Abstract
We have used a bioinformatic approach to predict the natural substrate proteins for the Escherichia coli chaperonin GroEL based on two simple criteria. Natural substrate proteins should contain binding motifs similar in sequence to the mobile loop peptide of GroES that displaces the binding motif during the chaperonin cycle. Secondly, each substrate protein should contain multiple copies of the binding motif so that the chaperonin can perform “work” on the substrate protein. To validate these criteria, we have used a database of 252 proteins that have been experimentally shown to interact with the chaperonin machinery in vivo. More than 80% are identified by these criteria. The binding motifs of all 79 proteins in the database with a known three-dimensional structure are buried (<50% solvent-accessible surface area) in the native state. Our results show that the binding motifs are inaccessible in the native state but become solvent-exposed in unfolded state, thus enabling GroEL to distinguish between unfolded and native states. The structures of the binding motif in the native states of the substrate proteins include α-helices, β-strands, and random coils. The diversity of secondary structures implies that there are large and varied conformational transitions in the recognition motifs after their displacement by the mobile loops of GroES.
Keywords: chaperonin, E. coli, natural substrates, recognition motif
It is illogical to employ the structural features of native proteins, with which the chaperonin GroEL does not interact, to define the recognition elements of incompletely folded proteins, with many, but not all, of which the chaperonin does interact. Instead, to determine what enables this interaction to occur, let us enumerate the characteristics of the chaperonin itself and of those incompletely folded substrate proteins with which it interacts.
An important property of the chaperonin GroEL is its ability to discriminate between the nonnative states of many proteins to which it binds and the native states of these proteins to which it does not bind (1, 2). It was suggested that the substrate protein binding motif (SPBM) with which GroEL interacts are accessible in the nonnative state of the substrate protein (SP) but become inaccessible during the transition to the native state (1). Until now, this conjecture has remained unproven because the nature of the SPBM has yet to be defined.
Except for small proteins, the stoichiometry of SP binding is one SP per chaperonin ring of seven subunits. To form a stable binary complex between GroEL and SP, each ring must contain three to four functional SP binding sites (3). Thus, SP binding is multivalent, which would require the presence of multiple SPBM in each SP.
An important feature of the chaperonin cycle is the dispersion of the SP binding sites (4). Recent evidence suggests that the chaperonin performs “work” on the SP during this movement of the SP binding sites (5), consistent with the iterative annealing mechanism (6). For work to be performed on the SP, it is axiomatic that the SP be bound at two or more sites, which in turn requires that the SP contain two or more SPBM. If the number of binding motifs is two, then for “work” to be performed during GroEL's allosteric changes two adjacent motifs should be separated minimally by a “spacer” of ≈10 residues (7).
The chaperonin GroEL is distinctly promiscuous. In vitro it can form stable binary complexes with 40% of the soluble proteins in Escherichia coli, provided that these are presented to GroEL in a nonnative state (2). Even artificial proteins composed of random sequences form stable binary complexes with GroEL (8). The SPBM must consequently be common and present in multiple forms in many proteins.
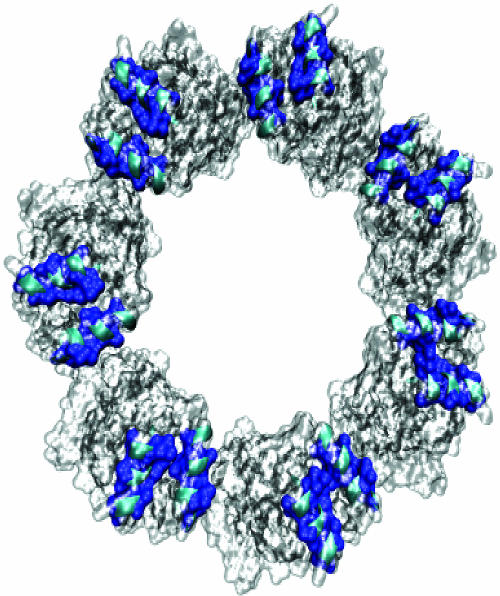
Crystallographic (9, 10) and mutational studies (3) show that the SP binding site is formed from the cleft between helices H and I in the apical domain of GroEL. This cleft is largely hydrophobic in nature (Fig. 1). In support of this, a study of peptides bound to this site on GroEL (11) showed that binding of the peptides correlated with their ability to form hydrophobic clusters as determined by retention time on reversed-phase HPLC.
