MS analysis confirmed that the three doublet peaks exhibiting retention times in the chromatographic trace of 22–23, 26–27, and 31 min correspond to TGs containing the immediate pheromone precursor, Δ10,12-hexadecadienoate (C16:2). These peaks are dramatically reduced in the pgACBP RNAi-treated samples (Fig. 5), further demonstrating that the necessary accumulation of pheromone precursors is inhibited with the introduction of pgACBP dsRNA. Similarly, in the chromatographic traces shown in Fig. 6, the peaks corresponding to the TGs eluting at 22 and 26 min (shown as peaks 3 and 4, respectively) are significantly smaller in the Bmpgdesat1 dsRNA-treated samples compared with the corresponding TG species in the control samples (for specific peak areas, see Table 1, which is published as supporting information on the PNAS web site). These results provide a strong indication that the availability of the C16:2 bombykol precursor for sequestration and subsequent accumulation within the TGs comprising the PG lipid droplets is severely limited after Bmpgdesat1 silencing. The increased area of peak 4a (predominantly C18:Acyls) in the Bmpgdesat1 silenced as compared with the control samples suggests that the decreased availability of C16:2 results in a greater percentage of the C18:Acyl pool being made available for incorporation into the lipid droplet TGs. These findings are in agreement with the in vitro conclusions of Moto et al. (6), namely that Bmpgdesat1 catalyzes the conversion of palmitate to Δ10,12-hexadecadienoic acid, and suggest that the reduced bombykol production in the Bmpgdesat1 dsRNA samples results from a C16:2-depleted biosynthetic pathway.
Fig. 5.
Effect of RNAi treatment on TG content. TG components from acetone-extracted PGs at 2 and 10 h postemergence ± RNAi treatment were separated by reversed-phase HPLC on a C22 docosil column. Note that the TG levels in the pgACBP knockout at 2 h postemergence are similar to those of controls at 10 h postemergence when the bombykol precursor stores have been depleted. Peaks were detected by using an evaporative light-scattering detector.
Fig. 6.
Silencing of Bmpgdesat1 decreases incorporation of C16:2 fatty acyl groups. TG components were separated on a C22 docosil column. Peaks were detected with an evaporative light-scattering detector, and identity of the TG fatty acyl components was confirmed by fast atom bombardment MS and tandem MS, as described (11).
Discussion
The extensive efforts of multiple labs studying numerous lepidopteran species have led to the development of a general scheme regarding the biosynthesis of C16:0/C18:0 fatty acyl-derived sex pheromones. Notably, the remarkable species specificity of the pheromone blends is derived from enzymatic modifications of the acetyl-CoA-derived hydrocarbon backbone (1–3). In recent years, the molecular identities of proteins believed to mediate the key steps of the pathways have been elucidated, albeit almost exclusively in a heterologous background. However, the functional relevance of these proteins has not been effectively demonstrated in vivo. The discovery that the functional expression of genes can be silenced by the introduction of the corresponding dsRNA has made it possible to unequivocally assign gene function and to determine how interconnected functionalities respond when an individual component is silenced.
Using this methodology, we examined the in vivo functional role of multiple proteins thought to be essential in the biosynthesis of bombykol, the principle sex pheromone component in B. mori. Injecting pupae 1 day after the larval-pupal molt with 1, 5, and 10 μg of dsRNAs corresponding to the full length cDNA of pgFAR, the ORFs of Bmpgdesat1, mgACBP, pgACBP, or nucleotides 379–796 of the PBANR ORF resulted in a dose-dependent knockdown of the corresponding gene transcripts (Fig. 3) as well as a reduction in bombykol production (Fig. 1). The knockdown efficacy, however, varied among the dsRNAs, with pgACBP exhibiting ≈77% reduction after 1-μg treatment, whereas 10 μg was needed to achieve a comparable level of reduction for PBANR. We speculated that this variation was the result of inefficient uptake of the dsRNAs by the pheromone-producing cells and not the result of poor suitability of the dsRNA itself, because PBANR expression was greatly reduced in BmN cells infected with baculovirus harboring the PBANR gene after incubation with PBANR dsRNA (Fig. 2). Disruption of pgACBP reduced the overall TG content of the cytoplasmic lipid droplets (Figs. 4 and 5), effectively limiting the magnitude of the precursor influx into the bombykol biosynthetic pathway, and as a consequence stalled bombykol production (Fig. 1). Silencing Bmpgdesat1 had no effect on droplet formation but did alter the fatty acyl composition of the TGs (Fig. 6), the direct result of which was decreased influx of the immediate bombykol precursor, Δ10,12-hexadecadienoic acid, into the pathway.
