Abstract
Clefts on protein surfaces are avoided by antigen-combining sites of conventional antibodies, in contrast to heavy-chain antibodies (HCAbs) of camelids that seem to be attracted by enzymes’ substrate pockets. The explanation for this pronounced preference of HCAbs was investigated. Eight single domain antigen-binding fragments of HCAbs (VHH) with nanomolar affinities for lysozyme were isolated from three immunized dromedaries. Six of eight VHHs compete with small lysozyme inhibitors. This ratio of active site binders is also found within the VHH pool derived from polyclonal HCAbs purified from the serum of the immunized dromedary. The crystal structures of six VHHs in complex with lysozyme and their interaction surfaces were compared to those of conventional antibodies with the same antigen. The interface sizes of VHH and conventional antibodies to lysozyme are very similar as well as the number and chemical nature of the contacts. The main difference comes from the compact prolate shape of VHH that presents a large convex paratope, predominantly formed by the H3 loop and interacting, although with different structures, into the concave lysozyme substrate-binding pocket. Therefore, a single domain antigen-combining site has a clear structural advantage over a conventional dimeric format for targeting clefts on antigenic surfaces.
Keywords: antibody–lysozyme structures, camel single domain antibody, enzyme inhibitor, epitope–paratope interactions
Six hypervariable antigen-binding loops constitute the antigen-combining sites of conventional antibodies. These loops, three (H1–H3) from the variable domain of the heavy chain (VH), three (L1–L3) from the variable domain of the light chain (VL) are juxtaposed forming a continuous surface (paratope) that is complementary to a surface on the antigen (epitope) (1). The paratope is essentially planar for protein antigens and forms a groove or cavity to interact with peptides and haptens (2, 3). The loops L1–L3 and H1–H2 fold into a limited number of canonical structure classes, determined by the loop length and the presence of conserved residues at key positions within the hypervariable and framework regions (4, 5). The extreme length and sequence variability of H3 makes the structure prediction of this loop extremely difficult (6).
The structures of antigen-binding sites and loops, as well as the canonical loop determining residues, are well established (1, 4, 5). In contrast, the elucidation of the molecular basis for the recognition of particular epitopes by antibodies remains a major challenge. Our knowledge and paradigms of protein–epitope recognition by antibodies is largely based on the analysis of the immune response toward hen egg white lysozyme (HEWL). This is due to the high antigenicity, the large number of natural variants of HEWL (7), and the availability of eleven different crystal structures of Fab or Fv antibody fragments (8–10) in complex with lysozyme, collected over the last two decades. Six structures represent Fabs or Fvs that are clearly clonally unrelated (8), and one additional structure involves an artificially assembled antigen-specific VH–VL pair (11).
Camelids possess a functional class of antibodies devoid of light chains (referred to as heavy-chain antibodies or HCAbs) (12, 13). The antigen-combining site of these heavy-chain antibodies is limited to only three hypervariable loops (H1–H3) provided by the N-terminal variable domain (VHH). The first crystal structures of VHHs revealed that the H1 and H2 loops are not restricted to the known canonical structure classes defined for conventional antibodies (14). The H3 loops of VHHs are on average longer than those of conventional antibodies (15). In one case, the H3 loop was shown to protrude from the remaining paratope and inserts into the active site cleft of HEWL (16–18). Furthermore, it seems that a large fraction of the dromedary HCAbs have a preference for binding into active sites of enzymes against which they were raised (19).
To elucidate the structural basis of the remarkable HCAb epitope preferences, we performed an in depth analysis of epitope recognition by HCAbs in analogy to the study done for conventional antibodies toward HEWL (7, 8). We immunized three different dromedaries with HEWL, isolated eight anti-HEWL VHHs via phage-display, and mapped their epitopes. These epitopes of the isolated VHHs cluster in two nonoverlapping regions with the vast majority (six of eight) binding into the enzyme’s active site cleft. The same proportion (≈85%) of active site binders was also observed within the polyclonal HCAb immune response toward HEWL. The crystal structures of six VHHs in complex with HEWL demonstrate that the paratopes are characterized by a variety of different structures, dominated by the H3 antigen-binding loop. Our results sharply contrast with the epitope preferences of murine anti-HEWL responses represented by the available Fab or Fv::lysozyme crystal structures (8), which display planar epitopes located at three patches outside the active site.
Results
Ontogeny of the Isolated VHHs.
Eight different anti-HEWL VHHs, originating from three different dromedaries were isolated from immune libraries. The hypervariable loops of these VHHs differ significantly in sequence (Fig. 5, which is published as supporting information on the PNAS web site). Especially the sequences and lengths (12–24 aa) of the H3 region vary considerably. VHHs, like VH domains, are assembled through the recombination of one variable (V), one diversity (D), and one joining (J) gene segment out of a pool that is present in the genome (20). A search in the VHH germ-line gene database (20) revealed that the V gene segments of these HEWL-binders most probably originated from either the cvhhp08 (AJ245114.1) or the cvhhp11 (AJ245117.1) germ-line genes. These V-genes are the most frequently used germ-line genes in the V-D-J recombination of VHHs (20). Nucleotide sequence identities between matured and germ-line V sequences range from 84.4% (D2-L31) to 94.1% (D2-L27), corresponding to 17–39 nucleotide substitutions (Fig. 5). These substitutions follow the characteristic pattern for a classical somatic hypermutation mechanism leading to antigen affinity maturation. The analysis further indicated that the J segments JH3 or JH5 were used in the V-D-J rearrangement to arrive at the VHHs (camelid JH germ-line sequences; V. K. Nguyen, personal communication).