Fig. 1.
Hydrophobic (blue) surfaces within the GroEL (gray) binding sites (top view) formed by helices (cyan) H (residues 234–243) and I (257–268). This image was created by using vmd (28) and pov-ray (www.povray.org).
During the chaperonin cycle the SP is transiently encapsulated (12) in the central cavity of GroEL underneath the GroES “lid.” During encapsulation the SP recognition motifs that were initially bound in the cleft between helices H and I are displaced by the mobile loop peptides of GroES (13). Because they share the same binding site, it is reasonable to suppose that the SPBM bears some structural resemblance to the mobile loop of GroES.
Recently we developed a bioinformatic approach to identify the natural SPs for GroEL, based on the idea that natural SPs are likely to contain patterns of residues similar to those found in the mobile loop of GroES (7). The method was validated by comparing the predicted results with experimentally determined natural SPs for GroEL (14). The precise number of natural SPs was shown to be a function of the number of contacts an SP makes with the apical domain (NC) and the number of binding sites (NB) in the GroEL with which it interacts. For known SPs for GroEL, we find 4 ≲ NC ≲ 5 and 2 ≤ NB ≤ 4. This sequence-based approach did not permit us to address the important question as to whether these multiple SPBM were accessible in the native structure. For that purpose we required a database of known GroEL SPs whose three-dimensional structure has been determined. Recently two data sets of natural chaperonin SPs with known structure were reported; one from Thermus thermophilus (15) and one from E. coli (16). We have used the larger E. coli data set to evaluate the accessibility of the SPBM. Here we report that all of the SPBM of natural SPs for GroEL, identified by our sequence-based approach, are inaccessible in the native structures of these proteins, further confirming the validity of this approach.
Results and Discussion
Conformational Change of GroES Mobile Loop upon Binding to GroEL.
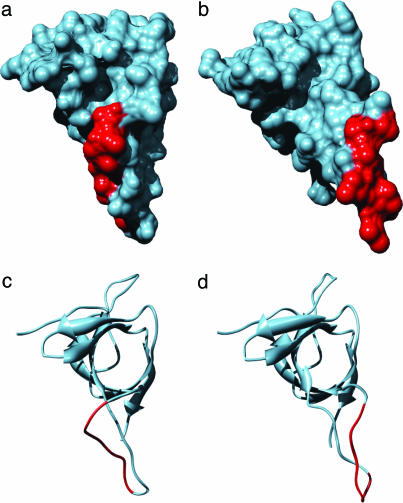
The flexible GroES mobile loop, shown in Fig. 2 c and d, binds to GroEL with NC = 6 contacts within the binding motif G_IVL_G_A. In addition to the large solvent-accessible surface (Eq. 1 in Methods) of the binding region (rs = 0.52), interaction with GroEL involves a conformational change of the mobile loop. The disordered mobile loop is structured upon binding between the crevices of helices H and I of GroEL. The large conformational change results in an optimal shape of the binding region (Fig. 2b) to fit in the groove-like binding site of GroEL. Binding of the GroES mobile loop results in an increase in surface area accessible to GroEL, rs = 0.68, of which 0.38 is buried at the GroEL–GroES interface.
Fig. 2.
GroES binding motif GGIVLTGAA, residues 23–31 (red), in the unbound [molecular surface representation (a) and ribbons (c)] and bound (b and d) structures. The images in Figs. 2 and 5 were produced by using the chimera program (29) based on solvent-excluded molecular surfaces determined by using the msms package (30).
SPBM from E. coli Are Inaccessible to Solvent in the Native State.
