Abstract
Most class I MHC ligands are generated from the vast majority of cellular proteins by proteolysis within the ubiquitin–proteasome pathway and are presented on the cell surface by MHC class I molecules. Here, we present the crystallographic analysis of yeast 20S proteasome in complex with the inhibitor homobelactosin C. The structure reveals a unique inhibitor-binding mode and provides information about the composition of proteasomal primed substrate-binding sites. IFN-γ inducible substitution of proteasomal constitutive subunits by immunosubunits modulates characteristics of generated peptides, thus producing fragments with higher preference for binding to MHC class I molecules. The structural data for the proteasome:homobelactosin C complex provide an explanation for involvement of immunosubunits in antigen generation and open perspectives for rational design of ligands, inhibiting exclusively constitutive proteasomes or immunoproteasomes.
Keywords: antigen presentation, IFN-γ, inhibition
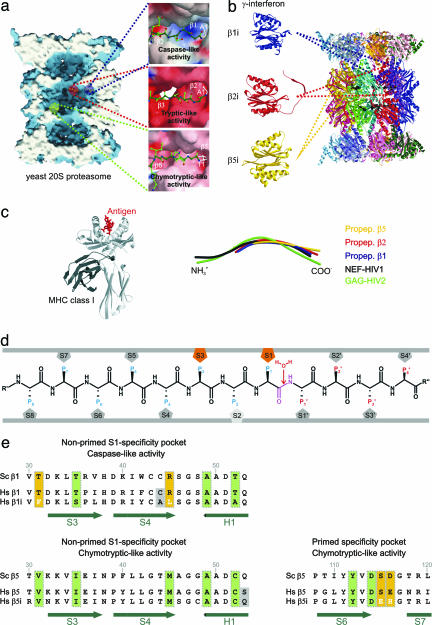
The ubiquitin–proteasome pathway generates peptide products with a narrow length distribution centered around 8–12 mers (1), a size suitable for binding to MHC class I molecules (2). This process allows CD8+ T lymphocytes to identify and eliminate cells that are synthesizing abnormal or “foreign” proteins, as may arise through mutations or infection by viruses (3). The 20S proteasome is the central component of this degradation system. It is composed of 28 subunits, which are arranged in four seven-membered rings that stack upon each other, yielding an α1–7β1–7β1–7α1–7 complex with twofold symmetry (4, 5). The two inner rings are solely built from β-type subunits, which form a hydrolytic chamber, whereas the α-rings are essential for assembly and substrate gating (1). Eukaryotic proteasomes contain three distinct proteolytically active sites, β1, β2, and β5 (4). Catalytic activities of these β-type subunits are associated with their N-terminal threonine residue, which acts as the nucleophile in hydrolysis, thus classifying proteasomes as members of the family of N-terminal nucleophilic hydrolases (6, 7). The maturation of active β-type subunits, which are synthesized as precursors, occurs by intrasubunit autolysis between residues Gly-1 and Thr-1 within the assembled particle, a process requiring a Gly-Thr site and catalytic residues (8, 9). Crystal structure analysis of autolysis-incompetent yeast proteasome mutants provided insights into the structure of the prosegments and helped to identify the three distinct nonprimed substrate-binding channels (10, 11). Although in vivo assays in eukaryotic cells have shown that proteasomes can cleave after most amino acids, cleavage preferences, termed caspase-, tryptic-, and chymotryptic-like activities, have been noted (12) and can roughly be assigned to the subunits β1, β2, and β5, respectively (Fig. 1a) (10). In vertebrates, specific catalytically active proteasomal β-type subunits, collectively referred to as immunosubunits (13), are found, which replace the constitutive proteolytically active subunits and considerably improve proteasome-dependent antigen processing. In addition, the immunosubunits control the number of cell-surface MHC class I molecules (14). IFN-γ induces the synthesis of the immunosubunits β1i, β2i, and β5i (Fig. 1b), which are cooperatively incorporated into nascent proteasomes, thereby replacing their homologues β1, β2, and β5 (15, 16) and generating immunoproteasomes. Thus, cytokine induction increases the functional diversity of the proteasome pool in cells, allowing them to produce a wider range of peptides for presentation to the immune system.