Kinetics and Affinity of the VHH–HEWL Interactions.
The kinetic and affinity constants of all VHH–HEWL interactions were determined by surface plasmon resonance (Fig. 6, which is published as supporting information on the PNAS web site). For the VHHs D3-L11, cAb-Lys-3, cAb-Lys-2, D2-L19, D2-L29, and D2-L24, the ka constants are within a range of 1 order of magnitude (0.21–5.4 × 106 M−1·s−1) and the kd values cover a range of two orders of magnitude (1.7–200 × 10−4 s−1). The affinity constants (KD) range from 77 pM to 70 nM. The kinetics of the D2-L27-HEWL interaction was too fast to measure. Nonlinear regression of the equilibrium binding response versus D2-L27 concentration yielded an affinity constant of 2.34 μM. The D2-L31–HEWL interaction exhibited complex binding kinetics consistent with an induced fit mechanism, with a global affinity constant of 69 nM. The ka, kd, and KD values for these VHH–HEWL interactions are within the range found for matured conventional antibody–antigen interactions (21).
Anti-HEWL VHHs Bind Predominantly to the Active Site Cleft of HEWL.
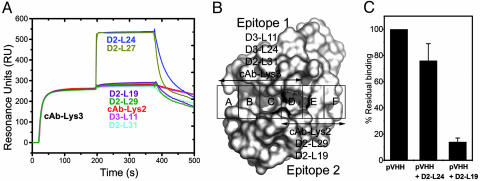
Using surface plasmon resonance (see Methods) we mapped the regions on the surface of HEWL that are recognized by our VHHs. The VHHs D2-L19, D2-L29, D2-L31, D3-L11, cAb-Lys-2, and cAb-Lys-3 (group 1) mutually exclude each other for antigen binding (Fig. 1A and Table 1, which is published as supporting information on the PNAS web site). The VHHs, D2-L24 and D2-L27 (group 2) also interfere with each other for antigen binding but remain competent to associate with HEWL in the presence of the VHHs of group 1 (Fig. 1A and Table 1). The small chemical compounds GlcNAc(β1–4)GlcNAc(β1–4)GlcNAc (NAG3) occupying the A-C substrate subsites of HEWL and Biebrich Scarlet, associating with the D-F carbohydrate-binding sites (22), inhibit the cAb-Lys-3, D2-L31, and D3-L11 interaction with HEWL. Only Biebrich Scarlet affects the HEWL binding of D2-L19, cAb-Lys-2, and D2-L29 (Fig. 1B and Table 1). Therefore, the assembly of all epitopes of group 1 VHHs covers a large part of the substrate-binding site of lysozyme.
Fig. 1.
Epitope mapping of the monoclonal and polyclonal VHHs. (A) HEWL binding of all VHHs in the presence of a saturating concentration of cAb-Lys-3 using a coinjection procedure. (B) Epitopes of HEWL active site binders based on the inhibition of binding by NAG3 and Biebrich Scarlet. (C) Residual HEWL binding of polyclonal VHH derived from the IgG3 fraction of dromedary D2. (Left) Binding in absence of competitor (set at 100%). (Center) Binding in presence of HEWL-saturating concentrations of D2-L24. (Right) Binding in presence of HEWL-saturating concentrations of D2-L19.
The dromedary HCAb isotypes, IgG2 and IgG3, are easily separated from conventional antibodies, IgG1 (12). A proteolytic digestion of these IgG3 antibodies (the most abundant HCAb isotype in the dromedary) allows the purification of a polyclonal VHH fraction (19). Such VHH pool of a dromedary immunized with HEWL was competed with D2-L19 (epitope group 1 member) or D2-L24 (group 2 member) for HEWL binding. Fig. 1C shows the percentage of binding in the absence (100%) or presence of monoclonal competitor, as a mean of three experiments at different concentrations of polyclonal serum VHHs. The group 2 and 1 VHHs inhibit the HEWL binding of the polyclonal VHH pool by ≈25% and ≈85%, respectively. Therefore, all HEWL-binding VHHs of IgG3 of dromedary D2 belong to one of the two observed complementation groups with the majority belonging to group 1. These epitope mapping experiments clearly show that the HCAb immune response is directed predominantly toward the active site of HEWL and that our set of monoclonal anti-HEWL VHHs contains a similar proportion of active cleft binders.
The VHH::HEWL Complex Structures.
