Abstract
Phosphorylation of a cluster of amino acids in the serum response factor (SRF) “MADS box” αI coil DNA binding domain regulated the transcription of genes associated with proliferation or terminal muscle differentiation. Mimicking phosphorylation of serine-162, a target of protein kinase C-α, with an aspartic acid substitution (SRF-S162D) completely inhibited SRF–DNA binding and blocked α-actin gene transcription even in the presence of potent myogenic cofactors, while preserving c-fos promoter activity because of stabilization of the ternary complex via Elk-1. Introduction of SRF-S162D into SRF null ES cells permitted transcription of the c-fos gene but was unable to rescue expression of myogenic contractile genes. Transition of proliferating C2C12 myoblasts to postfusion myocytes after serum withdrawal was associated with a progressive decline in SRF-S162 phosphorylation and an increase in α-actin gene expression. Hence, the phosphorylation status of serine-162 in the αI coil may constitute a novel switch that directs target gene expression into proliferation or differentiation programs.
Keywords: PKC, differentiation, α-actin, c-fos
Serum response factor (SRF) is a founding member of an ancient family of plant and animal DNA-binding proteins, whose relatives contain a highly conserved DNA-binding/dimerization domain of 90 aa termed the MADS box (1, 2). SRF is an obligatory transcriptional regulator of two classes of genes. One class comprises the immediate early genes, typified by c-fos and Egr1, which are transiently expressed in response to mitogenic signals (3). The other class comprises a large number of differentiation genes, typified by the skeletal, smooth, and cardiac muscle α-actins, which are activated in myocytes only after they cease proliferation and withdraw from the cell cycle (4, 5). How does SRF play such important roles in both proliferation and differentiation by regulating the transcription of functionally antagonistic sets of genes?
SRF functions as a master regulatory platform that directs different gene expression programs through combinatorial interactions with other transcription factors and cofactors. In replicating myoblasts, SRF recruits proteins having an Ets domain (Elk-1, Sap1, and Fli) and forms ternary complexes at the c-fos promoter (6–8). For the most part, Ets factor-associated ternary complexes do not play a role in regulating transcription of SRF-dependent myogenic gene targets (9, 10). Instead, upon serum withdrawal, SRF associates with coaccessory transfactors such as Nkx 2.5 (11), GATA4 (12), LIM-only factor CRP2 (13), myocardin (14–16), MAL (17), and MKL1 and MKL2 (18, 19) to activate transcription of SRF myogenic target genes, but does so only weakly in replicating myoblasts. What are the triggers that switch SRF from modulating the proliferation pathway to modulating the differentiation pathway? Because SRF isolated from cell extracts is highly phosphorylated in numerous sites (20–23), it is possible that site-specific phosphorylation is a mechanism for regulating the function of SRF. Thus far, studies of posttranslational modifications of SRF have yet to reveal specific alterations that actually control SRF’s ability to switch “on” and “off” myogenic gene expression. Here, we present a mechanism that facilitates strong repression of SRF-dependent myogenic differentiation genes through phosphorylation of a specific, evolutionarily conserved SRF residue in the MADS box, yet allows for activation of immediate early proliferation genes.
Results
Aspartate Substitution of Serine-162 in the MADS Box αI Coil Abolished SRF-c-fos Promoter DNA Binding.
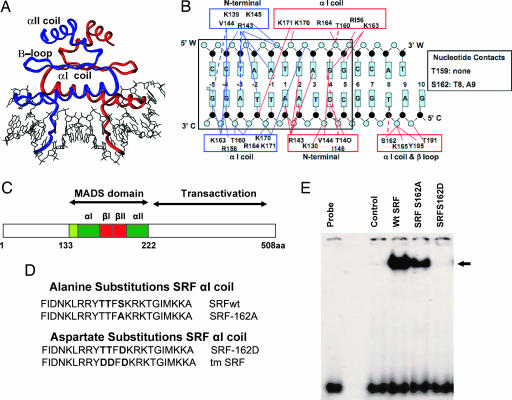
Previous experiments (24) and data from x-ray crystallographic analysis of SRF’s MADS box bound to its cognate serum response element (SRE) (ref. 25 and Fig. 1A and B) directed our attention to two potential phosphorylation sites (T159 and S162) closely grouped on the αI coil of the MADS box and well conserved across millennia from yeast to humans (Fig. 1 A–C). A monomer of SRF at S162 contacts T8 and A9 on the C strand, and phosphorylation of S162 was predicted to impede DNA binding directly by phosphate–phosphate repulsion and steric hindrance (Fig. 1B and T. Richmond, personal communication). We also noted that T159 did not make a significant contact on the CArG box, hence we focused on the role of S162 phosphorylation.
Fig. 1.
