Abstract
Free radical formation has been investigated in diverse experimental models of LPS-induced inflammation. Here, using electron spin resonance (ESR) and the spin trap α-(4-pyridyl-1-oxide)-N-tert-butylnitrone, we have detected an ESR spectrum of α-(4-pyridyl-1-oxide)-N-tert-butylnitrone radical adducts in the lipid extract of mouse skin treated with LPS for 6 h. The ESR spectrum was consistent with the trapping of lipid-derived radical adducts. In addition, a secondary radical-trapping technique using dimethyl sulfoxide (DMSO) demonstrated methyl radical formation, revealing the production of hydroxyl radical. Radical adduct formation was suppressed by aminoguanidine, N-(3-aminomethyl)benzylacetamidine (1400W), or allopurinol, suggesting a role for both inducible nitric oxide synthase (iNOS) and xanthine oxidase (XO) in free radical formation. The radical formation was also suppressed in iNOS knockout (iNOS−/−) mice, demonstrating the involvement of iNOS. NADPH oxidase was not required in the formation of these radical adducts because the ESR signal intensity was increased by LPS treatment in NADPH oxidase knockout (gp91phox−/−) mice as much as it was in the wild-type mouse. Nitric oxide (•NO) end products were increased in LPS-treated skin. As expected, the •NO end products were not suppressed by allopurinol but were by aminoguanidine. Interestingly, nitrotyrosine formation in LPS-treated skin was also suppressed by aminoguanidine and allopurinol independently. Pretreatment with the ferric iron chelator Desferal had no effect on free radical formation. Our results imply that both iNOS and XO, but neither NADPH oxidase nor ferric iron, work synergistically to form lipid radical and nitrotyrosine early in the skin inflammation caused by LPS.
Keywords: lipopolysaccharide, mouse skin
Lipopolysaccharide (LPS), an outer-membrane component of Gram-negative bacteria, interacts with CD14, which then presents LPS to the Toll-like receptor 4 (1, 2); filling of this receptor activates inflammatory gene expression through nuclear factor κB and mitogen-activated protein kinase signaling (3, 4). Over the years, LPS has frequently been used in experimental models of inflammation. The inflammatory response includes activation of free radical-generating enzymes in various types of cells that initiate host lipid peroxidation. For the detection and identification of these generated free radicals, the use of spin-trapping compounds appears to be the best method in vivo (5, 6). By using the electron spin resonance (ESR) spin-trapping technique, we have detected in vivo free radical production in lungs treated with LPS (7) or low-dose LPS plus diesel exhaust particles (8). In these articles, we have discussed the involvement of free radical-generating enzymes such as NADPH oxidase, xanthine oxidase (XO), and inducible nitric oxide synthase (iNOS).
NADPH oxidase is generally activated in neutrophils that infiltrate the skin (9). It is also present in keratinocytes (10, 11) and fibroblasts (12). XO is activated in 12-O-tetradecanoyl-13-phorbol acetate-treated murine skin (13) and in human skin keratinocyte cells irradiated with UV B (14). In addition, iNOS expression has been reported in LPS-treated skin; this phenomenon is known to proceed via the activation of nuclear factor κB (15, 16).
Thus, the activation of free radical-generating enzymes including NADPH oxidase, XO, and iNOS have been reported in various physiological and pathological models of skin. Although the expression or activity of these enzymes is well reported, the identification and detection of free radicals produced by these enzymes still remains to be resolved. To address this issue, we have used the ESR spin-trapping technique to examine whether LPS induces free radical production in vivo in the skin of control (C57BL/6J) mice, NADPH oxidase knockout (gp91phox−/−) mice, and iNOS knockout (iNOS−/−) mice. Allopurinol, a XO inhibitor, was used to explore the involvement of XO. We have also examined •NO oxidation products and nitrotyrosine formation in LPS-treated skin.