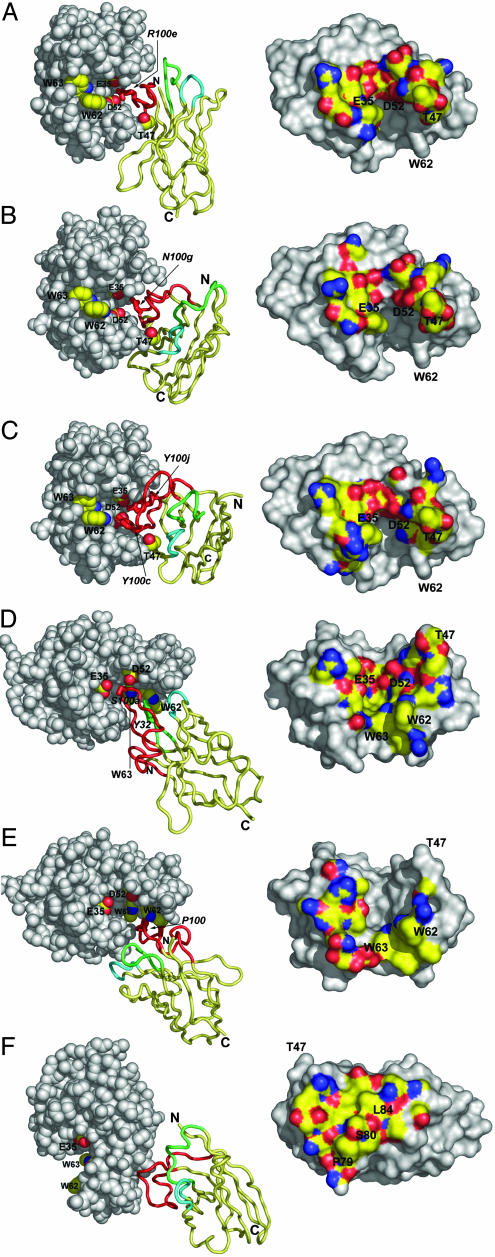
To allocate the different epitopes on HEWL at atomic resolution and to clarify the structural details of the paratope–epitope associations, we crystallized six VHH::HEWL complexes (Fig. 2). The structures were solved to a resolution varying between 1.4 and 2.1 Å and refined to Rfree factors between 0.196 and 0.239 (Table 2, which is published as supporting information on the PNAS web site).
Fig. 2.
The VHH::HEWL complex structures. The D2-L29::HEWL (A), D2-L19::HEWL (B), cAb-Lys-2::HEWL (C), cAb-Lys-3::HEWL (D), D3-L11::HEWL (E), and D2-L24::HEWL (F) complexes are shown. (Left) VHH::HEWL complexes are represented with HEWL molecules as gray space-filling models. The active site residues E35, D52, W62, and W63 are labeled and side-chain atoms color-coded: C, yellow; N, blue; and O, red, for all panels. For A–C, the residue T47 of HEWL is additionally colored and labeled. The VHHs are represented as ribbons, with the framework region painted in yellow and the H1, H2, and H3 loops painted in green, cyan, and red respectively. The most important amino acids that contact E35, D52, W62, or W63 of HEWL are represented as sticks and are labeled (italic). (Right) The surface on HEWL within 5-Å distance of the VHH is color-coded according to the atom type: C, yellow; N, blue; O, red; S, orange.
All VHHs possess the typical V Ig fold. The H1 loops adopt conformations that deviate significantly from the conventional canonical structure (Fig. 7A, which is published as supporting information on the PNAS web site), whereas the H2 loops adopt classical canonical structures (Fig. 7B). The H3 loops of all VHHs fold onto the five-stranded β-sheet and interact with residues of the framework 2 regions (Fig. 7C). This structural motif is a typical feature found in VHHs. It stabilizes the long H3 loops (23) and blocks access to the side of the domain corresponding to the VL interaction site of a conventional VH domain (13, 17).
The VHHs D2-L19, cAb-Lys-2, and D2-L29 contact highly overlapping epitopes (Fig. 2 Right), located at the D-F carbohydrate substrate subsites of HEWL, including the HEWL catalytic residues Glu-35L and Asp 52L (Fig. 2 A–C and Table 3, which is published as supporting information on the PNAS web site). D2-L29 buries the side chain of Thr-47L (located at the lower edge of the catalytic site) deeply between the H3 loop and the framework 2 region (Fig. 2A). This allows Arg100e of H3 to form a salt-bridge and two hydrogen bonds with Glu-35L and two with Asp-52L. D2-L19 and cAb-Lys-2 bury Thr-47L between their H3 and H2 loops (Fig. 2 B and C), which again allow residues of the H3 loops of both binders to make intimate contact with Glu-35L and Asp-52L (Fig. 2 B and C). The most remarkable difference between D2-L29 (Fig. 2A) and D2-L19/cAb-Lys-2 (Fig. 2 B and C) is found in the relative orientation to HEWL. In the former case, the H2 loop is more distant from the antigen and the H1 loop contacts the opposite side of the active site cleft. In the latter two cases, the HEWL is bound around the compact prolate VHH, without any involvement of the H1 loop for antigen binding.
The VHHs cAb-Lys-3 and D3-L11 target overlapping epitopes located at the A-D carbohydrate subsites of HEWL and include the residues Trp-62L and Trp-63L (Fig. 2 D and E and Table 3). The VHHs possess different loop structures and bind to their epitopes in a different orientation. The H1 and H2 loops of cAb-Lys-3 contact the W62L side of the catalytic cleft, whereas those of D3-L11 contact residues across the active site. The positioning of the antigen contacting residues results in a wedge-shaped paratope for both VHHs. Part of the H3 loop of cAb-Lys-3 even protrudes from the remaining paratope and penetrates more deeply into the active site cleft, allowing the side chain of Ser100a to contact the catalytic residues Asp-52L and Glu-35L.