SRF MADS box αI coil phospho-mimetic mutant SRF-S162D prevents SRF–DNA binding. (A) Schematic crystal structure of the SRF MADS box binding a CArG box (adapted from refs. 8 and 25) showed one SRF monomer in blue and the other SRF in red. (B) The MADS box αI coil contacts the CArG box with hydrogen-bond interactions shown as solid colored lines and hydrophobic contacts shown as broken colored lines that represent the two SRF monomers. Complementary DNA strands are labeled W and C. (C) Schematic representation of the MADS box functional regions. (D) Alanine and aspartate substitutions of the phosphorylatable residues in the MADS box αI coil is shown in bold. (E) EMSA with nuclear extracts of CV1 cells transfected with the indicated plasmids. The DNA probe was a c-fos SRE lacking an EBS. The control was a nuclear extract of cells transfected with empty vector. The arrow indicates the SRF–DNA complex. Identical results were obtained by using a skeletal α-actin SRE-1 probe (data not shown).
Phosphorylation of this residue on the αI coil was mimicked by substitution of S162 with aspartate, and absence of phosphorylation was achieved by substitution with alanine at the same site (Fig. 1D). SRF-S162D completely abolished SRF–DNA binding (Fig. 1E). In contrast, SRF-S162A decreased SRF–DNA binding only slightly.
Mimicking Phosphorylation of S162 Blocked Transactivation of Cardiac α-Actin and Smooth Muscle α-Actin (SMA) Promoters.
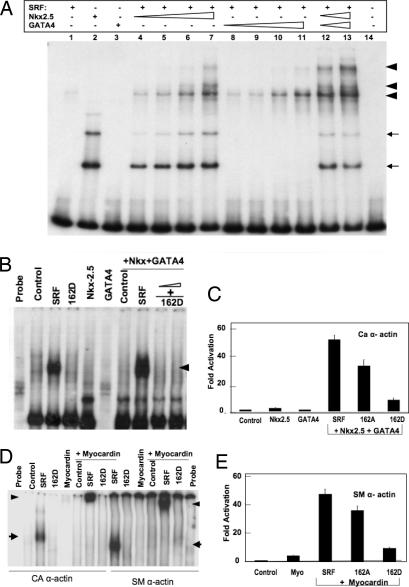
We explored the effects of mimicking phosphorylation of S162 on transactivation of myogenic genes in the presence or absence of key coactivators of SRF. We first asked whether disruption of SRF–DNA binding by SRF-S162D could be overcome by the presence of Nkx2.5 or GATA4 in the context of α-actin promoters. Addition of Nkx2.5 and GATA4 to a suboptimal concentration of SRF led to increased levels of higher-order SRF–DNA binding complexes, indicating occupation of ≈3 of the 4 CArG boxes in the cardiac α-actin promoter probe (Fig. 2A). SRF-S162D did not bind to the α-actin promoter and did not form higher-order complexes even with the addition of Nkx2.5 and GATA4 (Fig. 2B), indicating that inhibition of SRF–SRE binding by S162D cannot be rescued by these coactivators in the context of the α-actin promoter. SRF and S162A synergized with Nkx2.5 and GATA4 to enhance cardiac α-actin transcriptional activity very strongly in CV1 cells, whereas SRF-S162D greatly compromised cardiac α-actin promoter activity in the presence of these two coactivators (Fig. 2C). Myocardin alone did not bind DNA, but in the presence of SRF formed a ternary complex, as shown by EMSA with both a SMA promoter probe and a cardiac α-actin promoter probe (Fig. 2D). Once again, SRF-S162D blocked the formation of the ternary complex with myocardin in the context of both α-actin promoters. Consistent with these effects, wild-type SRF and SRF-S162A synergized with myocardin to enhance SMA transcriptional activity strongly, whereas SRF-S162D greatly compromised transactivation of the SMA promoter in the presence of myocardin (Fig. 2E).
Fig. 2.
SRF phospho-mimetic S162D blocked binding of SRF to α-actin promoters even in the presence of potent cofactors. (A) A subobptimal amount of wild-type SRF (lane 1) recruited Nkx2.5 and GATA4 and formed cooperative SRF–DNA binding multimeric complexes (arrowheads, lanes 4–13). Thin arrows indicate Nkx2.5 monomer (third arrow) and dimer (second arrow). (B) SRF nuclear extract (5 μg) formed a SRF–DNA complex with a 32P-labeled cardiac α-actin promoter (arrowhead). SRF-S162D did not bind to the cardiac α-actin promoter, even in the presence of Nkx 2.5 and GATA4. (C) SRF-S162D inhibited activation of the cardiac α-actin (CA α-actin) promoter, even in the presence of Nkx2.5 and GATA4. (D) SRF-S162D blocked formation of the SRF–DNA complexes, as shown by EMSA of nuclear extracts (5 μg), probed with a 32P-labeled cardiac α-actin promoter containing four SREs (Left) or a 32P-labeled SMA promoter containing two SREs (Right). Arrows indicate the SRF–DNA complex, and arrowheads indicate the ternary complex. (E) SRF-S162D was unable to activate the SMA promoter even in the presence of myocardin.