Depending on the species in question, PBAN regulates either the terminal reductase or a step(s) in or before fatty acid synthesis, although recent reports have suggested that lipolysis of PG TGs may also be PBAN-induced (10, 13, 24). We found that, at least in this species, PBAN does indeed drive lipase activity, because silencing PBANR prevented the depletion of the cytoplasmic lipid droplets (Fig. 4) that culminates in exhaustion of the bombykol precursors (Figs. 5 and 9). This provides clear evidence that the B. mori PBANR is the GPCR that mediates sex pheromone production and underscores the possibility that the B. mori PBANR and the H. zea PBANR (14) represent two distinct species-specific isoforms of the GPCR used to mediate sex pheromone biosynthesis. The structural differences associated with the 67-aa C-terminal extension present in the B. mori PBANR (9) but absent in the H. zea PBANR are reminiscent of the type I gonadotropin-releasing hormone receptors, in that the nonmammalian type I receptors possess a functionally important C-terminal tail that is missing in the mammalian isoforms (25, 26). They also may provide a basis for the different steps regulated by PBAN in the two species (i.e., in B. mori, the fatty acyl reductase and lipase steps versus steps involved in or before fatty acid synthesis in H. zea).
In adult females, expression of mgACBP and pgACBP is specific to the PG, with both transcripts exhibiting significant up-regulation 1 day before adult emergence (13). Because ACBP binds straight-chain (C14-C22) acyl-CoA esters with high affinity, effectively protecting them from hydrolysis (27–30), we speculated that their function within the PG is to serve as carriers or cellular deposits for the acyl-CoAs used in pheromone biosynthesis. The use of dsRNAs to disrupt pgACBP and mgACBP demonstrated that this is indeed the case, because loss of pgACBP function inhibited TG accumulation within the cytoplasmic lipid droplets (Fig. 4), effectively limiting the supply of precursors available for entry into the biosynthetic pathway (Fig. 5). The less-pronounced effects (i.e., moderate reduction in bombykol production and TG accumulation) observed with loss of mgACBP function may indicate inefficient uptake of the mgACBP dsRNA by the pheromone-producing cells or that the mgACBP sequence is unsuitable for this methodology. Alternatively, the high levels of the mgACBP transcript in the B. mori larval midgut (13) suggest that the poor knockdown effects could be an indication that the role of mgACBP in pheromone production is secondary. Given the expression profile of the Manduca sexta ACBP, which peaks during periods of active feeding and lipid transport from the midgut (31), it is likely that mgACBP functions in the transport of essential fatty acids derived from dietary lipids.