Results
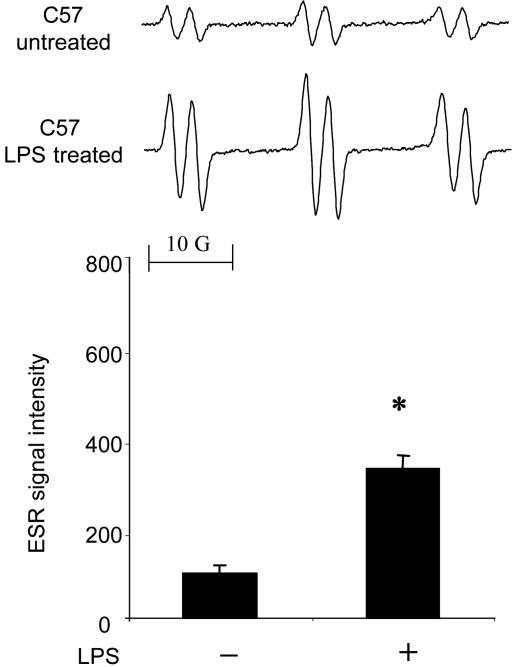
To determine whether LPS caused free radicals in the skin, we injected LPS (100 μg) s.c. into the dorsal area of mice. In controls, when mice were injected with α-(4-pyridyl-1-oxide)-N-tert-butylnitrone (POBN) without a prior LPS injection, a six-line ESR spectrum of unknown origin could be reproducibly detected in the skin extract (Fig. 1). Nonetheless, the 6-h prior treatment with LPS significantly increased the radical adduct formation (Fig. 1). The spectrum was simulated as a composite of a POBN spin adduct with hyperfine couplings of aN = 14.91 ± 0.04 G (1 G = 0.1 mT) and aβH = 2.45 ± 0.07 G, similar to those reported for the POBN radical adduct of a carbon-centered, lipid-derived radical (7).
Fig. 1.
Free radical generation in LPS-treated skin. C57BL/6J mice were treated with LPS (0.1 mg) for 6 h. (Upper) Representative ESR spectrum of POBN radical adducts detected in lipid extracts 30 min after POBN (20 mg) administration. (Lower) Results of relative levels of ESR signal intensity from pooled data. Values represent the mean ± SE derived from four independent experiments. ∗, P < 0.01 vs. untreated group.
Hydroxyl radical (•OH) can initiate lipid peroxidation by abstracting hydrogen. To investigate whether •OH is produced in the LPS-treated skin, the •OH scavenger dimethyl sulfoxide (DMSO) was also administered because it is well known that •OH is converted into the methyl radical, •CH3, via its reaction with DMSO (k = 7 × 109 M−1•s−1). In addition, •CH3 is converted to •OCH3 in the presence of O2 (17):
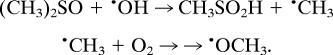 |
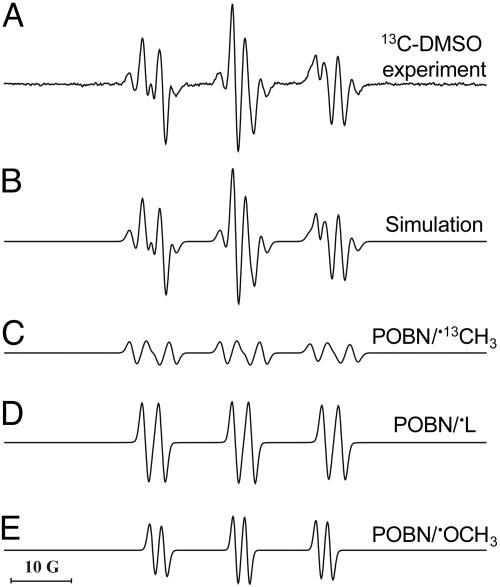
The •CH3 and •OCH3 are then detected as POBN adducts. POBN lipid radical did not appear to be increased by the presence of DMSO (data not shown). In the presence of 13C-labeled DMSO, the six-line signal was replaced by a 12-line signal whose computer simulation indicated the presence of three radical adducts (Fig. 2): POBN-13C-methyl, •CH3 (aN = 15.50 G, aβH = 2.43 G, and a13C = 4.45) (Fig. 2C); POBN-lipid, •L (aN = 14.90 G and aβH = 2.45 G) (Fig. 2D); and the POBN oxygen-centered •OCH3 adduct (aN = 13.61 G and aβH = 1.89 G) (Fig. 2E), in relative yields of 14%, 54%, and 32%, respectively. The in vitro experiment revealed that the two species •CH3 and •OCH3 are derived from DMSO (Table 1). These data strongly suggest that•OH is one of the radical species produced in LPS-treated skin.
Fig. 2.
Simulation of ESR spectra obtained by using 13C-labeled DMSO. C57BL/6J mice were treated with LPS (0.1 mg) for 6 h. (A) Representative ESR spectrum of POBN radical adducts detected in lipid extracts 30 min after POBN (20 mg) plus 13C-labeled DMSO administration. (B) Computer simulation of the spectrum in A. (C) Computer simulation of the POBN/•13CH3 radical adduct (14%). (D) Computer simulation of the spectrum from POBN/•L radicals (54%). (E) Computer simulation of the spectrum from POBN/•OCH3 radicals (32%).