Mimicking Phosphorylation of S162 in the MADS Box Permitted c-fos Promoter Activity.
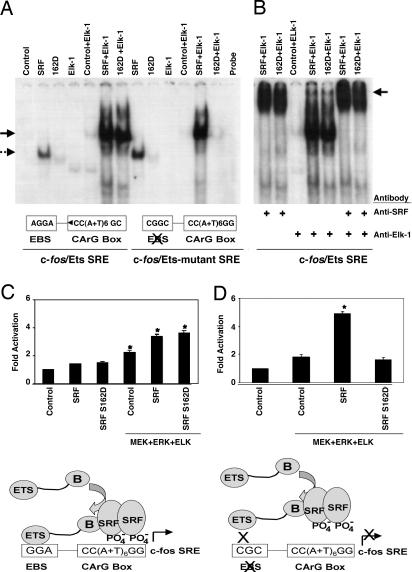
We next asked whether the disruption of SRF–DNA binding by SRF-S162D could be overcome by the presence of the ternary complex factor Elk-1 in the context of the c-fos promoter. SRF alone bound to the c-fos SRE probe and with Elk-1 formed a ternary complex (Fig. 3A). SRF-S162D displayed markedly diminished binding to the c-fos SRE, but in the company of Elk-1 was able to form a stable ternary complex. Mutation of the Ets binding site (EBS) motif adjacent to the c-fos SRE, to which Elk-1 binds, prevented Elk-1 from rescuing SRF-S162D–DNA binding, indicating that interaction with Elk-1 is necessary and sufficient for formation of a ternary complex involving the c-fos SRE and SRF-S162D. Mutation of the EBS did not prevent wild-type SRF from forming a ternary complex with Elk-1 and the SRE. Addition of SRF antibody to the ternary complex formed between the SRE and either wild-type SRF or SRF-S162D with Elk-1 caused a supershift to a slower migrating complex (Fig. 3B). When Elk-1 [activated by mitogen-activated protein kinase kinase (MEK) and extracellular regulated kinase (Erk)] was cotransfected with a wild-type c-fos promoter containing an intact EBS, both wild-type SRF and SRF-S162D enhanced c-fos promoter activity (Fig. 3C). However, SRF-S162D in the presence of Elk-1 could not enhance transcription of c-fos with a mutated EBS (Fig. 3D). Thus, SRF-S162D enhanced c-fos transcription, and Ets factor binding to the SRE and EBS was necessary for this activity.
Fig. 3.
SRF phospho-mimetic S162D transactivated the c-fos promoter by forming ternary complexes with an activated Ets factor bound to the adjacent EBS. (A) EMSA with the c-fos promoter showed that SRF-S162D did not bind to the c-fos SRE. SRF-S162D did form a ternary complex with a c-fos promoter in the presence of Elk-1, but not if the Elk-1 EBS had been mutated. Wild-type SRF formed a ternary complex with the c-fos promoter containing either intact or mutant EBS. (B) Anti-SRF antibody supershifted ternary complexes formed by wild-type SRF or SRF-S162D with Elk-1 (arrow). Anti-Elk-1 antibody also supershifted the ternary complex, but much less efficiently than the anti-SRF antibody. (C) Activated Elk-1 enhanced c-fos promoter activity via endogenous SRF (second control lane), exogenous SRF, and SRF-S162D. CV1 cells transiently transfected with a luciferase construct under the wild-type c-fos promoter and the indicated plasmids were treated with PMA for 30 min before lysis. Bars indicate fold increase in luciferase activity, and asterisks indicate significant (P < 0.05) increase over the corresponding basal levels. The schematic diagram illustrates how SRF could bind to the c-fos SRE and transactivate c-fos gene expression despite disruption of direct SRF–SRE binding caused by phosphorylation of S162. (D) In the presence of activated Elk-1, wild-type SRF enhanced transcriptional activity of the c-fos promoter bearing a mutated EBS, whereas SRF-S162D could not. The schematic diagram illustrates how c-fos transactivation by S162-phosphorylated SRF in the presence of Elk-1 was prevented by mutation of the EBS.
Introduction of SRF-S162D into SRF Null ES Cells Was Unable to Rescue the Expression of Muscle Contractile Protein Genes but Permitted the Activation of Immediate Early Genes.