Fig. 1.
Proteasomal proteolytically active sites involved in the generation of MHC class I ligands. (a) Surface representation of the yeast 20S proteasome in complex with propeptides, clipped along the cylindrical pseudo sevenfold symmetry axis. Accessible surfaces are depicted in blue, and the cutting surface is in white. Propeptides are shown as space-filling models in yellow and indicate the proteolytically different active sites. The various proteolytic active centers are marked in a specific color coding: blue, subunit β1; red, subunit β2; and green, subunit β5. Cleavage preferences, termed caspase-, tryptic-, and chymotryptic-like activity, are zoomed and illustrated as surfaces; propeptides are presented as ball-and-stick models. Surface colors indicate positive and negative electrostatic potential contoured from 15 kT/e (intense blue) to −15 kT/e (intense red). (b) Topology of the 28 subunits of the yeast 20S proteasome in ribbon presentation. IFN-γ-inducible mammalian subunits β1i, β2I, and β5i are modeled by the corresponding constitutive yeast subunits. (c Left) MHC class I molecule in complex with an antigen (c Right). Structural superposition of propeptides β1, β2, and β5 with NEF-HIV1 and GAG-HIV2 antigen bound to MHC class I molecules. (d) Standard orientation for peptide substrates bound to the proteasomal specificity pockets. Substrates are oriented from their N to their C terminus. The scissile peptide bond is shown in magenta, flanked by the nucleophilic water molecule, which is incorporated into the product during hydrolysis. Residues on the left side of the scissile peptide bond in substrates, generating the C-terminal part in the product, are termed P sites; residues on the right side are termed P′ sites. Specificity pockets, which are responsible for ligand stabilization, are termed S and S′ pockets, respectively (11). (e) Sequence alignment of the yeast and the human constitutive subunit and immunosubunit for subunit β1 (nonprimed S1 site, Upper) and subunit β5 (nonprimed S1 and primed substrate-binding channel, Lower). Conserved residues are marked by vertical green boxes, significant variations between human constitutive subunits and immunosubunits are highlighted by yellow boxes, and variations in residues in proximity to the specificity pockets are shown against a gray background. Secondary structure elements are indicated in green.
Results
Belactosin C, a natural product from Streptomyces sp. UCK14, exhibits antitumor activity (17), which has been shown to be significantly increased upon acetylation of the free amino group and esterification of the carboxyl group, as well as replacement of the ornithine moiety with lysine to furnish bis-benzyl-protected homobelactosin C (18). The latter shows IC50 values against human pancreoma and colon cancer cells at the low nanomolar level (19). The high antitumor activity of this compound is attributed to proteasomal inhibition (Fig. 2a).
Fig. 2.