The structure of the D2-L24–HEWL complex confirms the existence of an epitope, distinct from those of the active site binding group. This epitope is located outside of the active site and involves the short α-helix 79L-84L.
VHH::HEWL Complexes Versus Fv::Lysozyme Complexes.
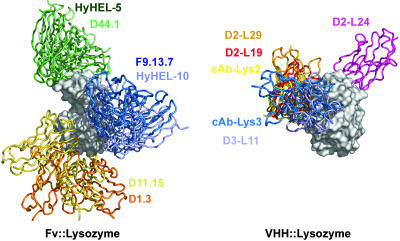
The HEWL superposition of six VHH::HEWL complexes gives a strikingly contrasting picture when compared to a corresponding superposition of the 6 representative Fv::lysozyme structures (Fig. 3) (8). The epitopes for the murine Fv’s cluster in three regions on the lysozyme surface, each of which involves patches outside the active site. The anti-HEWL VHHs cluster at two regions, one in the active site representing the majority of VHHs and one located outside the active site, representing the smaller group of anti-HEWL VHHs (represented by D2-L24, Fig. 3). To explain this remarkable difference in epitope recognition, we examined the structural and chemical features of the VHH::HEWL and Fv::HEWL interfaces. The statistical significance of the observed differences between the measured parameters of the different antibody classes was evaluated by using a Mann–Whitney U test.
Fig. 3.
Superposition of the antibody–lysozyme complexes for conventional antibodies (Left) and for VHHs (Right). The HEWL molecules (gray surfaces) are shown in the same orientation for both antibody classes. The antibody molecules are represented as colored ribbons and their identity is indicated in the same color.
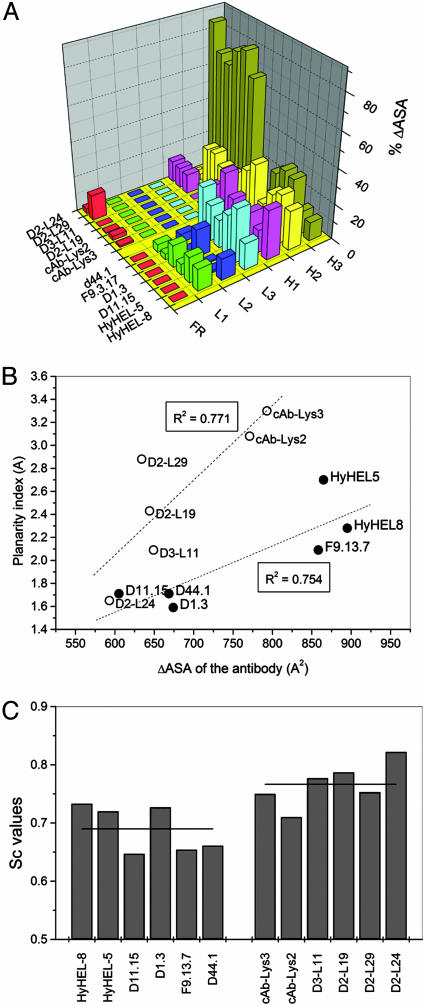
The ΔASA of the VHH or Fv and lysozyme upon complex formation, as well as the polarity of the interfaces (Tables 4 and 5, which are published as supporting information on the PNAS web site) is comparable (P > 0.1). Also, the number of contacts, hydrogen-bonds and ion-pairs (Tables 4–6, which are published as supporting information on the PNAS web site) do not differ significantly (P > 0.1) between VHH::HEWL and Fv::lysozyme complexes. In contrast to the anti-HEWL Fvs, where the paratope areas are more or less equally distributed over all six antigen-binding loops, the VHHs use predominantly their H3 loop for antigen interaction. The amino acids residing in the H3 loops of the VHHs dominate to a large extent the ΔASA of the VHH (≈60–80% of total interacting area) (Fig. 4A). In addition, the paratopes (and epitopes) of all VHHs, except D2-L24, deviate significantly from planarity. The ΔASAs of the VHH paratopes to HEWL are strongly dependent on the planarity index of the VHH paratopes (Fig. 4B), whereas for Fvs this dependency is less pronounced. Higher interface complementarities, as measured by the Sc parameter, further differentiate the VHH::HEWL interfaces from the Fv::lysozyme interfaces (P < 0.05) (Fig. 4C).
Fig. 4.
Structural and chemical differences between VHH::HEWL and Fv::lysozyme interfaces. (A) Contribution of each antigen-binding loop to the ΔASA of the paratope. (B) The planarity index of the VHH or Fv paratope versus their ΔASA. The dotted lines represent the linear trends for both types of antibodies (VHH or Fv) and the Pearson correlation coefficient is indicated. (C) The Sc values for all VHH::HEWL and Fv::lysozyme complexes. The horizontal line through the bars represents the average Sc value for Fvs and VHHs.
Discussion
Eight matured antigen-specific VHHs were isolated from three HEWL-immunized dromedaries by phage display. The VHH-HEWL kinetics and affinities are in the same range (i.e., nM) as those observed for the conventional Fab–HEWL interactions, and one VHH was found to interact via an induced fit mechanism. This finding shows that the VHH–HEWL interactions are as potent and display similar recognition mechanisms compared to Fab–lysozyme interactions.