Table 1.
Summary of hyperfine coupling constants (in G) for the POBN radical adducts detected in vitro and in vivo in lipid extracts of methanol plus chloroform
| Systems | POBN/•13CH3, G | POBN/•OCH3, G | POBN/•L, G |
|---|---|---|---|
| In vitro | |||
| 13C-DMSO + H2O2 + Fe2+ + POBN under N2 saturation | aN = 15.28 | Not detected | Not detected |
| aβH = 2.43 | |||
| a13C = 4.45 | |||
| 13C-DMSO + H2O2 + Fe2+ + POBN under O2 saturation | aN = 15.28 | aN = 13.90 | Not detected |
| aβH = 2.43 | aβH = 2.00 | ||
| a13C = 4.45 | |||
| In vivo | |||
| Skin + LPS + POBN | Not detected | Not detected | aN = 14.90 |
| aβH = 2.45 | |||
| Skin + LPS + 13C-DMSO + POBN | aN = 15.50 | aN = 13.70 | aN = 14.90 |
| aβH = 2.43 | aβH = 1.90 | aβH = 2.45 | |
| a13C = 4.45 | |||
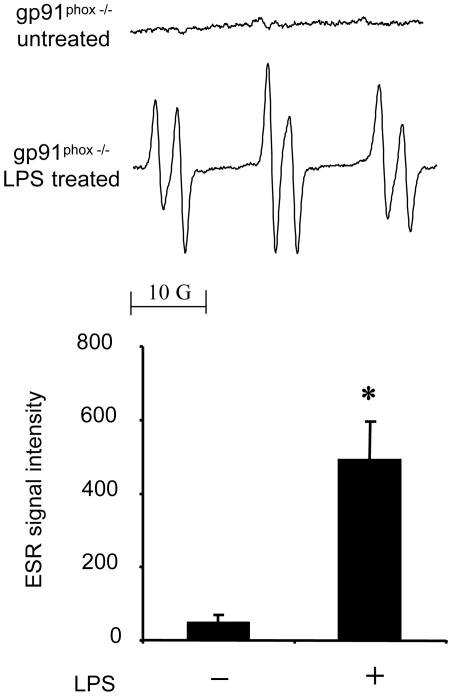
NADPH oxidase in neutrophils and/or macrophages is the major trigger of free radical production in the LPS-treated lung (7). To evaluate the role of NADPH oxidase, we investigated radical adduct formation in the LPS-treated skin of gp91phox−/− mice. In contrast to the lung, a 6-h LPS treatment of the skin significantly increased radical adduct formation in gp91phox−/− mice, just as in the wild type (Fig. 3). These data suggest that NADPH oxidase is not involved in LPS-induced radical adduct formation in the skin.
Fig. 3.
Free radical generation in LPS-treated skin of gp91phox−/− mice. gp91phox−/− mice were treated with LPS (0.1 mg). (Upper) Representative ESR spectrum of POBN radical adducts detected in lipid extracts 30 min after POBN (20 mg) administration. (Lower) Results of relative levels of ESR signal intensity from pooled data using isotopically normal DMSO. Values represent the mean ± SE derived from four independent experiments. ∗, P < 0.01 vs. untreated group.