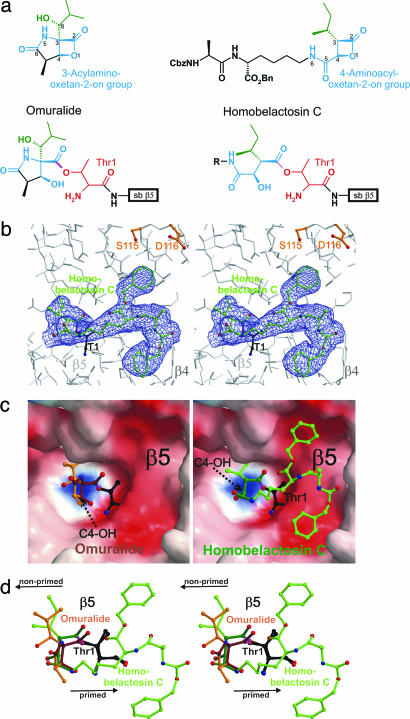
Homobelactosin C specifically binds to the chymotryptic-like active site by formation of an ester. (a) Chemical structures of omuralide and bisbenzyl-protected homobelactosin C in their native and bound conformation. The lead structure segments that in particular are involved in inhibitor binding are depicted in blue; Thr-1 of subunit β5 is in red. (b) Stereorepresentation of the chymotryptic-like active site of the yeast 20S proteasome (colored in gray) in complex with bisbenzyl-protected homobelactosin C (colored in green). Covalent linkage of the inhibitor with β5-Thr1Oγ is drawn in magenta. The electron density map (colored in blue) is contoured from 1σ around Thr-1 (colored in black) with 2Fo − Fc coefficients after twofold averaging. Temperature factor refinement indicates full occupancies of the inhibitor-binding site. The homobelactosin C derivative has been omitted for phasing. (c) Surface representation of the chymotryptic-like active site in complex with omuralide (depicted in brown, Left) and homobelactosin C (depicted in green, Right), covalently bound to Thr-1 (depicted in white). Note the overall similarity in the binding mode of both inhibitors but the different orientations of the generated C4-hydroxy group upon β-lactone ring opening (indicated by a black arrow). Surface colors indicate positive and negative electrostatic potential contoured from 15 kT/e (intense blue) to −15 kT/e (intense red). (d) Stereorepresentation of the superposition of lactacystin and bisbenzyl-protected homobelactosin C, including Thr-1 with respect to subunit β5. Omuralide is shown in brown, bisbenzyl-protected homobelactosin C is drawn in green, and the active site Thr-1 is in black. The superposition indicates that both inhibitors occupy the S1 specificity pocket in a unique way. The bisbenzyl-protected homobelactosin C is prolonged to the primed site. Nonprimed and primed sites are indicated by a black arrow.
Here, we describe the crystal structure of the bis-benzyl-protected homobelactosin C in complex with the yeast 20S proteasome. So far, all structural data reported on proteasomes in complex with diverse natural and synthetic ligands show characteristic binding only at the nonprimed sites of the proteasome (20). The homobelactosin C derivative discloses a high selectivity for the primed site of the chymotryptic-like active site. Covalent binding of the homobelactosin C derivative occurs by acylation of the free hydroxy group of the N-terminal Thr1Oγ of subunit β5 (Fig. 2b), in close analogy to what has been observed for omuralide (4). However, the hydrolysis of the β-lactone ring of each molecule has unique consequences. In the case of omuralide, the presence of the γ-lactam ring prevents free rotation about the C-3/C-4 bond, and the newly generated hydroxy group at C4 forms a hydrogen bond to the amino group of Thr-1. In contrast, the generated hydroxy group at C4 of bisprotected homobelactosin C points in the opposite direction and forms a hydrogen bond to Arg19O with a distance of 2.7 Å (Fig. 2c). A similar role is fulfilled by C-8OH of omuralide, although the atoms are not superpositioned. The amide nitrogen (NH-6) of the 4-aminocarbonyl side chain adopts a similar position to that of the C-4OH of omuralide, and the remainder of the side chain is bound to the primed site of the proteasome. Although both proteasome inhibitors, omuralide and homobelactosin C, are identically linked to the Thr1Oγ, they follow unique mechanisms to prevent cleavage of their newly formed ester bonds. In the case of omuralide, the generated C4-hydroxy group, which is fixed in position by the γ-lactam ring, occupies the position formerly taken by a nucleophilic water molecule in the unligated enzyme, thereby blocking the attack of the Thr1Oγ-CO ester bond (33). In the bisprotected homobelactosin C, the 4-aminocarbonyl side chain assumes this role. Omuralide has been found to specifically block the chymotryptic-like active site in accord with the neutral charge pattern of its S1 pocket. The structural superposition of bound omuralide and the homobelactosin C derivative shows a remarkable overlap of their P1 residues (Fig. 2d). The sterically demanding 4-aminocarbonyl side chain of the latter directs the inhibitor to the primed chymotryptic-like active site and prevents binding to the caspase- and tryptic-like active sites where it would cause a steric clash. It was shown that the benzyl ester derivative of belactosin A (IC50 = 48 nM) is five times more potent than belactosin A itself (19). The homobelactosin C derivative used here contains the same protective groups, which reveal characteristic hydrophobic interactions with protein residues (Fig. 2b), explaining the tighter binding. Because this binding pocket is negatively charged by the β5-Asp-114 and β5-Asp-116 moieties, appropriate positively charged substituents may further enhance binding.