We performed bioinformatic and structural analysis of the set of 252 E. coli proteins that have been suggested to interact with GroEL under normal cellular conditions (16). Our sequence-based method (7) identifies natural substrates of GroEL based on the hypothesis that they possess 2 ≤ NB ≤ 4 SPBM that contain 4 ≲ NC ≲ 6 GroES-like binding contacts (see Methods for details). Almost all of these proteins (94%) contain multiple copies of the core SPBM, namely the highly conserved GroES pattern G_IVL (NC = 4). With NC = 6, which is the number of contacts the mobile loop of GroES makes with GroEL, we find that ≈27% of the Kerner et al. substrates contain multiple GroES-type SPBM. More than 80% of these 252 proteins are identified as natural GroEL substrates according to the above (NB, NC) criterion. The remaining 20% proteins are depleted in motifs with NC = 5 or 6 (Table 1). Consequently, it is to be expected that this set of proteins containing shorter GroES-type motifs, NC = 4, would be more weakly bound to GroEL.
Table 1.
Natural substrates that interact with GroEL
The complete list of 284 proteins, including 32 nonspecific interacting SPs, with GroES-type SPBM is given in Table 4, which is published as supporting information on the PNAS web site. The number of motifs is calculated without regard to the separation between them.
*Minimum number of SPBM found in the protein sequence.
†Number of GroES-type binding contacts, NC.
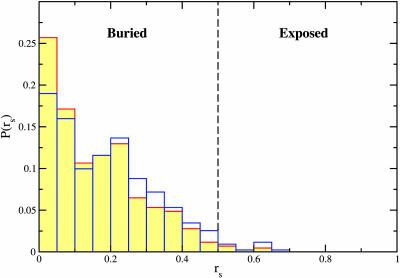
Analysis of the available native conformations of proteins with known three-dimensional structures in the Kerner et al. set (K3d), in conjunction with our bioinformatic analysis, sheds light on SP conformational requirements for interaction with GroEL. Because of the uncertainty in the prediction of structures based on homology modeling, we restricted ourselves to the subset of 79 proteins for which high-resolution structures are available. Recognition of protein substrates by GroEL requires that SPBM occupy a mostly hydrophobic groove in the binding site of one of the seven subunits of a GroEL ring. Formation of a single GroEL–SP interface involves NC = (4–6) SP residues making contact with GroEL. We infer that only fully solvent-exposed SPBM can interact most favorably with GroEL so that the thermodynamic stability criterion is satisfied. According to our criterion (see Methods), SPBM are GroEL-accessible if their average relative solvent-accessible surface area per residue (rs) exceeds 0.5. In Fig. 3we show the probability distribution, P(rs), of GroEL-accessible area of the putative SPBM found in 79 proteins in the K3d set. Table 5, which is published as supporting information on the PNAS web site, contains the precise values of rs and the identification of the SPBM for all of the SPs in the K3d set. Only 11 of 432 putative GroES-type SPBM, or 3%, are found to be GroEL-accessible in the native structure of these proteins. A subset of the GroEL-accessible SPBM is buried in the oligomeric interfaces, resulting in only 6 (1%) SPBM in oligomers capable of interacting with GroEL. Using these results, we assert that the SP conformations suitable for interaction with GroEL are those that contain multiple solvent-exposed SPBM. In agreement, none of the native structures in the K3d set satisfy this criterion for interaction with GroEL.
Fig. 3.
Probability distribution of average relative solvent-accessible surface area of GroES-type SPBM in native conformations of 79 proteins suggested to interact with GroEL (16). For biological oligomers, multiple chains are included (red contours) or excluded (blue).
Perturbation of SPBM Does Not Alter the GroEL Accessibility of SPBM in the Native State.
We consider the effect of variations in the GroES-type SPBM on the GroEL-accessibility of putative SPBM. Based on the sequence alignment of the bacteriophage T4 co-chaperonin Gp31 and GroES (17), we infer a Gp31-type SPBM G_IVL_K_A (P_HHH_+_H). The single substitution of a polar residue in the GroES-type motif by a charged residue dramatically reduces the number of long SPBM, i.e., those that contain NC = 5 or 6 contacts of the Gp31-type motif (P_HHH_+ and P_HHH_+_H, respectively). In the K3d set, we find 55 long putative Gp31-type SPBM compared with 171 GroES-type ones. Nevertheless, in both cases, only a few of the long putative binding regions are GroEL-accessible in the native structures of the K3d proteins, i.e., 8 (2%) Gp31-type patterns (in both oligomeric and monomeric structures) and 4 (1%) GroES-type patterns in monomeric structures (2% or 0% in oligomeric structures). Despite the larger solvent exposure of long Gp31-type patterns compared with the GroES-type patterns, only 20 of 435 (5%) of Gp31-type SPBM in the K3d set are GroEL-accessible in native monomeric structures and 17 (4%) in native oligomeric structures.
