Abstract
The dopamine (DA) transporter DAT1 is a major target bound by cocaine in brain. We examined the influence of functional genetic variants in DAT1 on cocaine addiction. Repeat polymorphisms, including a 30-bp variable-number tandem repeat (VNTR) in intron 8 (Int8 VNTR) with two common alleles, were genotyped in cocaine-dependent abusers (n = 699) and in controls with no past history of drug abuse (n = 866) from São Paulo, Brazil. Positive association was observed with allele 3 of the Int8 VNTR and cocaine abuse (allele odds ratio = 1.2, 95% confidence interval = 1.01–1.37, P = 0.036; 3/3 homozygote odds ratio = 1.45, 95% confidence interval = 1.18–1.78, P = 0.0008). Population stratification was assessed and did not affect the results. Haplotypic analyses using additional polymorphisms indicated that the Int8 VNTR is responsible for the observed association. Functional analyses in reporter–gene constructs, demonstrated that allele 3 mediates significant (P < 0.05) but small reduced expression compared with the “protective” allele 2. This difference increased when 1 and 10 μM cocaine was added to the cell culture (≈40% reduction of the 3 allele expression versus the 2 allele). The 3 allele also demonstrated ≈3-fold-increased expression over the 2 allele in response to KCl plus forskolin challenge. We demonstrate a robust association between cocaine dependence and a VNTR allele in SLC6A3, conferring a small but detectible effect, and we show that this VNTR may be functional. This study suggests that DAT1 gene variation may play a role in cocaine dependence etiology.
Keywords: addiction, genetics, SLC6A3
Dopamine (DA) is a key neurotransmitter in brain areas involved in movement and behavior, particularly reward pathways. The DA transporter DAT1 mediates the active reuptake of DA from the synapse and is a principal regulator of dopaminergic neurotransmission (1). Cocaine’s pleasurable and addictive effects are thought to be principally mediated through its blockage of DAT1, increasing substantially the concentration of extracellular DA, resulting in elevated stimulation of neurons in brain regions involved in reward and reinforcement behavior (2). In addition, family and twin studies suggest a substantial genetic component in the vulnerability of individuals to become dependent after exposure to cocaine (3–5). Thus, we hypothesized that polymorphic functional variants in the DAT1 gene may act to modify susceptibility for cocaine abuse and dependence.
The gene encoding DAT1 (gene symbol SLC6A3) consists of 15 exons spanning 60 kb on chromosome 5p15.32 [MIM *126455] (6). More than one hundred studies have assessed possible associations between variants in SLC6A3 and psychiatric disorders (for examples see refs. 7–9). However, the bulk of these studies examined only one polymorphism and have thus assessed only a portion of the phenotypic influence of genetic variation within the gene. Recent analyses using a larger selection of markers have shown a mixed pattern of functional variation and linkage disequilibrium (LD). They underline the necessity for thorough examination of the locus and the importance of identifying robust functional variants with replicable effects (10, 11).
The most commonly examined DAT1 gene polymorphism is a 40-bp variable-number tandem repeat (VNTR) in the 3′ UTR with repeat copy numbers ranging from 3 to 11 (6). Numerous studies have sought to assess the relationship between this 3′ UTR VNTR and clinical phenotypes thought to be related to DA dysfunction, such as attention deficit hyperactivity disorder (12), Parkinson’s disease (13), cocaine-induced paranoia (14), and methamphetamine-induced psychosis (15). However, the functional effect of this polymorphism is uncertain, because the data do not indicate a consistent effect of different alleles and genotypes on gene expression (8, 16, 17). One explanation for these variable results is that 3′ UTR VNTR may not be the major or an independent source of functional variation in the gene. Rather, it may functionally interact and/or be in LD with functional polymorphism motifs in and around the gene.
Our examination of the SLC6A3 genomic sequence revealed that the 3′ UTR VNTR is not unique. There are ≈15 other candidate simple tandem repeats and VNTRs in the introns of SLC6A3 with at least six repeat copies. Although single-nucleotide polymorphisms (SNPs) are markers of great utility in genetic studies, different alleles of a VNTR represent a very large physical and chemical change to a stretch of DNA sequence. They can act variously as (i) functional elements binding transcription factors and other proteins that inhibit or promote expression (18, 19); (ii) motif elements affecting the efficiency of mRNA splicing (20); (iii) elements having physical effects, such as varying the spacing between functional motifs or altering the structure and melting properties of DNA in their proximity. For these reasons we regard VNTRs as very good a priori functional candidates. We decided to characterize VNTR polymorphisms in the SLC6A3 by first investigating a possible relationship between these markers and susceptibility to cocaine dependence and then by examining functional effects of marker alleles found to be associated with the phenotype.
Greenwood and Kelsoe (11) reported a segmental pattern of LD within SLC6A3. A high preservation of LD was observed in the 5′ and 3′ regions, with little significant LD between them, probably because of a recombination hot spot located near the middle of the gene (introns 6–8). Based on these results, in addition to the 3′ UTR VNTR, an insertion deletion polymorphism in intron 14, and a VNTR 10-kb 3′ from the DAT gene, we selected a six-copy 30-bp VNTR located in intron 8 (Int8) for genotyping. This VNTR exists as just one perfect copy of the motif in the chimpanzee genome and is mildly conserved in rat and mouse, where this region of Int8 contains large, and presumably polymorphic, GA and CA repeats, respectively (http://genome.ucsc.edu).