Although the 4-aminocarbonyl side chain of the bisprotected homobelactosin C is not directly comparable to protein substrates, which would provide their distinct and characteristic structural elements for further stabilization, its binding mode indicates the preferred trajectory of ligand and substrate binding to the primed site of the proteasome.
IFN-γ-inducible exchange of proteasomal constitutive subunits by immunosubunits modulates the distribution of the generated peptides, thus providing fragments, which have an increased preference to bind to MHC class I molecules (21). Peptides effectively presented by MHC class I molecules usually have a size of eight to nine amino acids, which represents the typical product length generated by all 20S proteasomes after N-terminal trimming (22). Structural superposition shows that the conformation of MHC class I bound peptides matches quite well with the propeptides of the proteasomal active subunits (rms-bond deviation of polyAla <2.2 Å), which may be regarded as models for optimal substrates, suggesting coevolution of MHC molecules and proteasomal ligand-binding sites (Fig. 1d). Antigenic peptides exhibit predominantly basic or hydrophobic C-terminal anchor residues (23, 24), required for tight binding and stabilization of MHC class I molecules. The proteasomal chymotryptic- and tryptic-like active sites fulfill these criteria, but not subunit β1, which shows the most extensive exchanges of residues of the S1 specificity pocket in β1i: Thr-31 is replaced with phenylalanine and Arg-45 with leucine (Fig. 1e). Modeling experiments with wild-type yeast 20S proteasome show that these two substitutions reduce the size of the S1 pocket and suggest that the caspase-like activity is altered to a chymotryptic-like activity. This explains earlier results, showing that knockout mutants of subunit β1i have a modified viral-specific T cell response and modulated peptidase activity of the proteasome (25, 26). Similar modeling of subunits β2i and β5i does not indicate substantial modifications in the shape and specificities of their nonprimed S1 pockets (Fig. 1e). However, in vivo experiments in mice mutants lacking these immunosubunits show a severe defect in MHC class I presentation (27). Comparison of the primary sequences of human subunits β5 and β5i reveals a conspicuous alteration in the primed specificity channel: Ser-115 and Glu-116 of the constitutive subunit β5 are replaced by Glu and His in subunit β5i, respectively (Fig. 1e). These substitutions modify the size and polarity of the primed substrate-binding channel (Fig. 2b), which may have a significant effect on the substrate preference and cleavage pattern. Sequence alignment of mammalian β5 and β5i subunits shows strict conservation of these residues in the constitutive β5 subunits and minor species-specific differences in the immunosubunits β5i, indicating a conserved principle of modulating the primed substrate-binding channel. The putative primed substrate-binding channels in the β1 and β2 subunits modeled on the basis of subunit β5 show similar architectures and exhibit similar significant differences between constitutive subunits and immunosubunits. We suggest that the main effect of immunosubunit incorporation is a change of the cleavage pattern by β1i and of class I MHC ligand selection by the primed substrate-binding sites of all three subunits.