The VHHs recognize two independent regions on HEWL. One region is targeted by six of eight VHHs and is composed of two overlapping epitopes encompassing a large part of the substrate-binding pocket of lysozyme. The two remaining VHHs target a region located at the antipode of the active site. This set of anti-HEWL VHHs covers the epitope specificities of HCAbs in the serum of immunized dromedaries. Indeed, one monoclonal VHH (D2-L19) representative for the six active site binders could inhibit ≈85% of the HEWL binding of polyclonal VHHs prepared from serum IgG3 of HEWL-immunized dromedaries. A binder (D2-L24) of the other epitope-group blocked ≈25% of the polyclonal VHHs binding to HEWL (Fig. 1C). Therefore, the distribution of our eight VHHs over the two epitope groups is also present to the same proportion in the polyclonal VHH pool. Furthermore, the variety in sequence and length of the hypervariable loops of the eight VHHs proves that they are clonally unrelated and their broad range in affinity and kinetics argues against a biased selection during phage-display pannings.
Clearly, the largest fraction of the anti-HEWL VHHs interacts with the active site of the antigen. This HEWL epitope preference deviates strikingly with that of matured murine antibodies against lysozyme (Fig. 3). The murine humoral response is dominated by three noncontinuous and nonoverlapping regions, located on the surface of lysozyme outside of the active site (8, 24). However, the antibody HyHEL10 inhibits the hydrolytic activity of cell wall substrates of Micrococcus luteus by 95% (25). This inhibition results from a close contact between side chains at the periphery of the paratope with the catalytic cleft residues Trp-62L and Trp-63L, whereas the central part of the antibody–antigen interface is located further away from the substrate pocket. The VH domain of D11.15 binds also close to the borders of the active site cleft, although it does not penetrate into this cavity nor does it block the access to substrate binding (26). Hence, the mouse Fv::lysozyme paradigm fits perfectly with the proposal of conventional antibodies having paratopes that are essentially planar to interact with proteinaceous antigens (1–3, 27). It should be noted that most of the antibody structures in the protein database are murine antibodies and these antibodies, whose germ line genes for the H3 severely limit their length. Human and rabbit antibodies have, on average, longer H3 loops and, therefore, might have a higher frequency of producing convex paratopes. Indeed, structural data suggest that antiviral human antibodies (e.g., b12, 447–52D, 4E10, 2F5, 17b) also target clefts or canyons by using long protruding H3 loops (28).
Our set of VHH::HEWL structures reveals that different structural solutions exist for binding the active site of HEWL. Despite their different epitope preference to that of murine anti-HEWL antibodies, most of the interface characteristics are strikingly similar. In contrast, all VHHs, recognizing the HEWL active site, display a convex paratope. It is the compact prolate nature of the VHH domain (due to absence of VL) and the characteristic H3 loop folding over the VHH framework region that generates a considerably convex antigen-combing site, possibly enhanced by a protruding loop structure as observed for cAb-Lys-3 (17) or by protruding side chains. It further provides an interaction surface as large as that of a combined VH-VL pair while leaving a significantly smaller footprint on the antigen.
Assuming that the pronounced convex paratope of a VHH is a key element for the preferential recognition of the HEWL active site, other single domain antibody fragments should equivalently favor clefts on the surface of proteinaceous antigens. It was already reported that a considerable fraction of the dromedary HCAbs acts as potent α-amylase, carbonic anhydrasen or β-lactamase inhibitors (19, 29). The crystal structure of a VHH against α-amylase confirmed that the convex paratope of the binder inserts into the enzyme’s active site (30). In addition, a llama VHH seemed to competitively inhibit potato starch branching enzyme A (31). More strikingly, the single domain of an anti-HEWL HCAb from nurse shark (i.e., NAR) recognizes also the active site (32), where it overlaps with the epitope of cAb-Lys-3. Here as well, the long H3 loop provides a large convex antigen interaction surface. Furthermore, a competitive inhibitor for hepatitis C virus NS3 protease was derived from a synthetic library of “camelized” human VH domains (33). The unique convex paratope of single domain antibody fragments dominated by the H3 loop residues to participate in the catalytic site recognition offer a pharmacological benefit. It should be feasible to use the H3 antigen-binding loop as a lead to generate small peptide mimetics to develop agonists or antagonists against medically important targets.
Of note, VHHs also form planar paratopes as exemplified by the nonactive site binder D2-L24. Relatively small planar epitopes located outside clefts and enzyme active sites were already documented for several high-affinity VHH-antigen associations (34–37). Remarkably, the immunizations of dromedaries with bovine RNase A and human lysozyme failed so far to produce VHHs acting as competitive inhibitors (15, 19, 29). This absence might be explained, at least for these antigens, by a strong counterselection of self-epitopes and a redirection of the immune response toward planar epitopes. Indeed, the sequence comparison of bovine RNase A (38) and human lysozyme (39) with the equivalent dromedary enzymes (40, 41) revealed a strong conservation of the catalytic cleft residues.
Methods
Isolation of HEWL-Specific VHHs and Polyclonal Serum-VHH.