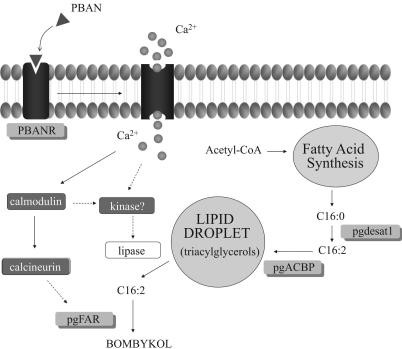
In summary, we have used RNAi-dependent gene silencing to demonstrate the in vivo relevance of the pgFAR, PBANR, Bmpgdesat1, and pgACBP gene products and confirmed their role during pheromonogenesis, thus allowing us to append our model of the bombykol biosynthetic pathway (Fig. 7). Binding of PBAN to PBANR triggers an influx of calcium that initiates a signal transduction cascade culminating in activation of the terminal reductive modification step, pgFAR, and the lipase that liberates C16:2 from the lipid droplet. Independent of PBAN action, the bifunctional Bmpgdesat1 catalyzes the reduction of acetyl-CoA-derived C16:0 (palmitate) to C16:2. The C16:2 is then incorporated into TGs by the actions of pgACBP and stored in the cytoplasmic lipid droplets. Furthermore, this work represents the initial phase of an ongoing study that proposes to use RNAi in conjunction with a PG-expressed-sequence tag database to systematically target the disruption of genes specifically expressed in the PG to facilitate molecular dissection of the bombykol biosynthetic pathway. That knowledge can then be applied to elucidating the molecular mechanisms underlying sex pheromone production in other lepidopterans.
Fig. 7.
Current model of the B. mori sex pheromone biosynthetic pathway. Gene products targeted for RNAi-mediated disruption are shown in the gray-shaded boxes. Potential sites of regulation are indicated by dashed lines.
Materials and Methods
Insects.
Larvae of the inbred P50 strain of B. mori, a gift from T. Shimada (University of Tokyo, Tokyo), were reared on mulberry leaves and maintained under a 16-h light:8-h dark photoperiod at 25°C. Pupal age was determined based on morphological characteristics as described (11).
Synthesis and Injection of dsRNA.
The templates for synthesis of dsRNAs corresponding to pgACBP, mgACBP, pgFAR, Bmpgdesat1, and PBANR were prepared by using gene-specific primers containing T7 polymerase sites and plasmids constructed previously (6, 7, 9, 13). The primer sets used are shown in Table 2, which is published as supporting information on the PNAS web site. PCR was performed by using KOD-Plus (Toyobo, Osaka) with the resulting products purified (Promega SV PCR purification kit) and used as templates to generate dsRNAs using the AmpliScribe T7 High Yield Transcription kit (Epicentre Technologies, Madison, WI), according to the manufacturer’s instructions. After synthesis, the dsRNAs were diluted with DEPC-treated H2O, the RNA concentrations measured (Abs260), and the products analyzed by gel electrophoresis to confirm annealing. Samples were diluted to the desired concentration (final volume 2–5 μl) and injected near the abdominal tip of 1-day-old pupae (i.e., pupae 1 day removed from the larval-pupal molt) using a 10-μl microsyringe (Hamilton). Control pupae were injected with 2 μl of DEPC-treated H2O alone or dsRNA corresponding to the enhanced GFP sequence (Clontech). After injection, pupae were maintained under normal conditions until adult emergence.
RT-PCR Analysis.
PGs were dissected from adult moths 3 h after emergence into insect Ringer’s solution [35 mM NaCl/36 mM KCl/12 mM CaCl2/16 mM MgCl2/274 mM glucose/5 mM Tris·HCl (pH 7.5)] and mechanically trimmed as described (32). Total RNA was isolated from the trimmed PGs by the method of Chomczynski and Sacchi (33), and first-strand cDNA synthesis was performed by using an RNA PCR kit (Takara Bio, Tokyo), according to the manufacturer’s instructions, with 500 ng of total RNA. Fragments of the genes of interest were amplified from oligonucleotide primer pairs designed from published sequences and electrophoresed on a 1.2% agarose gel in Tris/acetate/EDTA buffer.
In Vivo Bombykol Analysis.
Adult females were decapitated within 3 h of emergence and maintained at 25°C for 24 h. They were then injected with either 5 pmol (2 μl) of B. mori PBAN in PBS or PBS alone. Abdominal tips were dissected 90 min after injection, and bombykol production was measured by HPLC as described (34) by using a Senshu-PacNO2 column (Senshu Scientific, Tokyo).