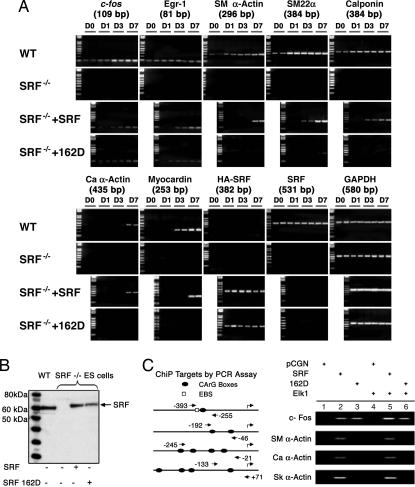
Homozygous SRF−/− mouse ES cells serve as a cellular model of the knockout animal, as they are unable to express SRF-dependent myogenic genes (26). There was no detectable expression of mouse endogenous SRF in SRF null ES cells (27), as tested by both RT-PCR (Fig. 4A) and immunoblot with anti-SRF antibody (Fig. 4B). There was also a complete block of transcription of both the SRF-regulated immediate early genes c-fos and Egr-1 and SRF-regulated terminal differentiation genes, such as the cardiac α-actin, skeletal α-actin, SMA, and calponin, in the SRF null ES cells. Myocardin expression was also absent in these cells. Transfection of wild-type human SRF into the SRF null ES cells restored transcriptional activity of these murine SRF gene targets. Transfection of SRF null ES cells with SRF- S162A also activated transcription of both c-fos and cardiac α-actin genes (results not shown). SRF-S162D restored transcriptional activity of the immediate early genes exclusively, that is, only those SRF-dependent genes that require ternary complex formation with an Ets factor. None of the SRF-dependent myogenic genes were transactivated after introduction of the SRF-162D mutant, indicating that phosphorylation of S162 in the MADS box α1 coil impedes activation of SRF-dependent myogenic gene programs without interfering with activation of SRF-dependent proliferation genes. To solidify this finding, chromatin immunoprecipitation assays were performed in the SRF null ES cells rescued with SRF or SRF-S162D, with or without Elk-1. The c-fos gene promoter was bound by SRF and SRF-S162D even without Elk-1 cotransfection, implying that endogenous Elk-1 is sufficient for induction of c-fos expression by both wild-type SRF and SRF-S162D (Fig. 4C). However, not even overexpression of Elk-1 could stabilize binding of SRF-S162D to the three myogenic α-actin promoters, because these promoters lack an EBS and hence cannot form a ternary complex with Elk.
Fig. 4.
Rescue of SRF null ES cells by SRF phospho-mimetic S162D permitted expression of SRF-regulated immediate early genes but not of SRF-regulated myogenic gene targets. (A) Wild-type ES cells expressed the immediate early genes c-fos and Egr-1, the terminal differentiation myogenic contractile genes SMA, SM22α, calponin, cardiac α-actin, and myocardin and endogenous (mouse) SRF, as shown by RT-PCR of these transcripts on the indicated days of cell culture (top row). SRF null ES cells failed to express any of these transcripts (second row). Introduction of HA-tagged wild-type human SRF induced expression of all of the immediate early genes and terminal differentiation myogenic contractile genes (third row). Introduction of HA-tagged SRF-S162D induced expression only of the immediate early genes (bottom row) but not of the SRF myogenic gene targets. (B) Immunoblot showing expression of recombinant wild-type SRF and SRF-S162D after transfection with the appropriate plasmids into the SRF null ES cells used in the chromatin immunoprecipitation (ChIP) assay shown in C. The control is human SRF overexpressed in wild-type ES cells; SRF proteins were identified by using anti-SRF antibody. (C) ChIP showed that SRF phospho-mimetic S162D could associate with the c-fos promoter in chromatin, but could not associate with the α-actin promoters. Both endogenous (lane 3) and exogenous (lane 6) Elk-1 stabilized the ternary complex with SRF-S162D on the c-fos promoter but did not stabilize SRF-162D binding to the α-actin promoters (lane 6). The schematic diagram shows the promoters of the target genes; arrows show location of PCR primers flanking the SREs amplified and electrophoresed on the right.
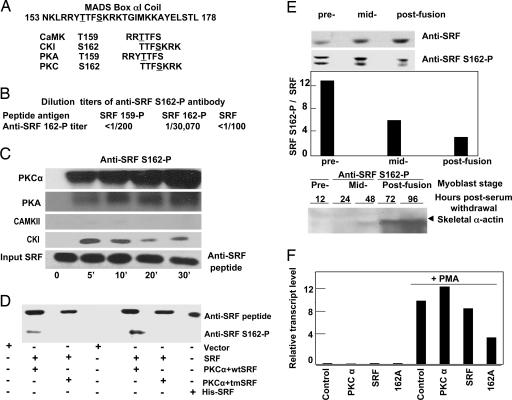
PKCα Highly Phosphorylated Serine-162.