Substrate Proteins with TIM-Barrel Fold Are Not Preferred by GroEL.
We focus on the suggested overexpression of (βα)8 TIM-barrel fold among the set of GroEL interactors (16). GroEL preference for a particular fold should be correlated with an excess of GroES-type motifs within this fold. Table 2shows the number of GroES-type motifs found in TIM-barrel GroEL interactors and in the overall set of suggested GroEL interactors (16). We find that the subset of 31 putative GroEL interactors assigned to the TIM-barrel fold class has an overall smaller number of predicted SPBM per protein than the overall set of 252 interactors. Although slightly more (8%) exact GroES SPBM (G_IVL_G_A) are found in the TIM-barrel protein set, shorter SPBM (G_IVL) are 40% more likely in the complete set of suggested GroEL interactors.
Table 2.
Binding motifs in substrate proteins identified in ref. 16
| Class of SPs | GroES pattern matches |
||
|---|---|---|---|
| 6 | 5 | 4 | |
| TIM-barrel class* | 36 (1.2)† | 75 (2.4) | 180 (5.8) |
| All‡ | 277 (1.1) | 729 (2.9) | 1,771 (7.1) |
The total number of SPs is 252.
*Includes only the 31 that have TIM-barrel architecture.
†The number in parentheses indicates the number of SPBM per protein in each set.
‡All 252 proteins are taken into account.
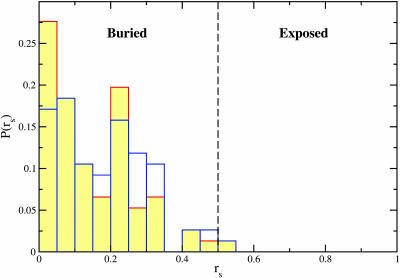
We examined the possibility that TIM-barrel protein SPBM, although not overexpressed, are more readily available for interaction with GroEL. For this purpose, we determined the accessibility of putative SPBM within the 18 TIM-barrel proteins with known native structure (TIM3d). The probability distribution of accessible area per motif in TIM3d, as in K3d, is dominated by motifs buried in the monomeric (oligomeric) structure (Fig. 4). A single SP binding motif is accessible to GroEL in TIM3d in monomeric (oligomeric) structures. In view of the paucity of SPBM on the surface of the native state conformation, we surmise that the native TIM-barrel fold does not increase the likelihood of interaction with GroEL. Denatured states of the TIM-barrel proteins capable of interacting with GroEL, i.e., those that expose multiple GroES-type SPBM, are likely to bear little structural similarity to the TIM-barrel class. In conclusion, our results suggest that there is no correlation between the TIM-barrel fold and the SP ability to interact with GroEL.
Fig. 4.
Same as Fig. 3 for proteins with known three-dimensional TIM-barrel native structure.
Other Known GroEL Substrates Bury SPBM in Their Native State.
We also examined the GroEL accessibility of binding motifs in the native structures among a set of known in vitro GroEL substrates (SP3d). In Table 3we show the number of GroES-type SPBM found in this set of SPs and the number of GroEL-accessible SPBM. All of the GroEL substrates considered contain multiple GroES-type SPBM (up to 11 in the case of Rubisco), which indicates a strong dependence of folding of these proteins on chaperonin assistance. Indeed, the propensity to undergo self-aggregation may be causally related to the number of SPBM. Both the cytoplasmic and mitochondrial malate dehydrogenase (cMDH and mMDH) contain six GroES SPBM. In contrast to mMDH, cMDH interacts only weakly in vitro with GroEL and does not require GroES for release from GroEL (18). It is likely that cMDH does not satisfy the kinetic criterion (see below). A careful analysis of the stability of cMDH–GroEL complex will be needed to resolve the differences between cMDH and mMDH preference for GroEL.
Table 3.