These markers were then genotyped in a sample of ≈700 cocaine abusers and ≈860 controls from Sao Paulo, Brazil. We also investigated potential functional effects of identified risk alleles, with the aim of providing a plausible rationale for the associations observed. Here we report that the 30-bp VNTR located in Int8 of the DAT1 gene is associated with cocaine abuse in the Brazilian population and, further, that the risk allele has a differential effect on reporter gene expression that is sensitive to stimuli, including cocaine.
Results
Initial Genotyping.
The allele counts and frequencies are indicated in Table 1 for the Int8 VNTR, Indel_14, the 3′ UTR VNTR, and the 37-bp VNTR. The genotyping failure rates were 4% for the Indel_14, 0.2% for the 3′ UTR, and 1% for the 37-bp VNTR. No significant case-control difference was observed for the 3′ UTR (genotype-wise: χ2 = 14.4; df = 14; P > 0.05) or 37-bp (χ2 = 6.4; df = 6; P > 0.05) VNTRs or Indel_14 (χ2 = 3.6; df = 2; P > 0.05) Association was found with cocaine abuse and the Int8 VNTR (χ2 = 30.7; df = 12; P = 0.002).
Table 1.
Number and frequency of the most common alleles (frequency > 10%) of the three VNTRs and the Indel_14 polymorphism in healthy controls and cocaine abusers in the Brazilian sample
| Alleles | No. of controls (%) | No. of cases (%) | P value | OR (95% CI) |
|---|---|---|---|---|
| Int8 VNTR | ||||
| 2 | 567 (33.1) | 408 (29.6) | 0.036 | 1.2 (1.01–1.37) |
| 3 | 1,145 (66.9) | 971 (70.4) | ||
| Total | 1,712 | 1,379 | ||
| Indel_14 | ||||
| 1 | 1,409 (85.1) | 1,169 (87.5) | >0.05 | 0.81 (0.66–1.01) |
| 2 | 247 (14.9) | 167 (12.5) | ||
| Total | 1,656 | 1,336 | ||
| 3′ UTR VNTR | ||||
| 9 | 464 (27.8) | 378 (28.1) | >0.05 | 0.98 (0.83–1.15) |
| 10 | 1,204 (72.2) | 968 (71.9) | ||
| Total | 1,668 | 1,346 | ||
| 37-bp VNTR | ||||
| 2 | 1,103 (65.9) | 944 (68.7) | >0.05 | 0.87 (0.75–1.02) |
| 3 | 572 (34.1) | 430 (31.3) | ||
| Total | 1,675 | 1,374 |
OR, odds ratio; CI, confidence interval.
Stratification analysis using different parameter options in the lpop program (43) consistently indicated that a three-population model best fitted the data (Akaike information criterion = 114907). Other models were also promising, such as a four-population model (Akaike information criterion = 114923). However, individuals in the three-population model were assigned to the classes with posterior probability greater than for the four-population model (data not shown). Moreover, posterior comparison demonstrated that this population substructure did not significantly differ among the cases and controls (χ2 = 3.8; df = 2; P = 0.15).
For Int8, allele and genotype-wise analyses were also assessed excluding the rare alleles and considering only genotypes 2/2, 2/3, and 3/3 (because they were >95% of the total alleles and genotypes observed). The individual probability of belonging to each of the three population groups indicated by lpop was used as a covariate in the regression analyses. The allele-wise odds ratio for the two most common alleles was 1.2, with a 95% confidence interval of 1.01–1.37 (P = 0.036), and the genotypic odds ratios considering a recessive model for the 3 allele, e.g., 3/3 genotype versus 2/3 and 2/2, was 1.45, with a 95% confidence interval of 1.18–1.78 (P = 0.0008).
Additional Genotyping.
Genotype and allele frequencies for the rs2963238, rs11564752, rs27048, rs6347, rs6876225, rs11564773, and rs1042098 SNPs did not show a significant difference between cases and controls (data not shown). In addition, both controls and cases were in Hardy–Weinberg equilibrium (P > 0.05) for all SNPs and VNTRs tested. Additional clinical variables such as type of cocaine used, age of onset, or sex did not show an association/interaction with the markers genotyped (data not shown).
LD and Haplotype Analysis.
The pairwise LD analyses revealed a pattern of disequilibrium along the SLC6A3 similar to that reported by Greenwood and Kelsoe (11) (Table 2). High levels of disequilibrium were observed between the two 3′ VNTRs and SNP rs1042098, the Indel_14 insertion/deletion, SNP rs115664773 in intron 14, together with SNP rs6876225 in intron 12 (D′ > 0.6; P < 0.0001), and between the two SNPs located in the intron 1 (D′ ≈ 0.6; P < 0.0001). In addition, the Int8 VNTR and SNPs in Int8 and exon 9 revealed a third region of the gene with significant level of disequilibrium (D′ > 0.7; P < 0.0001).
Table 2.