The MADS box αI coil sequence (amino acid 153-NKLRRYTTFSKRKTGIMKKAYELSTL-amino acid 178) contains predicted phosphorylation sites (ref. 28; net phos 2.0) of several kinases, including PKC isoforms known to play a role in regulating myogenesis (Fig. 5A). We generated a phospho-specific antibody targeting SRF-S162-P. Anti-phospho-S162 (anti-S162-P) detected SRF-S162-P at a maximum titer of 1/30,000 and showed minimal crossreactivity with SRF-T159-P (Fig. 5B). PKCα strongly phosphorylated S162 in vitro (Fig. 5C). PKCα also phosphorylated T159 in vitro, but about five times less potently than S162 (data not shown). The PKC activator phorbol 12-myristate 13-acetate (PMA) increased SRF-S162 phosphorylation in CV1 cells, as detected by anti-S162-P antibody (Fig. 5D). PKCα-induced SRF-S162 phosphorylation was absent in a mutant SRF with alanine substitutions at T159, T160, and S162, which also demonstrated that anti-S162-P does not crossreact with other epitopes on SRF.
Fig. 5.
PKCα strongly phosphorylated SRF serine-162. (A) Candidate protein kinases and their consensus phosphorylation sites on the α1 coil as predicted by the netphos 2.0 program. (B) Potency and crossreactivity of affinity-purified SRF-S162-P antibody against nonphosphorylated and SRF-T159- or SRF-S162-phosphorylated αI coil peptides. ELISA was used to measure potency and crossreactivity, as shown by the dilution titer of the antibodies. (C) In vitro phosphorylation of GST-SRF (residues 1–171) by purified PKCα, PKA, casein kinase I (CKI), and Ca2+/calmodulin-dependent PK II (CaMKII). Immunoblots were probed with anti-SRF-S162-P. (D) PKCα phosphorylated the MADS box αI coil in vivo. Shown are immunoblots of lysates of CV1 cells expressing PKCα with empty vector, wild-type SRF (wtSRF), or SRF with triple alanine substitutions for T159, T160, and T162 (tmSRF), with or without the addition of PMA, immunoprecipitated with HA-epitope antibody, and probed with anti-SRF or anti-SRF-S162-P. (E) SRF-S162 phosphorylation was highest during myoblast proliferation and declined during differentiation. (Top) Immunoblots of nuclear extracts of C2C12 myoblasts during replication (prefusion), shortly after withdrawal from replication (midfusion), and after terminal differentiation (postfusion). (Middle) Quantification of band intensity by densitometry, using the image quant 5.2 program. The y axis corresponds to the pixel value of the bands shown in Top, displayed as a ratio of immunoblottable SRF-S162-P to total SRF. (Bottom) The appearance of skeletal α-actin mRNA, indicated by blots of RNA isolated from C2C12 cells during myogenesis with a skeletal α-actin cDNA probe, correlated with the decline of SRF-S162 phosphorylation. (F) Endogenous c-fos mRNA levels were low in postdifferentiation C2C12 myotubes but increased after activation of PKC by 30 min of exposure to PMA; the effect of PMA was markedly blunted by preventing phosphorylation of S162 (i.e., S162A). These results are representative of two separate experiments.
Treatment with phorbol esters is known to repress cardiac α-actin transcription in primary muscle cultures (29), suggesting a role for PKCα in this phenomenon. To investigate whether the degree of S162 phosphorylation plays a role in the transition of α-actin transcription from a repressed state to an activated state during the process of myogenic differentiation, we tracked skeletal α-actin transcript levels in C2C12 cells at various stages after serum withdrawal. The highest level of SRF-S162 phosphorylation relative to the total amount of SRF was observed in replicating C2C12 cells cultured under high serum growth conditions; the level declined as differentiation progressed through midfusion to postfusion myotubes. The steady decline in the proportion of S162-phosphorylated SRF in the midfusion and postfusion stages correlated well with activation of the myogenic gene program, as shown by the appearance of skeletal α-actin transcripts (Fig. 5E). Furthermore, activation of PKC by brief (30 min) exposure to PMA in differentiated C2C12 cells resulted in a 70- to 100-fold increase in c-fos mRNA levels (Fig. 5F). This effect was significantly inhibited by single alanine substitution of S162, indicating that activation of PKCα, whose expression is normally suppressed in differentiated myotubes, is sufficient to induce c-fos expression in differentiated myotubes, and that phosphorylation of SRF-S162 (and the presence of the Ets factor) is required for this effect. Collectively, the data indicate a strong role for S162 phosphorylation (e.g., by PKCα activity) in shifting muscle cells toward the proliferation program and away from the differentiation program. Thus, SRF-S162 phosphorylation reflected a shift toward the proliferation program.