Known GroEL substrate proteins
| Protein | Swiss-Prot ID | PDB* | Organism | NB† | NM‡ | NO§ |
|---|---|---|---|---|---|---|
| Rubisco | RBL2_RHORU | 5RUB | Rhodospirillum rubrum | 11 | 0 | 0 |
| Bacterial luciferase α-chain | LUXA_VIBHA | 1LUC | Vibrio harveyi | 8 | 0 | 0 |
| Bacterial luciferase β-chain | LUXB_VIBHA | 1LUC | Vibrio harveyi | 4 | 0 | 0 |
| mMDH | MDHM_PIG | 1MLD | Sus scrofa | 6 | 0 | 0 |
| cMDH | MDHC_PIG | 5MDH | Sus scrofa | 6 | 1 | 0 |
| Subtilisin BPN′ precursor | SUBT_BACAM | 1GNS | Bacillus amyloliquefaciens | 4 | 0 | 0 |
| Barnase | RNBR_BACAM | 1A2P | Bacillus amyloliquefaciens | 2 | 0 | 0 |
| Dihydrofolate reductase | DYR_CHICK | 8DFR | Gallus gallus | 4 | 0 | 0 |
| Lysozyme C precursor | LYSC_CHICK | 1H87 | Gallus gallus | 3 | 0 | 0 |
| Rhodanese | THTR_BOVIN | 1BOI | Bos taurus | 6 | 0 | 0 |
| α-Lactalbumin precursor | LALBA_HUMAN | 1B9O | Homo sapiens | 5 | 1 | 1¶ |
| α-Lactalbumin precursor | LALBA_BOVIN | 1F6R | Bos taurus | 5 | 0 | 0‖ |
*Protein Data Bank ID code of substrate proteins.
†Number of GroES-type SPBM for which the GroEL-accessible area is determined.
‡Number of GroEL-accessible SPBM in monomeric native structures.
§Number of GroEL-accessible SPBM in oligomeric native structures.
¶A single chain is resolved in the crystal structure of the human α-lactalbumin, a heterodimer.
‖Six chains are present in the crystallographic asymmetric unit of the bovine α-lactalbumin, a heterodimer.
The analysis of solvent-accessible surface area of predicted SPBM indicates that they are drastically restricted from interacting with GroEL if the SP is in the native conformation. As shown in Table 3, most of the predicted SPBM are deeply buried in the monomeric structures (rs < 0.5). None of the oligomeric structures allow solvent exposure of the multiple SPBM. The reduced interaction with GroEL predicted for the folded SPs examined above is consistent with the observation that native dihydrofolate reductase (DHFR) is destabilized by GroEL (19).
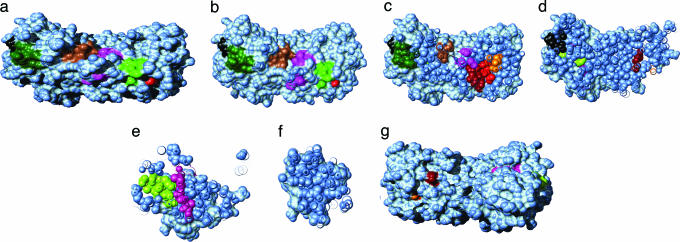
Graphical analysis of the native state of Rubisco (Fig. 5) clearly illustrates the GroEL-accessibility restrictions for residues that interact with GroEL. The SPBM with the largest solvent-accessible area, shown as contiguous-colored regions in Fig. 5 a and g, are the regions 86–90 (rs = 0.21; green), 97–105 (rs = 0.23; purple), 244–250 (rs = 0.27; dark green), and 268–274 (rs = 0.24; brown). None of these partially exposed SPBM are accessible to GroEL in the Rubisco native structure (rs < 0.5). By contrast, contiguous, non-SPBM, that have at least four residues on the Rubisco surface, expose significant surface area. For example, residues 31–37 have rs = 0.56, and residues 393–399 have rs = 0.35. Cross sections of the native structure shows that most of the binding motifs are deeply buried (Fig. 5 b–f). SPBM that are buried in Rubisco appear to be packed against each other in the native structure (Fig. 5c). These observations strongly suggest that Rubisco must denature or expose these motifs to be recognized by GroEL. In other words, the conformations recognized by GroEL are distinct from those in the folded structure.