The dromedary D0 immunization in Morocco and isolation of the cAb-Lys-3 and cAb-Lys-2 have been described elsewhere (42). Dromedary D2 and D3 immunizations in Dubai and phage-displayed libraries of their VHHs (19) yielded six lysozyme-specific VHHs: D2-L19, D2-L24, D2-L29, D2-L27, D2-L31, and D3-L11. All VHH genes were recloned in a pHEN06 vector (29) for expression with a His-6 tail. The recombinant proteins were produced in Escherichia coli, and purified according to described protocols (29).
A polyclonal pool of monomeric VHHs was prepared from the serum HCAbs. The HCAbs were first purified from dromedary D2 (bleeding at day 54 in the immunization program; ref. 19) by differential adsorption on protein A/G chromatography (29). This IgG3 HCAb isotype was incubated with Staphylococcus aureus V8 endogluproteinase (Boehringer Mannheim) according to Lauwereys et al. (19) to prepare the monomeric polyclonal VHH.
Kinetic and Affinity Measurements of the VHH–HEWL Interaction.
The kinetic constants of the VHH–HEWL interactions were determined by surface plasmon resonance (Biacore 3000). A CM5 chip with ≈100 pg/mm2 (i.e., 100 resonance units) of immobilized HEWL was prepared according to published protocols (43). Association traces of five VHH concentrations (from 2000 to 3 nM depending on the affinity) and a zero concentration (buffer) were recorded for 3 min in PBS, pH 7.4/0.005% Tween 20/3 mM EDTA. Dissociation of the complexes was followed for 10–20 min. A 30-s injection of 100 mM glycine·HCl (pH 1.5) regenerated the surface. Curves obtained after subtraction of the reference and buffer signals were fitted to a 1:1 Langmuir binding model with biaeval 3.1 (BIAcore). The KD value of the D2-L27 was determined by nonlinear regression of the equilibrium binding signals versus the VHH concentration. The D2-L31–HEWL interaction was consistent with a model describing a conformational change upon binding.
Epitope Mapping and Interference of Binding by NAG3 and Biebrich Scarlet.
The epitopes of all VHHs were mapped on BIACORE 3000 and IAsys. For this purpose, a CM5 chip with ≈300 pg/mm2 (300 resonance units) or an IAsys dextran curvette with ≈3,000 pg/mm2 of immobilized HEWL was prepared according to published protocols (18, 43). VHH–HEWL binding for each VHH in the collection of HEWL specific binders was examined in the presence of a different VHH member or in the presence of the small lysozyme inhibitors GlcNAc(β1–4)GlcNAc(β1–4)GlcNAc (NAG3) and Biebrich Scarlet. This was accomplished by using a coinjection procedure. Hereby, a HEWL saturating concentration of the VHH or small molecule was injected, followed by the injection of an identical sample supplemented with a competing VHH. After regeneration of the surface, the competing VHH alone was applied. The binding levels of the competing VHH for both injections were then compared. Signal differences between both injections of at least 50% were considered to originate from competition for overlapping epitopes. The concentrations of the inhibitors NAG3 (KD = 0.01 mM; ref. 22) or Biebrich Scarlet (KD = 0.13 mM; ref. 22) were competitive (KD1/KD2 = C1/C2) with those of the VHH–HEWL association.
Structure Solution of the VHH–HEWL Complexes.
X-ray diffraction data of crystals of the D3-L11::HEWL, D2-L29::HEWL and D2-L27::HEWL complexes (crystallization conditions described in Supporting Text, which is published as supporting information on the PNAS web site) were obtained by using European Molecular Biology Laboratory (EMBL) beamlines at the Deutsches Elektronen Synchrotron (DESY) synchrotron facility (Hamburg, Germany) or at the European Synchrotron Radiation Facility (ESRF, Grenoble, France). Data were processed with denzo and scalepack (44). For the D3-L11::HEWL and D2-L24::HEWL data, amore (45) generated clear molecular replacement solutions for HEWL and a CDR-deleted model of cAb-RN05 [Protein Data Bank (PDB) ID code 1BZQ] (34) or cAb-Lys-3 (PDB ID code 1JTT) (16). For the data of D2-L29::HEWL, phaser (46) generated clear molecular replacement solutions for HEWL and a CDR-deleted model of cAb-An33 (PDB ID code 1YC7). The complete VHH sequences were built in the electron densities. Simulated annealing and refinement protocols of cns (47) refined the structures. Structure determinations of cAb-Lys-3::HEWL (PDB ID code 1JTT), cAb-Lys-2::HEWL (PDB ID code 1RJC), and D2-L19::HEWL (PDB ID code 1RI8) were reported elsewhere (16, 43). Space groups, unit cells, data collection, and refinement statistics are presented in Table 2.
Analysis of the VHH::HEWL and Fab::HEWL Interfaces.