Methods
Crystals of 20S proteasome from Saccharomyces cerevisiae were grown in hanging drops at 24°C (28) and incubated for 60 min with homobelactosin C (10 mM in DMSO), which was synthesized as described (18). The protein concentration used for crystallization was 40 mg/ml in 10 mM Tris·HCl (pH 7.5) and 1 mM EDTA. Drops contained 3 μl of protein and 2 μl of reservoir solution [30 mM MgOAc/100 mM Mes (pH 7.2)/10% of 2-methyl-2,4-pentanediol (MPD)]. The space group of proteasomal complex crystals belongs to P21 with cell dimensions of a = 135.5 Å, b = 301.1 Å, c = 144.4 Å, and β = 112.8°. Data to 3.1 Å were collected by using synchrotron radiation with λ = 1.05 Å at the BW6-beamline at Deutsches Elektronen Synchrotron (Hamburg, Germany). Crystals were soaked in a cryoprotecting buffer (30% MPD/20 mM MgOAc/100 mM Mes, pH 6.9) and frozen in a stream of liquid nitrogen gas at 90 K (Oxford Cryosystems, Oxford, U.K.). X-ray intensities were evaluated by using the denzo program package, and data reduction was performed with scalepack (29, 30). Anisotropy of diffraction was corrected by an overall anisotropic temperature factor, comparing observed and calculated structure amplitudes by using the program cns (31). A total of 2,018,345 reflections yielded 209,872 unique reflections (99.2% completeness). The corresponding Rmerge was 10.1% at 3.1-Å resolution (43.6% for the last-resolution shell). Electron density was improved by averaging and back-transforming 10 times over the twofold noncrystallographic symmetry axis using the program package main (32). Conventional crystallographic rigid body and positional and temperature factor refinements were carried out with cns by using the yeast 20S proteasome structure as starting model (4). For model building, the program main was used. The structure was refined to a crystallographic R factor of 22.0% (free R factor 24.4%) with rms deviations from target values of 0.008 Å for bonds and 1.35° for angles. Modeling experiments were performed by using the coordinates of yeast 20S proteasome with the program main.
Acknowledgments
The help of G. Bourenkow (Deutsches Elektronen Synchrotron, BW6, Hamburg, Germany) with synchrotron data collection is gratefully acknowledged. We thank B. Potts (Nereus Pharmaceuticals, San Diego) for sharing her extensive chemical knowledge in the field of proteasome inhibition with us.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: Coordinates and diffraction data for the structure reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2FNY).
References
- 1.Groll M., Bochtler M., Brandstetter H., Clausen T., Huber R. Chembiochem. 2005;6:222–256. doi: 10.1002/cbic.200400313. [DOI] [PubMed] [Google Scholar]
- 2.Michalek M. T., Grant E. P., Gramm C., Goldberg A. L., Rock K. L. Nature. 1993;363:552–554. doi: 10.1038/363552a0. [DOI] [PubMed] [Google Scholar]
- 3.Cresswell P., Bangia N., Dick T., Diedrich G. Immunol. Rev. 1999;172:21–28. doi: 10.1111/j.1600-065x.1999.tb01353.x. [DOI] [PubMed] [Google Scholar]
- 4.Groll M., Ditzel L., Lowe J., Stock D., Bochtler M., Bartunik H. D., Huber R. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 5.Unno M., Mizushima T., Morimoto Y., Tomisugi Y., Tanaka K., Yasuoka N., Tsukihara T. Structure (London) 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 6.Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 7.Brannigan J. A., Dodson G., Duggleby H. J., Moody P. C., Smith J. L., Tomchick D. R., Murzin A. G. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 8.Ditzel L., Huber R., Mann K., Heinemeyer W., Wolf D. H., Groll M. J. Mol. Biol. 1998;279:1187–1191. doi: 10.1006/jmbi.1998.1818. [DOI] [PubMed] [Google Scholar]