Fig. 5.
Putative GroEL SPBM (shown as contiguous-color regions) in the native conformation of Rubisco (light blue). (a and g) Front (back) view of the protein in a solvent-accessible surface area representation. (b–f) Cross sections, 10 Å apart, of the protein perpendicular to direction of observation in a. Molecular movies of the Rubisco surface and cross sections are available at www.biotheory.umd.edu/supplementary/motifs.html.
Conclusions
An important implication of our findings is that there is an ensemble of disordered conformations that can be recognized by GroEL. Exposure of deeply buried segments of folded SPs necessarily results in expansion of the polypeptide. As a result, the ensemble of such misfolded structures must have conformational diversity that is a characteristic of random coils or molten globules. Indeed, an NMR analysis of an SP bound to GroEL (20) concluded that the conformation of the SP included random coil conformations devoid of stable higher-order structure. Instead, the SP is characterized as a dynamic ensemble of randomly structured conformers, which implies that SPBM is not composed of higher-order structures, as has been suggested (14, 16).
In addition to exposure of buried SPBM, binding to GroEL may also require unfolding of the native secondary structure. The unfolding requirement arises from the possible incompatibility of the native secondary structure with conformational restrictions imposed by helices H and I of GroEL. For example, preferred conformations of peptides bound to GroEL appear to be extended and/or loop structures (9, 13, 21), whereas native conformations are disordered (9, 13, 22). Examination of known GroEL SPs (Table 3) suggests that putative SPBM have diverse native secondary structures. For example, in the native structure of Rubisco, SPBM are found in β-strand (regions 23–27, 81–85, 124–128, 157–165, and 256–260), helix (97–105, 181–189, 207–211, 244–260, and 268–274), and random coil (86–90) structures. In the native structure of the bacterial luciferase α-chain, SPBM 4–8, 38–46 73–81, 142–148, and 169–173 are β-strands, and SPBM 55–63, 124–128, and 180–184 are helical. In the mMDH, SPBM 38–42, 101–107, 154–158, and 226–230 are helical, 109–113 is a random coil, and 266–270 is a β-strand. The diversity of native secondary structures of the predicted SPBM suggests that SPBM undergo large conformational transitions upon binding to GroEL. Moreover, the potential incompatibility between the secondary structures of bound SP and the native fold implies that GroEL does not bias the SP to reach the native state.
The SP–GroEL complex must satisfy two necessary conditions (22). The SP–GroEL interaction must be strong enough so that the capture process results in a stable complex. However, the stability of SP–GroEL should not be so great as to make the subsequent release of the SP, which is facilitated by allosteric transitions in GroEL, improbable. Marginal stability of the SP–GroEL complex is satisfied if the interaction strength per residue, ε, is in the range kBT ≲ ε ≲ 2kBT (23). For efficient function of the chaperonin machinery the rate of SP binding to GroEL must exceed the rate of SP aggregation. In addition, the time needed for burying the exposed putative SPBM during monomeric folding should be longer than the time scale for SP–GroEL binding. It is likely that some percentage of natural SPs do not satisfy either the thermodynamic stability requirement nor the kinetic criterion. Such kinetic considerations may be the underlying reason that our structure-based approach fails to discriminate between isoforms of the same protein [e.g., cMDH and mMDH, E. coli MDH and mMDH (24)].
Methods
Identification of GroEL Natural Substrates and SPBM.