Several interface parameters were calculated for all VHH::HEWL complexes, and with identical settings for the six representative Fab/Fv::HEWL structures: F9.13.7::GEL (PDB ID code 1FBI); D11.15::JEL (PDB ID code 1JHL); D1.3::HEWL (PDB ID code 1VFB); D44.1::HEWL (PDB ID code 1MLC); HyHEL-5::HEWL (PDB ID code 3HFL); and HyHEL-8::HEWL (PDB ID code 1NDG). The change in solvent accessible surface area (ΔASA) as well as the polar fraction of the ΔASA of the paratopes and epitopes was calculated by the protein interaction server (2). The server also provided the epitope/paratope planarity index, defined as the root mean square deviation of the epitope/paratope atoms from the best-fit plane through their coordinates. contact from the ccp4 software (48), tabulated the antibody–lysozyme atom pairs within a contact distance of 4 Å, of which (oppositely) charged pairs were considered to form salt bridges. hbplus (49) identified potential antibody–lysozyme hydrogen bonds and bridging solvent molecules. The paratope–epitope complementarities were estimated by calculating the Sc values (50) for solvent-excluded antibody::lysozyme complexes. The Mann–Whitney U test, a nonparametric alternative for the parametric t test, implemented in spss for windows (release 12.0, 2003, SPSS Inc., Chicago), tested the significance of observed differences in interfaces parameters between VHH::HEWL and Fv::lysozyme interfaces. Figures were prepared with pymol (www.pymol.org). Amino acid sequences were numbered according to Kabat et al. (51).
Supplementary Material
Acknowledgments
We acknowledge D. Maes for running the statistical significance tests. This research was sponsored by Fonds voor Wetenschappelijk Onderzoek–Vlaanderen, Vlaams Interuniversitair Instituut voor Biotechnologie, and Geconcerteerde Onderzoeks Actie (Research Council, Vrije Universiteit Brussel). We acknowledge the use of the synchrotron beam time at European Molecular Biology Laboratory (EMBL) beam lines at the DORIS storage ring (Hamburg, Germany) and EMBL beam lines at ESRF (Grenoble, France).
Abbreviations
- VH
heavy-chain variable domain
- VL
light-chain variable domain
- HEWL
hen egg white lysozyme
- HCAb
heavy-chain antibody
- VHH
variable domain of a heavy-chain antibody.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1ZVY, 1ZVH, and 1ZV5).
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Padlan E. A. Adv. Protein Chem. 1996;49:57–133. doi: 10.1016/s0065-3233(08)60488-x. [DOI] [PubMed] [Google Scholar]
- 2.Jones S., Thornton J. M. Proc. Natl. Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacCallum R. M., Martin A. C., Thornton J. M. J. Mol. Biol. 1996;262:732–745. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- 4.Al Lazikani B., Lesk A. M., Chothia C. J. Mol. Biol. 1997;273:927–948. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- 5.Chothia C., Lesk A. M., Tramontano A., Levitt M., Smith-Gill S. J., Air G., Sheriff S., Padlan E. A., Davies D., Tulip W. R., et al. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 6.Morea V., Tramontano A., Rustici M., Chothia C., Lesk A. M. J. Mol. Biol. 1998;275:269–294. doi: 10.1006/jmbi.1997.1442. [DOI] [PubMed] [Google Scholar]
- 7.Smith-Gill S. J. EXS. 1996;75:277–300. doi: 10.1007/978-3-0348-9225-4_15. [DOI] [PubMed] [Google Scholar]
- 8.Bentley G. A. EXS. 1996;75:301–319. doi: 10.1007/978-3-0348-9225-4_16. [DOI] [PubMed] [Google Scholar]
- 9.Cauerhff A., Goldbaum F. A., Braden B. C. Proc. Natl. Acad. Sci. USA. 2004;101:3539–3544. doi: 10.1073/pnas.0400060101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Li H., Yang F., Smith-Gill S. J., Mariuzza R. A. Nat. Struct. Biol. 2003;10:482–488. doi: 10.1038/nsb930. [DOI] [PubMed] [Google Scholar]
- 11.Ay J., Keitel T., Kuttner G., Wessner H., Scholz C., Hahn M., Hohne W. J. Mol. Biol. 2000;301:239–246. doi: 10.1006/jmbi.2000.3971. [DOI] [PubMed] [Google Scholar]
- 12.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E. B., Bendahman N., Hamers R. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 13.Muyldermans S., Cambillau C., Wyns L. Trends Biochem. Sci. 2001;26:230–235. doi: 10.1016/s0968-0004(01)01790-x. [DOI] [PubMed] [Google Scholar]
- 14.Decanniere K., Muyldermans S., Wyns L. J. Mol. Biol. 2000;300:83–91. doi: 10.1006/jmbi.2000.3839. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen V. K., Desmyter A., Muyldermans S. Adv. Immunol. 2001;79:261–296. doi: 10.1016/s0065-2776(01)79006-2. [DOI] [PubMed] [Google Scholar]
- 16.Decanniere K., Transue T. R., Desmyter A., Maes D., Muyldermans S., Wyns L. J. Mol. Biol. 2001;313:473–478. doi: 10.1006/jmbi.2001.5075. [DOI] [PubMed] [Google Scholar]