RNAi in Cell Culture.
Recombinant PBANR with an N-terminal His tag was generated from pIB/PBANR (9) by using pFastBac-HT according to the manufacturer’s instructions for the Bac-to-Bac baculovirus expression system (GIBCO/BRL). The resulting plasmid, pFB/PBANR, was sequentially digested with RsrII and SpeI, and the resulting fragment was ligated into predigested (XbaI/SmaI) BmNPV transfer vector, pBm031. The resulting transfer vector, pBm-PBANR, was cotransfected into cultured B. mori BmN cells with linearized BmNPV genome using Lipofectamine (GIBCO/BRL). After 5 days, recombinant virus, BacPBANR, was plaque-purified and propagated on BmN cells. For PBANR expression, a monolayer of BmN cells was infected with 100 μl of BacPBANR for 1 h. At 3 h.p.i., cells were treated overnight at 27°C with cellfectin (Invitrogen) ± 50 nM PBANR dsRNA, rinsed, and maintained at 27°C until 48 h.p.i. Cells were harvested, and aliquots (10 μl) of total cell extracts were analyzed by 10% SDS/PAGE and then transferred to a nitrocellulose membrane. The blot was probed with an anti-His antibody (Amersham Pharmacia), incubated with goat anti-rabbit IgG conjugated to horseradish peroxidase, and developed by using an enhanced chemiluminescence plus detection kit (Amersham Pharmacia Biotech). To confirm silencing of the recombinant PBANR gene, BmN cells were treated as described above until 48 h.p.i., at which time they were incubated with a rhodamine red-labeled analog of PBAN as described (9) to assess the effects of RNAi treatment on ligand binding.
Microscopic Examination of Cytoplasmic Lipid Droplets.
Abdominal tips were dissected and mechanically trimmed from normal, decapitated, and RNAi-treated females. The excised glands were fixed with a 4% formalin/PBS solution and stained with Nile red (a fluorescent probe for intracellular neutral lipids; Molecular Probes), as described (10). Fluorescence microscopy was performed with an Olympus (Melville, NY) BX-60 system equipped with a PM-30 exposure unit and a BH20-RFL-T3 light source (×400). Nile red imaging was done with a 330- to 385-nm band-pass excitation filter, a 400-nm dichroic mirror, and a 420-nm long-pass barrier filter (Olympus cube WU). Images were processed and merged by using photoshop cs (Adobe Systems, San Jose, CA).
Analysis of Lipid Droplet Components.
Ten trimmed PGs were prepared from the desired stages of female moths and dipped in 100 μl of acetone for 10 min at room temperature. The dried acetone extracts were dissolved in n-hexane and loaded on either a Senshu-Pak PEGASIL-Silica 120–5 column (Senshu Science, Tokyo; 4.6 mm inner diameter × 250 mm) equilibrated with n-hexane/acetic acid (99/1) or a SSC-C22 docosil column (Senshu Science; 4.6 mm inner diameter × 250 mm; pore size, 300 μm) equilibrated with acetonitrile/ethanol (6/4). TG components were separated as described (11) and detected by using an evaporative light-scattering detector (model 75; SEDEX, Sedere, France). Fatty acyl groups in the TGs were identified from Fast Atom Bombardment MS, and tandem MS analyses performed by using a JEOL (Tokyo) JMS HX/HX-110A tandem mass spectrometer as described (11).
Supplementary Material
Acknowledgments
We thank Shinji Atsusawa and Masaaki Kurihara for technical assistance, Dr. Toshihiro Nagamine for assistance with virus construction, and Dr. Yasuaki Esumi for help with the MS analyses. This work was supported by the Bioarchitect Research Program and the Chemical Biology Research Program from RIKEN and by Ministry of Education, Culture, Sports, Science and Technology of Japan grants-in-aid for Scientific Research (B) (17380041).