Pairwise LD values: absolute value of D′
| Absolute value of D′ | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.637 | 0.206 | 0.317 | 0.173 | 0.102 | 0.029 | 0.126 | 0.44 | 0.392 | 0.586 | |
| 2 | 0.187 | 0.136 | 0.17 | 0.065 | 0.246 | 0.063 | 0.015 | 0.005 | 0.102 | ||
| 3 | 0.82 | 0.72 | 0.286 | 0.17 | 0.101 | 0.117 | 0.103 | 0.041 | |||
| 4 | 0.745 | 0.077 | 0.475 | 0.272 | 0.534 | 0.441 | 0.368 | ||||
| 5 | 0.059 | 0.32 | 0.293 | 0.537 | 0.412 | 0.311 | |||||
| 6 | 0.887 | 0.97 | 0.05 | 0.438 | 0.742 | ||||||
| 7 | 0.799 | 0.197 | 0.215 | 0.238 | |||||||
| 8 | 0.189 | 0.35 | 0.692 | ||||||||
| 9 | 0.885 | 0.667 | |||||||||
| 10 | 0.726 |
Marker’s names and locations: 1, rs11564752 (intron 1); 2, rs2963238 (intron 1); 3, rs27048 (Int8); 4, Int8 VNTR (Int8); 5, rs6347 (exon 9); 6, rs6876225 (intron 12); 7, Indel_14 (intron 14); 8, rs11564773 (intron 14); 9, rs1042098 (3′ UTR); 10, 3′ UTR VNTR (3′ UTR); 11, 37-bp VNTR (3′ UTR).
Haplotype analyses including all markers demonstrated a trend (P < 0.1) for association with cocaine abuse [likelihood ratio test (LRT) = 24.23; df = 16; P = 0.08]. However, when the Int8 VNTR was dropped from the analyses, no trend was observed (LRT = 23.752; df = 18; P = 0.163).
Based on the LD results, we then analyzed haplotypes within each of these regions separately and tested their interaction with the Int8 VNTR by removing and then adding it to the analysis. Haplotype 1 consists of six markers in the 3′ region of the gene (3′ UTR through intron 12), haplotype 2 includes the three markers between Int8 and exon 9, and haplotype 3 comprises the two SNPs in the intron1. Haplotype 1 did not show significant association with cocaine abuse (LRT = 12.75; P = 0.12); however, when the Int8 VNTR was added to the haplotype analyses a positive association became evident (LRT = 20.40; P = 0.008). Moreover, analyses of haplotype 2 (which includes the Int8 VNTR) demonstrated an association between these markers (LRT = 14.77; P = 0.02) and cocaine abuse, but when the Int8 VNTR was dropped from the haplotype no association was observed with haplotype 2 (LRT = 2.33; P = 0.49). Haplotype 3 did not demonstrate a significant association with cocaine abuse (LRT = 0.31; P = 0.85), but when the Int8 VNTR was added a positive association was detected (LRT = 10.55; P = 0.03).
Sequence Analysis.
We have sequenced the seven alleles found for the Int8 VNTR. Upon analysis it appeared that all of the alleles are composed of variable numbers of an a repeat motif and one b repeat motif, which has two single-base-pair changes from the a repeat. (Fig. 1). Moreover, to investigate whether there were any sequence differences between the Int8 VNTR alleles in terms of the sequence of the repeat unit, two pools containing PCR products of 25 individuals homozygous for the 2 and 3 alleles were sequenced in addition to three individual homozygotes for each allele. The reads were generated on an Applied Biosystems 3730 DNA Analyzer platform and analyzed by using sequencher software, Version 4.0.5.
Fig. 1.
The sequences shown represent the flanking sequences of the Int8 VNTR and the 3 allele, which is six copies of the repeat (Upper) and the 2 allele, which is five copies of the repeat (Lower). All of the alleles found for this polymorphism are composed of variable numbers of an a repeat motif and one b repeat motif, which has two single-base-pair changes from the a repeat.
Functional Results.
Because the location of the Int8 VNTR was within the gene, away from the 5′ or 3′ regions, we used a modified Renilla vector with a unique intronic Asc1 site to allow us to examine potentially interesting intronic regulatory domains. The Int8 VNTR, when cloned in this domain and transfected into the dopaminergic SN4741 cell line, which expresses DAT (21), demonstrated a reproducible and statistically significant increase at baseline (Student’s t test = 2.64; df = 27; P = 0.013) in reporter gene expression supported by the 2 allele of the Int8 VNTR versus that supported by the 3 allele. In both cases expression levels were <50% of the unmodified Renilla vector. Additionally, to examine potential effects on heteronuclear RNA/mRNA stability, the Int8 VNTR alleles were also inserted 5′ in pGLP3 luciferase vector and transfected in JAR cells. This experiment demonstrated repression of transcription, with the vectors showing <50% of unmodified vector expression (data not shown).
We then tested the response of the Int8–VNTR 2 and Int8–VNTR 3 alleles to distinct challenges in the form of (i) cocaine, (ii) KCl, and (iii) KCl and forskolin by adding these chemical stimuli to the cell culture medium as outlined. Here we summarize the conditions and results obtained for each stimulus.
Two concentrations of cocaine hydrochloride (Sigma), 1 and 10 μM, were added to the cell culture medium. In both cases, the Int8 VNTR 3 allele vector demonstrated an ≈40% reduction in expression (t = 4.0 and t = 6.04; df = 16; P = 0.001 and P < 0.001) versus the 2 allele, with a slightly increased effect for the 10 μM concentration. Cocaine has been observed to modulate many neuronal genes, in part through activation of specific transcription factors pathways including those regulated by members of the AP1 family such as c-fos and c-jun (22, 23).
In cultured cells, it has been shown that increasing extracellular K+ leads to an increase in cytosolic Ca2+ and results in enhanced (Cre-dependent) transcription (24, 25). We induced Ca2+ signaling by depolarizing the membrane potential of the cell by adding KCl (40 mM) to the medium. This exposure resulted in increased expression from the allele 3 vector versus the 2 allele (t = −3.0; df = 16; P = 0.008).