- 17.Desmyter A., Transue T. R., Ghahroudi M. A., Thi M. H., Poortmans F., Hamers R., Muyldermans S., Wyns L. Nat. Struct. Biol. 1996;3:803–811. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 18.Transue T. R., De Genst E., Ghahroudi M. A., Wyns L., Muyldermans S. Proteins. 1998;32:515–522. doi: 10.1002/(sici)1097-0134(19980901)32:4<515::aid-prot9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 19.Lauwereys M., Arbabi G. M., Desmyter A., Kinne J., Holzer W., De Genst E., Wyns L., Muyldermans S. EMBO J. 1998;17:3512–3520. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen V. K., Hamers R., Wyns L., Muyldermans S. EMBO J. 2000;19:921–930. doi: 10.1093/emboj/19.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundberg E. J., Mariuzza R. A. Adv. Protein Chem. 2003;61:119–160. doi: 10.1016/s0065-3233(02)61004-6. [DOI] [PubMed] [Google Scholar]
- 22.Holler E., Rupley J. A., Hess G. P. Biochemistry. 1975;14:1088–1094. doi: 10.1021/bi00676a032. [DOI] [PubMed] [Google Scholar]
- 23.Bond C. J., Marsters J. C., Sidhu S. S. J. Mol. Biol. 2003;332:643–655. doi: 10.1016/s0022-2836(03)00967-7. [DOI] [PubMed] [Google Scholar]
- 24.Davies D. R., Cohen G. H. Proc. Natl. Acad. Sci. USA. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kam-Morgan L. N., Smith-Gill S. J., Taylor M. G., Zhang L., Wilson A. C., Kirsch J. F. Proc. Natl. Acad. Sci. USA. 1993;90:3958–3962. doi: 10.1073/pnas.90.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chitarra V., Alzari P. M., Bentley G. A., Bhat T. N., Eisele J. L., Houdusse A., Lescar J., Souchon H., Poljak R. J. Proc. Natl. Acad. Sci. USA. 1993;90:7711–7715. doi: 10.1073/pnas.90.16.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laskowski R. A., Luscombe N. M., Swindells M. B., Thornton J. M. Protein Sci. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardoso R. M., Zwick M. B., Stanfield R. L., Kunert R., Binley J. M., Katinger H., Burton D. R., Wilson I. A. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Conrath K. E., Lauwereys M., Galleni M., Matagne A., Frere J. M., Kinne J., Wyns L., Muyldermans S. Antimicrob. Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desmyter A., Spinelli S., Payan F., Lauwereys M., Wyns L., Muyldermans S., Cambillau C. J. Biol. Chem. 2002;277:23645–23650. doi: 10.1074/jbc.M202327200. [DOI] [PubMed] [Google Scholar]
- 31.Jobling S. A., Jarman C., Teh M. M., Holmberg N., Blake C., Verhoeyen M. E. Nat. Biotechnol. 2003;21:77–80. doi: 10.1038/nbt772. [DOI] [PubMed] [Google Scholar]
- 32.Stanfield R. L., Dooley H., Flajnik M. F., Wilson I. A. Science. 2004;305:1770–1773. doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- 33.Martin F., Volpari C., Steinkuhler C., Dimasi N., Brunetti M., Biasiol G., Altamura S., Cortese R., De Francesco R., Sollazzo M. Protein Eng. 1997;10:607–614. doi: 10.1093/protein/10.5.607. [DOI] [PubMed] [Google Scholar]
- 34.Decanniere K., Desmyter A., Lauwereys M., Gharoudi M. A., Muyldermans S., Wyns L. Structure (London) 1999;7:361–370. doi: 10.1016/s0969-2126(99)80049-5. [DOI] [PubMed] [Google Scholar]
- 35.Desmyter A., Decanniere K., Muyldermans S., Wyns L. J. Biol. Chem. 2001;276:26285–26290. doi: 10.1074/jbc.M102107200. [DOI] [PubMed] [Google Scholar]
- 36.Dumoulin M., Last A. M., Desmyter A., Decanniere K., Canet D., Larsson G., Spencer A., Archer D. B., Sasse J., Muyldermans S., et al. Nature. 2003;424:783–788. doi: 10.1038/nature01870. [DOI] [PubMed] [Google Scholar]
- 37.Loris R., Marianovsky I., Lah J., Laeremans T., Engelberg-Kulka H., Glaser G., Muyldermans S., Wyns L. J. Biol. Chem. 2003;278:28252–28257. doi: 10.1074/jbc.M302336200. [DOI] [PubMed] [Google Scholar]
- 38.Barnard E. A. Annu. Rev. Biochem. 1969;38:677–732. doi: 10.1146/annurev.bi.38.070169.003333. [DOI] [PubMed] [Google Scholar]
- 39.McKenzie H. A. EXS. 1996;75:365–409. [PubMed] [Google Scholar]
- 40.Beintema J. J. FEBS Lett. 1985;185:115–120. doi: 10.1016/0014-5793(85)80752-3. [DOI] [PubMed] [Google Scholar]
- 41.Irwin D. M. J. Mol. Evol. 1995;41:299–312. [PubMed] [Google Scholar]
- 42.Ghahroudi M. A., Desmyter A., Wyns L., Hamers R., Muyldermans S. FEBS Lett. 1997;414:521–526. doi: 10.1016/s0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 43.De Genst E., Silence K., Ghahroudi M. A., Decanniere K., Loris R., Kinne J., Wyns L., Muyldermans S. J. Biol. Chem. 2005;280:14114–14121. doi: 10.1074/jbc.M413011200. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Navaza J. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- 46.Storoni L. C., McCoy A. J., Read R. J. Acta Crystallogr. D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 47.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Baily S. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 49.McDonald I. K., Thornton J. M. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 50.Lawrence M. C., Colman P. M. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 51.Kabat E. A., Wu T. T., Perry H. M., Gottesman K. S., Foeler C. Sequences of Proteins of Immunological Interest. Washington, DC: U.S. Public Health Service; 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.