Discussion
A long-standing conundrum in the field of myogenesis concerns the mechanisms whereby SRF could regulate both replication- and differentiation-dependent genes with apparent antagonistic processes. We suggest a model in which phosphorylation of serine-162 in the SRF MADS box αI coil is a key determinant of the switch in SRF-dependent gene expression between cell replication and differentiation. The MADS box serves as a regulatory nexus that mediates interactions between SRF and a myriad of transcription cofactors, hence differential phosphorylation of MADS box residues could direct specific SRF transcriptional complexes toward the expression of either proliferation or differentiation genes. Most SRF accessory proteins that have been identified as coregulators of c-fos induction are at endpoints of growth factor-induced signal transduction cascades. However, the SREs of cardiomyogenic genes recruit different collateral factors, such as Nkx2.5 and GATA4, that strongly enhance SRF–DNA binding affinity, permitting the formation of higher-order DNA-binding complexes at relatively low SRF levels (30). Similarly, smooth muscle gene SREs recruit the LIM protein CRP2, which bridges SRF and GATA factors through interaction with the MADS αI coil (13), and myocardin, which competes with the Ets factors that interact with the MADS αII coil (16). All of these myogenic cofactors greatly enhance SRF transactivation activity. Thus, the MADS box region is likely to be an important site for the receipt of crucial intracellular signals that enable SRF to recruit specific cofactors to their respective DNA binding sites.
What are the upstream signals that could induce these functionally important covalent modifications in the MADS box αI coil? The PKC family has been implicated in the control of proliferation and differentiation of muscle cells (29, 31–33). Growth factor-stimulated PKC pathways converge on the c-fos SRE to promote myoblast proliferation (34), and by phosphorylating a conserved site on myogenin, a basic helix–loop–helix protein, also de-repress the myogenic differentiation program (35). Capiati et al. (32, 33) have shown that PKCα is involved mainly in the proliferation process in myoblasts. Phosphorylation of S162, as mimicked by replacement of this residue by aspartate, prevents binding of SRF to the c-fos SRE in the absence of an Ets factor protein, but SRF–SRE binding is restored to normal in the presence of Elk-1, which forms a ternary complex with SRF on the c-fos SRE. Wang et al. (16) have demonstrated that myocardin and the Ets factor Elk-1 compete for interaction with common sites on the SRF αII coil to control smooth muscle gene expression. This observation supports a model in which growth factor stimulation leads to phosphorylation of S162 in the MADS box in a manner that suppresses the expression of muscle differentiation genes, while coordinately permitting or increasing the association between Ets factors and SRF to up-regulate c-fos transcription. This model is also supported by the present data from the SRF null ES cell “rescue” experiments. Upon transfection with wild-type SRF or SRF-S162D, the SRF null ES cells express only the proliferation program genes, such as c-fos and Egr1, whereas the expression of muscle differentiation genes, such as the cardiac α-actin, skeletal α-actin, and SMA, remain suppressed. Thus, phosphorylation of the highly conserved serine-162 residue in the αI coil of the SRF MADS box represents another pathway that regulates SRF activity to modulate gene expression during cellular replication and differentiation. This mechanism is a key element in a novel switch that could direct target gene expression into proliferation or differentiation programs.
Experimental Procedures
Expression Vectors and Reporter Genes.
The luciferase reporter plasmids for α-actin gene promoters and SRF expression vectors have been described (13). The c-fos promoter construct (pwtGL3) and the mutant c-fos promoter (ppm18GL3) were gifts of Ron Prywes (Columbia University, New York; ref. 36). SRF MADS box mutants were made by PCR mutagenesis and cloned into the pCGN vector as described (24). Construction of a GST mouse myocardin fusion protein in pGEX-4T-1 was as described (14). Maltose binding protein-tagged Nkx2.5 and GATA4 constructs were as described (37). pCMV5 Elk-1 and the Elk-1-histidine fusion protein DNA vectors were provided by Peter Shaw (University of Nottingham, Nottingham, U.K.) and Andrew Sharrocks (University of Manchester, Manchester, U.K.). Metallothionein-PKCα was from Stuart Yuspa (National Cancer Institute, Bethesda). The PKA clone, MT-CEVa, was from Stanley McKnight (University of Washington, Seattle). MEK-1 and Erk-1 in the vector pMT3-HA were from Zhijun Luo (Boston University Medical Center, Boston). GST-SRF (amino acid residues 1–171) in pGEX 4T3 was as described (24).
Phosphorylation of the SRF MADS Box, Assayed by Phospho-Specific Antibodies.