Abbreviations
- PG
pheromone gland
- Bmpgdesat1
B. mori PG Z11/Δ10,12 desaturase
- pgFAR
PG fatty acyl reductase
- PBAN
pheromone biosynthesis activating neuropeptide
- GPCR
G protein-coupled receptor
- TG
triacylglycerol
- pgACBP
PG acyl-CoA-binding protein
- mgACBP
midgut ACBP
- PBANR
PBAN receptor
- RNAi
RNA interference
- DEPC
diethyl pyrocarbonate
- h.p.i.
hours postinfection
- bombykol
(E,Z)-10,12-hexadecadien-1-ol.
- C16:2
Δ10,12-hexadecadienoate.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jurenka R. A. In: Insect Pheromone Biochemistry and Molecular Biology: The Biosynthesis and Detection of Pheromones and Plant Volatiles. Blomquist G. J., Vogt R. G., editors. Oxford, U.K.: Elsevier Academic; 2003. pp. 53–80. [Google Scholar]
- 2.Rafaeli A. Int. Rev. Cytol. 2002;213:49–91. doi: 10.1016/s0074-7696(02)13012-9. [DOI] [PubMed] [Google Scholar]
- 3.Tillman J. A., Seybold S. J., Jurenka R. A., Blomquist G. J. Insect Biochem. Mol. Biol. 1999;29:481–514. doi: 10.1016/s0965-1748(99)00016-8. [DOI] [PubMed] [Google Scholar]
- 4.Ando T., Hase T., Funayoshi A., Arima R., Uchiyama M. Agric. Biol. Chem. 1988;52:141–147. [Google Scholar]
- 5.Fonagy A., Yokoyama N., Okano K., Tatsuki S., Maeda S., Matsumoto S. J. Insect Physiol. 2000;46:735–744. doi: 10.1016/s0022-1910(99)00162-6. [DOI] [PubMed] [Google Scholar]
- 6.Moto K., Suzuki M. G., Hull J. J., Kurata R., Takahashi S., Yamamoto M., Okano K., Imai K., Ando T., Matsumoto S. Proc. Natl. Acad. Sci. USA. 2004;101:8631–8636. doi: 10.1073/pnas.0402056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moto K., Yoshiga T., Yamamoto M., Takahashi S., Okano K., Ando T., Nakata T., Matsumoto S. Proc. Natl. Acad. Sci. USA. 2003;100:9156–9161. doi: 10.1073/pnas.1531993100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rafaeli A., Jurenka R. A. In: Insect Pheromone Biochemistry and Molecular Biology: The Biosynthesis and Detection of Pheromones and Plant Volatiles. Blomquist G. J., Vogt R. G., editors. Oxford, U.K.: Elsevier Academic; 2003. pp. 107–136. [Google Scholar]
- 9.Hull J. J., Ohnishi A., Moto K., Kawasaki Y., Kurata R., Suzuki M. G., Matsumoto S. J. Biol. Chem. 2004;279:51500–51507. doi: 10.1074/jbc.M408142200. [DOI] [PubMed] [Google Scholar]
- 10.Fonagy A., Yokoyama N., Matsumoto S. Arthropod Struct. Dev. 2001;30:113–123. doi: 10.1016/s1467-8039(01)00027-5. [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto S., Fonagy A., Yamamoto M., Wang F., Yokoyama N., Esumi Y., Suzuki Y. Insect Biochem. Mol. Biol. 2002;32:1447–1455. doi: 10.1016/s0965-1748(02)00065-6. [DOI] [PubMed] [Google Scholar]
- 12.Yoshiga T., Okano K., Mita K., Shimada T., Matsumoto S. Gene. 2000;246:339–345. doi: 10.1016/s0378-1119(00)00047-0. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto S., Yoshiga T., Yokoyama N., Iwanaga M., Koshiba S., Kigawa T., Hirota H., Yokoyama S., Okano K., Mita K., et al. Insect Biochem. Mol. Biol. 2001;31:603–609. doi: 10.1016/s0965-1748(00)00165-x. [DOI] [PubMed] [Google Scholar]