We identify the natural substrates of GroEL (7) based on the hypothesis that they contain SPBM that are similar to those in the GroES mobile loop (7) that bind to GroEL (Fig. 2 c and d). The disordered GroES mobile loop undergoes a large conformational change that results in six of its residues forming contacts with the two helices (H and I) in the GroEL binding site (13). Two residues are in contact if the closest distance between heavy atoms is <4 Å and the contact surface area is at least 20 Å2. The resulting GroES binding pattern is G_IVL_G_A, where “_” represents any residue. Because strong conservation of chemical character of residues is important for chaperonin function (25) we assume that the general pattern of residues that is recognized by GroEL should be classified by using four residue types, namely hydrophobic (H), polar (P), positively charged (+), and negatively charged (−). Using one-letter code for amino acids, the hydrophobic (H) residues are C, F, I, L, W, V, M, Y, and A, the polar (P) residues are G, P, N, T, S, Q, and H, positively charged (“+”) residues are R and K, and the negatively charged (“−”) are D and E. The resulting GroES-like SPBM is P_HHH_P_H. Variations in the length of this motif are allowed to account for dispersion of the stabilizing interaction between SPBM and GroEL. Based on the conservation behavior of residues in the GroES motif (25), which indicates that the core residues are P_HHH, we consider motifs of length NC = 5 (P_HHH), 7 (P_HHH_P), and 9 (P_HHH_P_H). To determine the likely natural substrates, we use the additional criterion that stringent substrate proteins must form contacts with multiple GroEL binding sites (3). Thus, natural SPs are those that contain multiple (NB ≥ 2) GroES-type SPBM.
GroEL-Accessible Surface Area.
The stability requirement of the SP–GroEL complex is satisfied if the residues in the SPBM have a large solvent-accessible surface area. We use the Lee and Richards algorithm to calculate the solvent-accessible area per residue (26):
where as is the accessible surface area of amino acid i (of type X) in the polypeptide chain, and a0 (X) is the reference accessible surface in an extended conformation Gly–X–Gly (27). The accessible surface area per SPBM is
 |
where N = 5, 7, 9 (corresponding to NC = 4, 5, 6) is the binding motif length. Several residues in the SPBM must expose significant surface area to solvent to allow the formation of multiple GroEL–GroES-type binding contacts (NC ≥ 4). In the GroES native structure (J. F. Hunt, personal communication), the accessible surface area of the binding region GGIVLTGAA is rs = 0.52. To identify GroEL-accessible SPBM, we use a similar criterion rs ≥ 0.5.
The use of rs ≥ 0.5 is also consistent with the requirement of marginal stability of GroEL–SP complex for efficient function. The GroEL–SP interaction must be strong enough so that the capture process results in a stable complex. However, the complex should not be hyperstable, which would render the release of SP improbable. Using theoretical arguments it has been shown that the SP–GroEL interaction strength per residue (εs) of ≈2 kBT satisfies the marginal stability requirement. Assuming NC = 6 and the surface tension γ ≈ 25 cal/(mol·Å2), the surface-area exposure based on marginal stability is εs/γ ≈ 290 Å2. This estimate is within a factor of 1.5 of the surface area exposed by the mobile loop of GroES. The approximate energetic criterion further supports our choice of rs.
Protein Sets Analyzed.
We analyze sequences and known three-dimensional structures of E. coli proteins suggested by Kerner et al. (16) to interact with GroEL. The GroEL interactome, available at http://pedant.gsf.de, contains 252 proteins of which 79 have known three-dimensional structure. We also analyzed a set of known GroEL substrates (shown in Table 3) for which the three-dimensional structures are known.
Supplementary Material
Acknowledgments
We thank Changbong Hyeon and Lee Woodcock for help with molecular graphics. This work was supported in part by National Institutes of Health Grant 1R01GM067851-01 (to G.H.L. and D.T.) and the Intramural Research Program of the National Heart, Lung, and Blood Institute/National Institutes of Health.