Forskolin (10 μM), a membrane-permeable activator of adenylate cyclase, and K+ (40 mM) were coadministered to the cell culture medium. In this system, the Int8–VNTR 3 allele demonstrated a large increase (6-fold) in reporter gene expression in response to synergistic challenge over the 2 allele (t = −8.95; P < 0.001; df = 8.882 as equal variances not assumed in this case). We have previously shown that these stimuli act synergistically on gene expression supported by reporter gene constructs both in clonal cell lines and in organotypic CNS cultures (24, 26, 27).
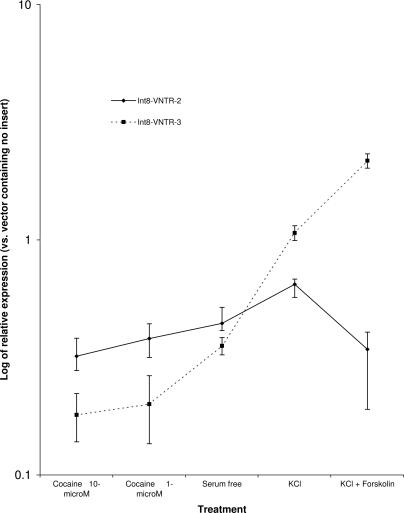
These data are graphed in Fig. 2, and relative expression level values are shown in Table 3, which shows the relative effect of cocaine and the other stimuli on the expression of the vectors, with the vector containing the 3 allele showing an increased response to stimuli compared with the 2 allele at every treatment. In addition, for every treatment the 3 allele shows a significant change (P < 0.05) from baseline conditions, whereas for the 2 allele the only significant differences observed were for the KCl and 10 μM cocaine treatments (data not shown). These changes were significantly smaller than the effects observed for the 3 allele in both cases (see above).
Fig. 2.
Differential expression of Int8 VNTR alleles when exposed to cocaine at baseline and chemical stimuli.
Table 3.
Observed levels of expression of allele 2 and allele 3 of Int8 VNTR in the intronic reporter construct relative to vector without insertion
| Treatment | Allele 2 | Allele 3 |
|---|---|---|
| KCl plus forskolin | 0.89 ± 0.36 | 1.78 ± 0.33 |
| KCl | 0.78 ± 0.19 | 1.02 ± 0.11 |
| No drug | 0.37 ± 0.03 | 0.28 ± 0.05 |
| Cocaine (1 μM) | 0.35 ± 0.04 | 0.18 ± 0.02 |
| Cocaine (10 μM) | 0.32 ± 0.02 | 0.17 ± 0.01 |
Data are shown ± SD.
Discussion
We examined a total of 699 cocaine abusers and 866 controls and identified a positive association with alleles and genotypes of the 30-bp VNTR in Int8 of the DAT1 gene, the Int8 VNTR. Association and haplotype analyses using other polymorphisms in and near the gene (two VNTRs, one Indel, and seven SNPs) indicate that the Int8 VNTR is the variant responsible for the observed association despite significant LD existing between it and some of the other variants. Moreover, stratification analyses indicated a similar distribution of population substructure in the cases and controls, and the association remained positive after covarying for substructure in the regression analyses. This result suggests that population stratification is not responsible for the observed association.
Is the VNTR polymorphism in Int8 functional? Our experiments show that the risk allele 3 exhibits differential and increased responses to stimuli in comparison to the protective allele 2. Specifically, we demonstrated a differential effect on reporter gene expression supported by the Int8–VNTR 3 allele when placed in the intron of an expression vector and transfected into a mouse dopaminergic (substantia nigra-derived) cell line, SN4741, expressing the DAT1 (21). When challenged with the addition of KCl and forskolin to the cell culture medium, the 3 allele demonstrated an increased regulation of reporter gene expression (six times normal regulation), whereas the 2 allele did not exhibit any change or changed only slightly in comparison. We previously used the synergistic activation of K+-induced depolarization and forskolin to modulate the effects on expression of a variety of distinct transcriptional regulatory domains (24, 26–28). But, perhaps of more relevance, the effects observed were opposite when the cells were exposed to cocaine, with decreased expression of the 3 allele and little or no response of allele 2. From Fig. 3 a consistent effect can be observed with the 3 allele mediating large differential responses to stimuli, whereas the expression of the 2 allele remaining (comparatively) constant across the treatments.
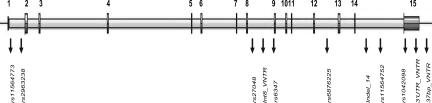
Fig. 3.
Schematic diagram of the DAT1 gene (SLC6A3) indicating the position of the markers analyzed in this study.
These expression data indicate that the Int8 VNTR sequence domain supports differential gene expression dependent on both the allele analyzed and the stimulus applied to the cell. These are the characteristics of a stimulus-sensitive regulatory domain whose function is determined by the binding of sequence-specific nucleic acid binding proteins. Because the alleles support differential expression we must assume that the difference in primary sequence as defined by the copy number is allowing differential binding of transcription factors, although we have not yet identified these. Even though it appears that the common Int8 VNTR alleles differ by only one perfect repeat unit, it has been demonstrated that the on/off rate for transcription factors in recognizing their cognate binding sites in tandem repeats can be affected by the copy number of those repeats (29, 30).