- 14.Choi M. Y., Fuerst E. J., Rafaeli A., Jurenka R. Proc. Natl. Acad. Sci. USA. 2003;100:9721–9726. doi: 10.1073/pnas.1632485100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabrick J. A., Kanost M. R., Baker J. E. J. Insect Sci. 2004;4:15. doi: 10.1093/jis/4.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bettencourt R., Terenius O., Faye I. Insect Mol. Biol. 2002;11:267–271. doi: 10.1046/j.1365-2583.2002.00334.x. [DOI] [PubMed] [Google Scholar]
- 17.Rajagopal R., Sivakumar S., Agrawal N., Malhotra P., Bhatnagar R. K. J. Biol. Chem. 2002;277:46849–46851. doi: 10.1074/jbc.C200523200. [DOI] [PubMed] [Google Scholar]
- 18.Quan G. X., Kanda T., Tamura T. Insect Mol. Biol. 2002;11:217–222. doi: 10.1046/j.1365-2583.2002.00328.x. [DOI] [PubMed] [Google Scholar]
- 19.Hirai M., Terenius O., Li W., Faye I. Insect Mol. Biol. 2004;13:399–405. doi: 10.1111/j.0962-1075.2004.00497.x. [DOI] [PubMed] [Google Scholar]
- 20.Vermehren A., Trimmer B. A. J. Neurobiol. 2005;62:289–298. doi: 10.1002/neu.20088. [DOI] [PubMed] [Google Scholar]
- 21.Harborth J., Elbashir S. M., Vandenburgh K., Manninga H., Scaringe S. A., Weber K., Tuschl T. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 22.Arziman Z., Horn T., Boutros M. Nucleic Acids Res. 2005;33:W582–W588. doi: 10.1093/nar/gki468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holen T., Amarzguioui M., Wiiger M. T., Babaie E., Prydz H. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster S. P. J. Insect Physiol. 2001;47:433–443. doi: 10.1016/s0022-1910(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 25.McArdle C. A., Franklin J., Green L., Hislop J. N. J. Endocrinol. 2002;173:1–11. doi: 10.1677/joe.0.1730001. [DOI] [PubMed] [Google Scholar]
- 26.Pawson A. J., Katz A., Sun Y. M., Lopes J., Illing N., Millar R. P., Davidson J. S. J. Endocrinol. 1998;156:R9–R12. doi: 10.1677/joe.0.156r009. [DOI] [PubMed] [Google Scholar]
- 27.Mogensen I. B., Schulenberg H., Hansen H. O., Spener F., Knudsen J. Biochem. J. 1987;241:189–192. doi: 10.1042/bj2410189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen J. T., Faergeman N. J., Kristiansen K., Knudsen J. Biochem. J. 1994;299:165–170. doi: 10.1042/bj2990165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen J. T., Rosendal J., Knudsen J. Biochem. J. 1993;292:907–913. doi: 10.1042/bj2920907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhuiyan A. K., Pande S. V. Mol. Cell. Biochem. 1994;139:109–116. doi: 10.1007/BF01081733. [DOI] [PubMed] [Google Scholar]
- 31.Snyder M. J., Antwerpen R. V. Cell Tissue Res. 1997;288:177–184. doi: 10.1007/s004410050804. [DOI] [PubMed] [Google Scholar]
- 32.Ozawa R., Matsumoto S. Insect Biochem. Mol. Biol. 1996;26:259–265. doi: 10.1016/0965-1748(95)00088-7. [DOI] [PubMed] [Google Scholar]
- 33.Chomczynski P., Sacchi N. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto S., Kitamura A., Nagasawa H., Kataoka H., Orikasa C., Mitsui T., Suzuki A. J. Insect Physiol. 1990;36:427–432. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.