- 9.Heinemeyer W., Fischer M., Krimmer T., Stachon U., Wolf D. H. J. Biol. Chem. 1997;272:25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- 10.Groll M., Heinemeyer W., Jager S., Ullrich T., Bochtler M., Wolf D. H., Huber R. Proc. Natl. Acad. Sci. USA. 1999;96:10976–10983. doi: 10.1073/pnas.96.20.10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schechter I., Berger A. Biochem. Biophys. Res. Commun. 1968;32:898–902. doi: 10.1016/0006-291x(68)90326-4. [DOI] [PubMed] [Google Scholar]
- 12.Orlowski M., Cardozo C., Michaud C. Biochemistry. 1993;32:1563–1572. doi: 10.1021/bi00057a022. [DOI] [PubMed] [Google Scholar]
- 13.Aki M., Shimbara N., Takashina M., Akiyama K., Kagawa S., Tamura T., Tanahashi N., Yoshimura T., Tanaka K., Ichihara A. J. Biochem. (Tokyo) 1994;115:257–269. doi: 10.1093/oxfordjournals.jbchem.a124327. [DOI] [PubMed] [Google Scholar]
- 14.Gaczynska M., Rock K. L., Goldberg A. L. Nature. 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 15.Ortiz-Navarrete V., Seelig A., Gernold M., Frentzel S., Kloetzel P. M., Hammerling G. J. Nature. 1991;353:662–664. doi: 10.1038/353662a0. [DOI] [PubMed] [Google Scholar]
- 16.Frentzel S., Pesold-Hurt B., Seelig A., Kloetzel P. M. J. Mol. Biol. 1994;236:975–981. doi: 10.1016/0022-2836(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 17.Asai A., Hasegawa A., Ochiai K., Yamashita Y., Mizukami T. J. Antibiot. 2000;53:81–83. doi: 10.7164/antibiotics.53.81. [DOI] [PubMed] [Google Scholar]
- 18.Larionov O. V., de Meijere A. Org. Lett. 2004;6:2153–2156. doi: 10.1021/ol049409+. [DOI] [PubMed] [Google Scholar]
- 19.Asai A., Tsujita T., Sharma S. V., Yamashita Y., Akinaga S., Funakoshi M., Kobayashi H., Mizukami T. Biochem. Pharmacol. 2004;67:227–234. doi: 10.1016/j.bcp.2003.08.035. [DOI] [PubMed] [Google Scholar]
- 20.Groll M., Huber R. Biochim. Biophys. Acta. 2004;1695:33–44. doi: 10.1016/j.bbamcr.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 21.Driscoll J., Brown M. G., Finley D., Monaco J. J. Nature. 1993;365:262–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 22.Beninga J., Rock K. L., Goldberg A. L. J. Biol. Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 23.Silver M. L., Parker K. C., Wiley D. C. Nature. 1991;350:619–622. doi: 10.1038/350619a0. [DOI] [PubMed] [Google Scholar]
- 24.Engelhard V. H. Curr. Opin. Immunol. 1994;6:13–23. doi: 10.1016/0952-7915(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 25.Boes B., Hengel H., Ruppert T., Multhaup G., Koszinowski U. H., Kloetzel P. M. J. Exp. Med. 1994;179:901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sibille C., Gould K. G., Willard-Gallo K., Thomson S., Rivett A. J., Powis S., Butcher G. W., De Baetselier P. Curr. Biol. 1995;5:923–930. doi: 10.1016/s0960-9822(95)00182-5. [DOI] [PubMed] [Google Scholar]
- 27.Fehling H. J., Swat W., Laplace C., Kuhn R., Rajewsky K., Muller U., von Boehmer H. Science. 1994;265:1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 28.Groll M., Huber R. Methods Enzymol. 2005;398:329–336. doi: 10.1016/S0076-6879(05)98027-0. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z., Borek D., Majewski W., Minor W. Acta Crystallogr. A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 31.Brünger A., Adams P., Clore G., DeLano W., Gros P., Grosse-Kunstleve R., Jiang J., Kuszewski J., Nilges M., Pannu N., et al. Acta Crystallogr. D. 1998;1:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Turk D. PhD thesis. Munich: Technische Universität München; 1992. [Google Scholar]
- 33.Groll M., Huber R., Potts B. J. Am. Chem. Soc. 2006 doi: 10.1021/ja058320b. in press. [DOI] [PubMed] [Google Scholar]