Given the differential response characteristics of the reporter gene containing the Int8 VNTR in reporter constructs and the neurochemical and neurotoxic effects of cocaine, we theorize that people possessing the 3/3 genotype will exhibit a differential response via altered DAT1 gene expression when exposed to cocaine. However, we have only shown this differential regulation to cocaine on the individual intronic VNTR domains, out-with their genomic context. Precise molecular characterization of the role that specific VNTR domains, such as Int8, play in endogenous regulation of DAT1 is not within the scope of this study. This experiment would potentially require complex reporter gene constructs that span the whole gene to encompass the many regulatory domains both 5′ and 3′ (31, 32). Nevertheless, what we were able to ascertain about the regulatory properties of the Int8–VNTR domain clearly suggests a mechanism by which this polymorphism can be associated with cocaine abuse by altering the tissue-specific or stimulus-inducible expression of the DAT1 gene. A previous study investigating the effect of cocaine abuse in the postmortem human brain demonstrated that, in cocaine users, DAT1 levels and DA uptake were elevated in the ventral striatum when compared with control subjects, providing further evidence of adaptation in DA uptake after chronic cocaine exposure (33).
It is notable that, to our knowledge, this is the first medium-to-large-scale study of the role of DAT1 gene variants in cocaine abuse both in terms of the sample size examined and the number of polymorphisms genotyped. There have been relatively few genetic association studies of cocaine abuse, only one of which (14) looked at the role of a DAT1 polymorphism in a sample of 102 cocaine abusers through genotyping the 3′ UTR VNTR. In that study, Gelernter et al. (14) failed to find an association with cocaine abuse but did suggest a role for DAT1 3′ UTR VNTR alleles in cocaine-induced paranoia, which was not measured in our sample. Other molecular genetic studies that have examined cocaine abuse as the primary phenotype have focused principally on the serotonin transporter (34–36), DA β-hydroxylase (37), and DA receptors (38).
To conclude, we found a robust association between cocaine abuse and a functional VNTR allele in the DAT1, conferring a small but detectible effect. Whereas a false positive finding due to population stratification is a potential concern for this type of study, this was addressed by stratification analysis. This indicated that there was population substructure, which was similarly distributed in the cases and controls. Covarying for this substructure in the association test did not change the results. The demonstration of differential functional effects for the risk allele, including an increased inhibitory effect on reporter gene expression when exposed to cocaine, indicates a possible functional role for this polymorphism in the response to cocaine. In addition, the haplotype and other genetic analyses in the sample support the idea that the Int8–VNTR is the relevant functional polymorphic motif in this sample. However, both the functional and genetic case-control work will require replication. If true, additional studies of drug abuse phenotypes other than cocaine abuse will also be necessary to explore the role of this polymorphism in addictive processes in general.
Methods
Association Study.
Patients.
Six hundred ninety-nine cocaine abusers (668 males, 31 females, mean age 26.7 years, SD = 7.2) were ascertained (39, 40). The study group consisted of drug users who were in treatment from August 1997 to October 1998 in one outpatient and six inpatient units located in the city of São Paulo, Brazil. Inclusion criteria were as follows: age 18 years and older, a history of cocaine abuse, and drug treatment at the selected centers. Individuals with another psychiatric diagnosis, such as psychosis, or a chronic physical illness such as diabetes or other metabolic disorders were excluded. All current cocaine users were then interviewed by using a structured interview to collect data on sociodemographic characteristics, data on sexual behaviors, and a drug-use profile. All subjects satisfied an ICD10 diagnosis of cocaine dependence (41). Blood samples were collected from all participants for genetic and other analyses. A total of 63.8% of the participants reported having smoked cocaine (crack) over the previous month, and 51.5% had snorted cocaine over the same period. The overall lifetime prevalence for heroin use in the sample was <5% (47).
Controls.
Eight hundred sixty-six healthy controls (592 males, 274 females, mean age 31.7 years, SD = 9.9) were recruited from the Blood Transfusion Unit of the Hospital das Clinicas, Faculty of Medicine, University of São Paulo. Each blood donor was screened by using a short questionnaire investigating contagious diseases and the use of any kind of drug. Subjects with a past history of drug abuse or with recent use of an illegal drug were excluded. During the act of donation a short interview was conducted, and subjects with a lifetime history of a psychiatric disorder requiring admission to a hospital or suffering from a psychiatric condition at time of interview were excluded.
Ethics.
All of the subjects included in this study gave written informed consent, and this project was approved by the Ethical Committee of the Federal University of São Paulo and other relevant ethics committees.
Selection of Genetic Markers.
VNTRs.
In silico analyses of publicly available human DAT1 sequence (http://genome.ucsc.edu, July 2003 release, NCBI Version 31) revealed a large number of genomic VNTRs in introns. The 40-bp 3′ UTR VNTR (designated here as 3′ UTR) and a VNTR in Int8 of the gene were selected for the investigation.
To help estimate haplotype patterns of this region, we also selected a novel VNTR (37-bp VNTR) located ≈10 kb 3′ to the 3′ UTR VNTR and a putatively functional 15-bp insertion/deletion polymorphism located in intron 14 of the gene (Indel_14) (11). The 37-bp VNTR is described as 6.1 copies of a 37-bp repetitive element polymorphism, and the Int8 is described also as 6.1 copies, but of a 30-bp element in the Simple Repeat table of the Human Genome in the University of California (Santa Cruz) Golden Path database (http://genome.ucsc.edu) (42). Alleles of the Int8 and 37-bp VNTRs were called according to their relative size (smallest = allele 1), and the coding system using the range from 3 to 11 copies was applied for the 40-bp 3′ UTR VNTR (6).
SNPs.