Synthesis of anti-SRF peptide antibody, anti–SRF threonine 159 phospho-specific antibody, and anti-SRF serine-162 phospho-specific antibody have been described (24). Briefly, the antibodies were raised in rabbits against the following peptides representing amino acids 152–169 of SRF, DNKLRRYTTFSKRKTGIM, DNKLRRYT(-PO4)TFSKRKTGIM, and DNKLRRYTTFS(-PO4)KRK TGIM (Bethyl Laboratories, Montgomery, TX), and affinity-purified before use. PKCα or empty vector was cotransfected with pCGN, wild-type SRF, or triple-alanine mutant SRF (SRF-T159A/SRF-T160A/SRF-S162A) into CV1 cells by using Lipofectamine (Invitrogen) according to the manufacturer’s protocol. The cells were treated with 100 nM PMA for 30 min before cell lysis in buffer containing 50 mM Tris·Cl (pH 7.5), 500 mM NaCl, 0.5% Triton X-100, 2 mM sodium vanadate, 50 mM NaF, and protease inhibitors. Recombinant hemagglutinin (HA)-tagged SRF proteins were immunoprecipitated from clarified lysate by using 12CA5 antibody (Roche Applied Science), subjected to SDS/PAGE, and immunoblotted by using anti-SRF peptide antibody or anti-SRF-S162-P.
Preparation of Nuclear Extracts and EMSAs.
Nuclear fractions were prepared according to the protocol of Dignam et al. (38), and some cells were treated with PMA (100 nM) before harvest. Nuclear preparations contained 50 mM sodium fluoride and 2 mM sodium vanadate as phosphatase inhibitors and were normalized for protein concentration. For SRF–DNA binding in EMSA, the following DNA probes were used: cardiac α-actin complete promoter, nucleotides −315 to −58; c-fos SRE, ACACAGGATGTCCATATTAGGACATCTGC; c-fos SRE lacking the complete EBS GGATGTCCATATTAGG; c-fos SRE with EBS mutation, ACACcGGcTGTCCATATTAGGACATCTGC; SMA, nucleotides −141 to +1; and skeletal α-actin SRE1 nucleotides −98 to −76. Purified Elk-1 protein was first phosphorylated by using p42 mitogen-activated protein kinase or Erk-1 and MEK-1 in the presence of 1 mM cold ATP. EMSA was performed as described (13, 24).
Acknowledgments
We thank Dr. Susan Samson for help with quantitative RT-PCR and Dr. Timothy J. Richmond for helpful discussions regarding the SRF crystal structure. Support was provided by American Heart Association Beginning Grant-in-Aid 0465066Y (to D.I.) and National Institutes of Health Grants R01DK-59537 and R01HL-73696 (to A.B.), P01HL-49953 (to R.J.S. and E.N.O.), HL-079628 (to R.J.S.), and P01-HL75380 and R01-HL56915 (to M.S.P.).
Abbreviations
- SRF
serum response factor
- SRE
serum response element
- SMA
smooth muscle α-actin
- EBS
Ets binding site
- MEK
mitogen-activated protein kinase kinase
- Erk
extracellular regulated kinase
- PMA
phorbol 12-myristate 13-acetate
- HA
hemagglutinin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sommer H., Beltran J. P., Huijser P., Pape H., Lonnig W. E., Saedler H., Schwarz-Sommer Z. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharrocks A. D., Gille H., Shaw P. E. Mol. Cell. Biol. 1993;13:123–132. doi: 10.1128/mcb.13.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Treisman R. EMBO. J. 1995;14:4905–4913. doi: 10.1002/j.1460-2075.1995.tb00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boxer L. M., Prywes R., Roeder R., Kedes L. Mol. Cell. Biol. 1989;9:515–522. doi: 10.1128/mcb.9.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow K.-L., Schwartz R. J. Mol. Cell. Biol. 1990;10:528–538. doi: 10.1128/mcb.10.2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Treisman R. Curr. Opin. Genet. Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 7.Ling Y., West A. G., Roberts E. C., Lakey J. H., Sharrocks A. D. J. Biol. Chem. 1998;273:10506–10514. doi: 10.1074/jbc.273.17.10506. [DOI] [PubMed] [Google Scholar]
- 8.Hassler M., Richmond T. J. EMBO J. 2001;20:3018–3028. doi: 10.1093/emboj/20.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gineitis D., Treisman R. J. Biol. Chem. 2001;276:24531–24539. doi: 10.1074/jbc.M102678200. [DOI] [PubMed] [Google Scholar]