Abbreviations
- cMDH
cytoplasmic malate dehydrogenase
- mMDH
mitochondrial malate dehydrogenase
- SP
substrate protein
- SPBM
substrate protein binding motif
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Goloubinoff P., Gatenby A. A., Lorimer G. H. Nature. 1989;337:44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- 2.Viitanen P. V., Gatenby A. A., Lorimer G. H. Protein Sci. 1992;1:363–369. doi: 10.1002/pro.5560010308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farr G. W., Furtak K., Rowland M. R., Ranson N. A., Saibil H. R., Kirchhausen T., Horwich A. L. Cell. 2000;100:561–573. doi: 10.1016/s0092-8674(00)80692-3. [DOI] [PubMed] [Google Scholar]
- 4.Lorimer G. H. Nature. 1997;388:720–723. doi: 10.1038/41892. [DOI] [PubMed] [Google Scholar]
- 5.Motojima F., Chaudhry C., Fenton W. A., Farr G. W., Horwich A. L. Proc. Natl. Acad. Sci. USA. 2004;101:15005–15012. doi: 10.1073/pnas.0406132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Todd M. J., Lorimer G. H., Thirumalai D. Proc. Natl. Acad. Sci. USA. 1996;93:4030–4035. doi: 10.1073/pnas.93.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stan G., Brooks B. R., Lorimer G. H., Thirumalai D. Protein Sci. 2005;14:193–201. doi: 10.1110/ps.04933205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoki K., Motojima F., Taguchi H., Yomo T., Yoshida M. J. Biol. Chem. 2000;275:13755–13758. doi: 10.1074/jbc.275.18.13755. [DOI] [PubMed] [Google Scholar]
- 9.Chen L. L., Sigler P. B. Cell. 1999;99:757–768. doi: 10.1016/s0092-8674(00)81673-6. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q., Buckle A. M., Fersht A. R. J. Mol. Biol. 2000;304:873–881. doi: 10.1006/jmbi.2000.4276. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z., Feng H., Landry S. J., Maxwell J., Gierasch L. M. Biochemistry. 1999;38:12537–12546. doi: 10.1021/bi991070p. [DOI] [PubMed] [Google Scholar]
- 12.Weissman J. S., Hohl C. M., Kovalenko O., Kashi Y., Chen S., Braig K., Saibil H. R., Fenton W. A., Horwich A. L. Cell. 1995;83:577–587. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]
- 13.Xu Z., Horwich A. L., Sigler P. B. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 14.Houry W. A., Frishman D., Eckerskorn C., Lottspeich F., Hartl F. U. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- 15.Shimamura T., Yokoyama A. K. K., Masui R., Murai N., Yoshida M., Taguchi H., Iwata S. Structure. 2004;12:1471–1480. doi: 10.1016/j.str.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 16.Kerner M. J., Naylor D. J., Ishihama Y., Maier T., Chang H., Stines A. P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Hartl F. U. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 17.Hunt J. F., van der Vies S. M., Henry L., Deisenhofer J. Cell. 1997;90:361–371. doi: 10.1016/s0092-8674(00)80343-8. [DOI] [PubMed] [Google Scholar]
- 18.Staniforth R. A., Cortés A., Burston S. G., Atkinson T., Holbrook J. J., Clarke A. R. FEBS Lett. 1994;344:129–135. doi: 10.1016/0014-5793(94)00348-3. [DOI] [PubMed] [Google Scholar]
- 19.Viitanen P. V., Donaldson G. K., Lorimer G. H., Lubben T. H., Gatenby A. A. Biochemistry. 1991;30:9716–9723. doi: 10.1021/bi00104a021. [DOI] [PubMed] [Google Scholar]
- 20.Horst R., Bertelsen E. B., Wider J. F. G., Horwich A. L., Thrich K. W. Proc. Natl. Acad. Sci. USA. 2005;102:12748–12753. doi: 10.1073/pnas.0505642102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buckle A. M., Zahn R., Fersht A. R. Proc. Natl. Acad. Sci. USA. 1997;94:3571–3575. doi: 10.1073/pnas.94.8.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stan G., Brooks B. R., Thirumalai D. J. Mol. Biol. 2005;350:817–829. doi: 10.1016/j.jmb.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Orland H., Thirumalai D. J. Phys. 1997;7:533–560. [Google Scholar]
- 24.Tieman B. C., Johnston M. F., Fisher M. T. J. Biol. Chem. 2001;276:44541–44550. doi: 10.1074/jbc.M106693200. [DOI] [PubMed] [Google Scholar]
- 25.Stan G., Thirumalai D., Lorimer G. H., Brooks B. R. Biophys. Chem. 2003;100:453–467. doi: 10.1016/s0301-4622(02)00298-3. [DOI] [PubMed] [Google Scholar]
- 26.Lee B., Richards F. M. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 27.Miller S., Janin J., Lesk A. M., Chothia C. J. Mol. Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 28.Humphrey W., Dalke A., Schulten K. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 29.Pettersen E., Goddard T., Huang C., Couch G., Greenblatt D., Meng E., Ferrin T. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 30.Sanner M., Olson A., Spehner J. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.