In a previous study analyzing variation within DAT1, Greenwood et al. (10) selected a number of SNPs that span the gene from the distal promoter through the 3′ UTR that are suitable for LD analyses. We genotyped seven of these SNPs: rs2963238 (C/A) and rs11564752 (G/T) in intron 1, rs27048 (A/G) in Int8, rs6347 (A/G) in exon 9, rs6876225 (A/C) in intron 12, rs11564773 (A/G) in intron 14, and rs1042098 in the 3′ UTR of the gene. See Fig. 3 for a diagram with the location of all markers.
Markers for Stratification Analysis.
We selected 17 SNPs and seven highly polymorphic microsatellite markers that exhibit large allele frequency differences among the three main Brazilian ancestral populations (Europeans, Africans, and Native Americans; M. Shriver, personal communication). Genotyping of all SNPs selected for this study was performed blind to status by using an amplifluor assay and was performed under contract by K-Biosciences (Cambridge, U.K.). Primers and conditions for the microsatellites used can be obtained upon request.
Statistical Analyses.
Population substructure in the study sample was studied by using the program lpop (43) to define clusters of ancestry of similar individuals by using multilocus genotypes. To identify the best model for the Brazilian population, we performed runs combining various available parameters in the program and different numbers of genetic clusters (K = 1 to K = 10) represented by the individuals genotyped.
Genotype and allele frequencies were compared by using a χ2 test, and P values were assessed by using spss version 10.0 and checked by means of simulation (clump Version 2.2) (44). The role of other clinical variables was tested with ANOVA or a nonparametric test as appropriate. genecounting Version 2.0 (refs. 44 and 45; www.mds.qmw.ac.uk/statgen/dcurtis/software.html) and whap (http://pngu.mgh.harvard.edu/~purcell/whap) were used to estimate haplotype frequencies. Odds ratios and 95% confidence intervals were derived from logistic regression. In addition, pairwise LD was calculated by using the LD pairs program from the gc utilities package (45). Hardy–Weinberg equilibrium was tested by calculating a χ2 statistic with one degree of freedom for the SNPs and by using clump, which uses simulations of the data to generate a distribution of χ2 statistics to assess the significance of the test-derived χ2 for the multiallelic markers tested.
Expression Analyses.
Cell growth.
SN4741 is a mouse substantia nigra-derived dopaminergic neuronal cell line and was cultured at 33°C, 5% CO2, in DMEM with Glutamax supplemented with 10% FCS, 1% glucose, and penicillin–streptomycin. After transfection, cells were incubated in the same conditions (46). JAR cells were used for the pGL3p luciferase vector (Promega) transfections and were similarly cultured.
Construction of reporter gene constructs.
We inserted the two common Int8 allelic variants into the intron of the Renilla vector phRLsv40 (Promega) using a plasmid containing a single AscI linker within its intronic region to allow for cloning of test-postulated intronic regulatory domains. The primers were modified (forward, 5′-TTGGCGCGCCGCTTGGGGAAGGAAGGG-3′; reverse, 5′-TTGGCGCGCCGTGTGCGTGCATGTGG-3′) to include Asc1 restriction sites enabling direct ligation of the PCR product into phRLsv40 (Promega). All plasmids were confirmed for validity and directionality by DNA sequencing.
Transfection.
Cells (≈1 × 104) were plated onto 24-well plates 24 h before transfection with Transfast (Promega). Transfections were optimized according to the manufacturer’s instructions. In brief, reporter plasmid (0.5 μg per well) was mixed with serum-free cell medium (200 μl per well), and Transfast was added at a ratio of 2:1 (3 μl per well). Cells were then washed twice with PBS, and DNA medium mixture was added for 1 h, after which 1 ml of relevant cell medium containing 10% FBS was added.
Reporter gene assay.
After 48 h cells were washed twice with PBS and then lysed with Passive Lysis buffer (Promega). After 15 min of agitation at room temperature, the cell lysate was centrifuged briefly at 10,000 × g. Supernatants were assayed for reporter gene expression by using the relevant Promega assay system. Supernatant (20 μl) was added to 100 μl of assay reagent in opaque 96-well plates, and the light emission was measured over a given time interval with the Life Sciences Labsystems Luminoskan (model RT). Where appropriate, cells were serum-deprived for 1 h and incubated for 24 h in normal cell medium, during which they were exposed to a potassium-evoked depolarization, a potassium-evoked depolarization combined with forskolin, cocaine hydrochloride (Sigma), or not exposed. Forskolin was used at 10 μM final concentration. Depolarization of SN4741 cells was achieved by using final concentrations of 10 mM CaCl2 and 40 mM KCl. The cells were also exposed to cocaine hydrochloride at concentrations of 1 μM and 10 μM. Luciferase/Renilla results were normalized to total protein concentration, which was measured with the BCA protein assay kit (Pierce) in accordance with the manufacturer’s instructions. Results are means ± SE of three or more experiments performed in triplicate using cells of the same or similar passage number.
Acknowledgments
We thank all of the patients and control subjects who took part in this study, which would not have been possible without them. G.B. is a Medical Research Council Bioinformatics Training Fellow. This work was supported in part by Fundação Para o Amparo à Pesquisa do Estado de São Paulo Grant 99/04678-2 (to H.V. and R.L.).