- 10.Murai K., Treisman R. Mol. Cell. Biol. 2002;22:7083–7092. doi: 10.1128/MCB.22.20.7083-7092.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C. Y., Schwartz R. J. Mol. Cell. Biol. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belaguli N. S., Sepulveda J. L., Nigam V., Charron F., Nemer M., Schwartz R. J. Mol. Cell. Biol. 2000;20:7550–7558. doi: 10.1128/mcb.20.20.7550-7558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang D. F., Belaguli N. S., Iyer D., Roberts W. B., Wu S., Dong X., Marx J. G., Moore M. S., Beckerle M. C., Majesky M. W., Schwartz R. J. Dev. Cell. 2003;4:107–108. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 14.Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang D., Li S., Hockemeyer D., Sutherland L., Schratt G., Richardson R. A., Nordheim A., Olson E. N. Proc. Natl. Acad. Sci. USA. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z., Wang D.-Z., Hockemeyer D., McAnally J., Nordheim A., Olson E. N. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 17.Miralles F., Posern G., Zaromytidou A. I., Treisman R. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 18.Cen B., Selvaraj A., Burgess R. C., Hitzler J. K., Morris S. W., Prywes R. Mol. Cell. Biol. 2003;23:6597–6608. doi: 10.1128/MCB.23.18.6597-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selvaraj A., Prywes R. J. Biol. Chem. 2003;278:41977–41987. doi: 10.1074/jbc.M305679200. [DOI] [PubMed] [Google Scholar]
- 20.Manak J. R., de Bisschop N., Kris R. M., Prywes R. Genes Dev. 1990;4:955–967. doi: 10.1101/gad.4.6.955. [DOI] [PubMed] [Google Scholar]
- 21.Marais R. M., Hsuan J. J., McGuigan C., Wynne J., Treisman R. EMBO J. 1992;11:97–105. doi: 10.1002/j.1460-2075.1992.tb05032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miranti C. K., Ginty D. D., Huang G., Chatila T., Greenberg M. E. Mol. Cell. Biol. 1995;15:3672–3684. doi: 10.1128/mcb.15.7.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heidenreich O., Neininger A., Schratt G., Zinck R., Cahill M. A., Engel K., Kotlyarov A., Kraft R., Kostka S., Gaestel M., Nordheim A. J. Biol. Chem. 1999;274:14434–14443. doi: 10.1074/jbc.274.20.14434. [DOI] [PubMed] [Google Scholar]
- 24.Iyer D., Belaguli N. S., Fluck M., Rowan B. G., Wei L., Weigel N. L., Booth F. W., Epstein H. F., Schwartz R. J., Balasubramanyam A. Biochemistry. 2003;42:7477–7486. doi: 10.1021/bi030045n. [DOI] [PubMed] [Google Scholar]
- 25.Pellegrini L., Tan S., Richmond T. J. Nature. 1995;376:490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- 26.Arsenian S., Weinhold B., Oelgeschlager M., Ruther U., Nordheim A. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du K. L., Ip H. S., Li J., Chen M., Dandre F., Yu W., Lu M., Owens G. K., Parmacek M. S. Mol. Cell. Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blom N., Gammeltoft S., Brunak S. J. Mol. Biol. 1999;294:135–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Y. Y., Schwartz R. J., Crow M. T. J. Cell Biol. 1991;115:745–754. doi: 10.1083/jcb.115.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sepulveda J. L., Vlahopoulos S., Iyer D., Belaguli N. S., Schwartz R. J. J. Biol. Chem. 2002;277:25775–25782. doi: 10.1074/jbc.M203122200. [DOI] [PubMed] [Google Scholar]
- 31.Gliki G., Wheeler-Jones C., Zachary I. Cell Biol. Int. 2002;26:751–759. doi: 10.1016/s1065-6995(02)90926-1. [DOI] [PubMed] [Google Scholar]
- 32.Capiati D. A., Tellez-Inon M. T., Boland R. L. Mol. Cell Endocrinol. 1999;153:39–45. doi: 10.1016/s0303-7207(99)00093-3. [DOI] [PubMed] [Google Scholar]
- 33.Capiati D. A., Limbozzi F., Tellez-Inon M. T., Boland R. L. J. Cell. Biochem. 1999;74:292–300. [PubMed] [Google Scholar]
- 34.Soh J.-W., Lee E. H., Prywes R., Weinstein I. B. Mol. Cell. Biol. 1999;19:1313–1324. doi: 10.1128/mcb.19.2.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L., Zhou J., James G., Heller-Harrison R., Czech M. P., Olson E. N. Cell. 1992;71:1181–1194. doi: 10.1016/s0092-8674(05)80066-2. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Falasca M., Schlessinger J., Malstrom S., Tsichlis P., Settleman J., Hu W., Lim B., Prywes R. Cell Growth Differ. 1998;9:513–522. [PubMed] [Google Scholar]
- 37.Sepulveda J. L., Belaguli N. S., Nigam V., Chen C.-Y., Nemer M., Schwartz R. J. Mol. Cell. Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dignam J. D., Lebovitz R. M., Roeder R. G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]