Abbreviations
- DA
dopamine
- DAT
DA transporter
- VNTR
variable-number tandem repeat
- SNP
single-nucleotide polymorphism
- LD
linkage disequilibrium
- Int8
intron 8
- LRT
likelihood ratio test.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Giros B., Caron M. G. Trends Pharmacol. Sci. 1993;14:43–49. doi: 10.1016/0165-6147(93)90029-j. [DOI] [PubMed] [Google Scholar]
- 2.Ritz M. C., Lamb R. J., Goldberg S. R., Kuhar M. J. Science. 1987;237:1219–1223. doi: 10.1126/science.2820058. [DOI] [PubMed] [Google Scholar]
- 3.Bierut L. J., Dinwiddie S. H., Begleiter H., Crowe R. R., Hesselbrock V., Nurnberger J. I., Jr., Porjesz B., Schuckit M. A., Reich T. Arch. Gen. Psychiatry. 1998;55:982–988. doi: 10.1001/archpsyc.55.11.982. [DOI] [PubMed] [Google Scholar]
- 4.Kendler K. S., Prescott C. A. Br. J. Psychiatry. 1998;173:345–350. doi: 10.1192/bjp.173.4.345. [DOI] [PubMed] [Google Scholar]
- 5.Merikangas K. R., Stolar M., Stevens D. E., Goulet J., Preisig M. A., Fenton B., Zhang H., O’Malley S. S., Rounsaville B. J. Arch. Gen. Psychiatry. 1998;55:973–979. doi: 10.1001/archpsyc.55.11.973. [DOI] [PubMed] [Google Scholar]
- 6.Vandenbergh D. J., Persico A. M., Hawkins A. L., Griffin C. A., Li X., Jabs E. W., Uhl G. R. Genomics. 1992;14:1104–1106. doi: 10.1016/s0888-7543(05)80138-7. [DOI] [PubMed] [Google Scholar]
- 7.Ueno S., Nakamura M., Mikami M., Kondoh K., Ishiguro H., Arinami T., Komiyama T., Mitsushio H., Sano A., Tanabe H. Mol. Psychiatry. 1999;4:552–557. doi: 10.1038/sj.mp.4000562. [DOI] [PubMed] [Google Scholar]
- 8.Martinez D., Gelernter J., Abi-Dargham A., van Dyck C. H., Kegeles L., Innis R. B., Laruelle M. Neuropsychopharmacology. 2001;24:553–560. doi: 10.1016/S0893-133X(00)00216-5. [DOI] [PubMed] [Google Scholar]
- 9.Kirley A., Lowe N., Hawi Z., Mullins C., Daly G., Waldman I., McCarron M., O’Donnell D., Fitzgerald M., Gill M. Am. J. Med. Genet. 2003;121B:50–54. doi: 10.1002/ajmg.b.20071. [DOI] [PubMed] [Google Scholar]
- 10.Greenwood T. A., Alexander M., Keck P. E., McElroy S., Sadovnick A. D., Remick R. A., Shaw S. H., Kelsoe J. R. Mol. Psychiatry. 2002;7:165–173. doi: 10.1038/sj.mp.4000958. [DOI] [PubMed] [Google Scholar]
- 11.Greenwood T. A., Kelsoe J. R. Genomics. 2003;82:511–520. doi: 10.1016/s0888-7543(03)00142-3. [DOI] [PubMed] [Google Scholar]
- 12.Chen C. K., Chen S. L., Mill J., Huang Y. S., Lin S. K., Curran S., Purcell S., Sham P., Asherson P. Mol. Psychiatry. 2003;8:393–396. doi: 10.1038/sj.mp.4001238. [DOI] [PubMed] [Google Scholar]
- 13.Le Couteur D. G., Leighton P. W., McCann S. J., Pond S. Movement Disorders. 1997;12:760–763. doi: 10.1002/mds.870120523. [DOI] [PubMed] [Google Scholar]
- 14.Gelernter J., Kranzler H. R., Satel S. L., Rao P. A. Neuropsychopharmacology. 1994;11:195–200. doi: 10.1038/sj.npp.1380106. [DOI] [PubMed] [Google Scholar]
- 15.Ujike H., Harano M., Inada T., Yamada M., Komiyama T., Sekine Y., Sora I., Iyo M., Katsu T., Nomura A., et al. Pharmacogenomics J. 2003;3:242–247. doi: 10.1038/sj.tpj.6500189. [DOI] [PubMed] [Google Scholar]
- 16.Miller G. M., Madras B. K. Mol. Psychiatry. 2002;7:44–55. doi: 10.1038/sj.mp.4000921. [DOI] [PubMed] [Google Scholar]
- 17.Fuke S., Suo S., Takahashi N., Koike H., Sasagawa N., Ishiura S. Pharmacogenomics J. 2001;1:152–156. doi: 10.1038/sj.tpj.6500026. [DOI] [PubMed] [Google Scholar]
- 18.Lesch K., Balling U., Gross J., Strauss K., Wolozin B. L., Murphy D. L., Riederer P. J. Neural Trans. Gen. 1994;95:157–162. doi: 10.1007/BF01276434. [DOI] [PubMed] [Google Scholar]
- 19.Contente A., Dittmer A., Koch M. C., Roth J., Dobbelstein M. Nat. Genet. 2002;30:315–320. doi: 10.1038/ng836. [DOI] [PubMed] [Google Scholar]
- 20.Hui J., Reither G., Bindereif A. RNA. 2003;9:931–936. doi: 10.1261/rna.5660803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michelhaugh S. K., Fiskerstrand C., Lovejoy E., Bannon M. J., Quinn J. P. J. Neurochem. 2001;79:1033–1038. doi: 10.1046/j.1471-4159.2001.00647.x. [DOI] [PubMed] [Google Scholar]
- 22.Hope B., Kosofsky B., Hyman S. E., Nestler E. J. Proc. Natl. Acad. Sci. USA. 1992;89:5764–5768. doi: 10.1073/pnas.89.13.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moratalla R., Vickers E. A., Robertson H. A., Cochran B. H., Graybiel A. M. J. Neurosci. 1993;13:423–433. doi: 10.1523/JNEUROSCI.13-02-00423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobson S. P., Quinn J. P., Morrow J. A., Mulderry P. K. Neurosci. Lett. 1994;167:19–23. doi: 10.1016/0304-3940(94)91018-9. [DOI] [PubMed] [Google Scholar]
- 25.Tabuchi A., Sakaya H., Kisukeda T., Fushiki H., Tsuda M. J. Biol. Chem. 2002;277:35920–35931. doi: 10.1074/jbc.M204784200. [DOI] [PubMed] [Google Scholar]
- 26.Morrison C. F., McAllister J., Dobson S. P., Mulderry P. K., Quinn J. P. Mol. Cell. Neurosci. 1994;5:165–175. doi: 10.1006/mcne.1994.1018. [DOI] [PubMed] [Google Scholar]
- 27.Walker P. D., Andrade R., Quinn J. P., Bannon M. J. J. Neurochem. 2000;75:882–885. doi: 10.1046/j.1471-4159.2000.0750882.x. [DOI] [PubMed] [Google Scholar]
- 28.Morrison C. F., McAllister J., Lyons V., Chapman K., Quinn J. P. Neurosci. Lett. 1994;181:117–120. doi: 10.1016/0304-3940(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 29.Quinn J. P., Holbrook N., Levens D. Mol. Cell. Biol. 1987;7:2735–2744. doi: 10.1128/mcb.7.8.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levens D., Howley P. M. Mol. Cell. Biol. 1985;5:2307–2315. doi: 10.1128/mcb.5.9.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacKenzie A., Payne C., Boyle S., Clarke A. R., Quinn J. P. Mol. Cell. Neurosci. 2000;16:620–630. doi: 10.1006/mcne.2000.0902. [DOI] [PubMed] [Google Scholar]
- 32.MacKenzi A., Quinn J. P. Neuropeptides. 2004;38:1–15. doi: 10.1016/j.npep.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Mash D. C., Pablo J., Ouyang Q., Hearn W. L., Izenwasser S. J. Neurochem. 2002;81:292–300. doi: 10.1046/j.1471-4159.2002.00820.x. [DOI] [PubMed] [Google Scholar]
- 34.Patkar A. A., Berrettini W. H., Hoehe M., Hill K. P., Sterling R. C., Gottheil E., Weinstein S. P. Addict. Biol. 2001;6:337–345. doi: 10.1080/13556210020077064. [DOI] [PubMed] [Google Scholar]
- 35.Patkar A. A., Berrettini W. H., Hoehe M., Hill K. P., Gottheil E., Thornton C. C., Weinstein S. P. Psychiatr. Genet. 2002;12:161–164. doi: 10.1097/00041444-200209000-00007. [DOI] [PubMed] [Google Scholar]
- 36.Patkar A. A., Berrettini W. H., Hoehe M., Thornton C. C., Gottheil E., Hill K., Weinstein S. P. Psychiatry Res. 2002;110:103–115. doi: 10.1016/s0165-1781(02)00098-7. [DOI] [PubMed] [Google Scholar]
- 37.Cubells J. F., Kranzler H. R., McCance-Katz E., Anderson G. M., Malison R. T., Price L. H., Gelernter J. Mol. Psychiatry. 2000;5:56–63. doi: 10.1038/sj.mp.4000657. [DOI] [PubMed] [Google Scholar]
- 38.Comings D. E., Gonzalez N., Wu S., Saucier G., Johnson P., Verde R., MacMurray J. P. Mol. Psychiatry. 1999;4:484–487. doi: 10.1038/sj.mp.4000542. [DOI] [PubMed] [Google Scholar]
- 39.Turchi M. D., Diaz R. S., Martelli C. M. T., Sabino E. C., da Silva W. P., Filho O. F., Laranjeira R. R., Busch M. P., Castelo A. J. Acquired Immune Defic. Syndr. 2002;30:527–532. doi: 10.1097/00126334-200208150-00009. [DOI] [PubMed] [Google Scholar]
- 40.Messas G., Meira-Lima I., Turchi M., Franco O. F., Guindalini C., Castelo A., Laranjeira R. R., Vallada H. Psychiatr. Genet. 2005;15:171–174. doi: 10.1097/00041444-200509000-00006. [DOI] [PubMed] [Google Scholar]
- 41.World Health Organization. The ICD10 Classification of Mental and Behavioural Disorders: Diagnostic Criteria for Research. Geneva: W.H.O.; 1993. [Google Scholar]
- 42.Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell S., Sham P. Hum. Hered. 2004;58:93–107. doi: 10.1159/000083030. [DOI] [PubMed] [Google Scholar]
- 44.Sham P. C., Curtis D. Ann. Hum. Genet. 1995;59:97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhao J. H. Bioinformatics. 2004;20:1325–1326. doi: 10.1093/bioinformatics/bth071. [DOI] [PubMed] [Google Scholar]
- 46.Son J. H., Chun H. S., Joh T. H., Cho S., Conti B., Lee J. W. J. Neurosci. 1999;19:10–20. doi: 10.1523/JNEUROSCI.19-01-00010.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guindalini C., Vallada H., Breen G., Laranjeira R. BMC Public Health. 2006;6:10. doi: 10.1186/1471-2458-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]