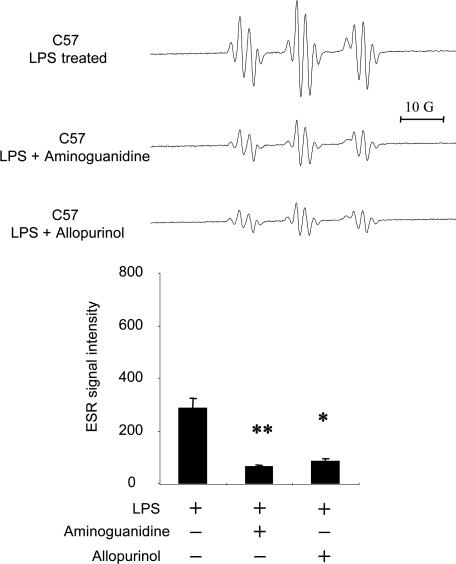
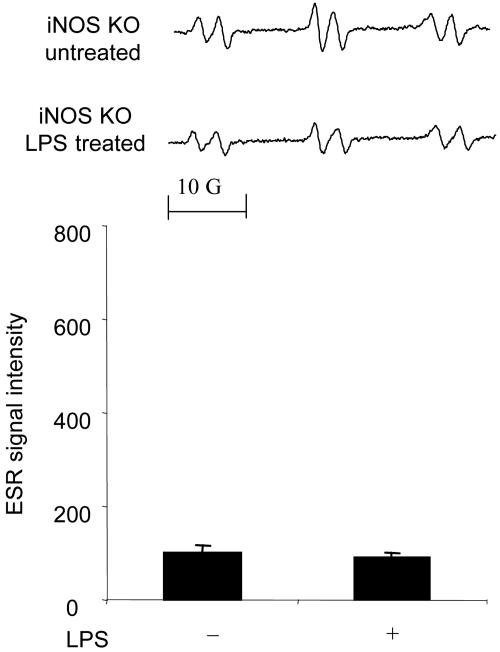
Because iNOS and/or XO (8) were considered to be other free radical generators of LPS-induced radical adduct formation in skin, we examined whether the iNOS inhibitors aminoguanidine and N-(3-aminomethyl)benzylacetamidine (1400W) and allopurinol, a noncompetitive inhibitor of XO, would inhibit the LPS-induced increase in the ESR signal. As shown in Fig. 4, aminoguanidine (0.1 mg) suppressed radical adduct formation in skin treated with LPS (Fig. 4). 1400W had the same inhibitory effect as aminoguanidine (data not shown). Allopurinol (0.1 mg) also significantly suppressed LPS-induced radical adduct formation (Fig. 4 Lower). In addition, to determine whether allopurinol and aminoguanidine affected the yields of each of the radicals trapped in the presence of DMSO differently, we performed experiments with C13-DMSO and the two inhibitors aminoguanidine and allopurinol. As shown in Fig. 4 Upper, the line shape of each spectrum was not changed, thus indicating that the ratio of the three components of the spectrum shown in Fig. 2 remained the same. The above results indicate that in skin, allopurinol and aminoguanidine independently suppressed LPS-induced radical adduct formation. Thus, both XO and iNOS are essential free radical generators in LPS-treated skin. To further confirm the requirement of iNOS in free radical production, we used iNOS−/− mice. No increase in radical adduct formation was observed in the LPS-treated skin of iNOS−/− mice (Fig. 5).
Fig. 4.
Effects of aminoguanidine and allopurinol on free radical generation in LPS-treated skin. C57BL/6J mice were treated with LPS (0.1 mg), LPS (0.1 mg) plus aminoguanidine (0.1 mg), and LPS (0.1 mg) plus allopurinol (0.1 mg) for 6 h. (Upper) Representative ESR spectrum of POBN radical adducts detected in lipid extracts 30 min after POBN/13C-DMSO (20 mg) administration. (Lower) Results of relative levels of ESR signal intensity from pooled data. Values represent the mean ± SE derived from four independent experiments. ∗, P < 0.01 vs. untreated group; ∗∗, P < 0.001 vs. untreated group.
Fig. 5.
Free radical generation in LPS-treated skin of iNOS−/− mice. iNOS−/− mice were treated with LPS (0.1 mg). (Upper) Representative ESR spectrum of POBN radical adducts detected in lipid extracts 30 min after POBN (20 mg) administration. (Lower) Results of relative levels of ESR signal intensity from pooled data. Values represent the mean ± SE derived from four independent experiments. No statistically significant difference was found.
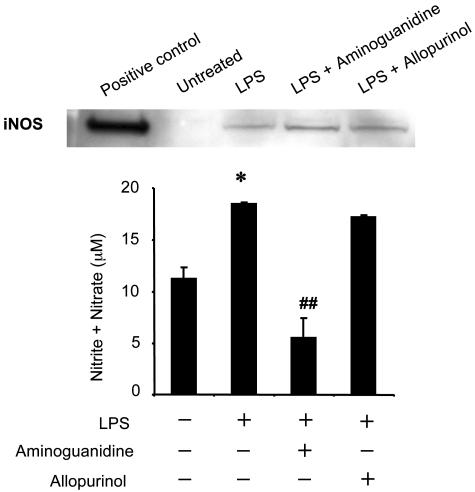
We then examined iNOS protein expression and nitric oxide end products in skin tissue. iNOS protein expression was undetectable in untreated skin but easily detected in LPS-treated skin (Fig. 6Upper). The detected iNOS protein expression was not altered by aminoguanidine (1 mg) or allopurinol (1 mg). The nitrite-plus-nitrate levels were increased in LPS-treated skin (Fig. 6 Lower); these increased levels were not affected by allopurinol (1 mg) but were suppressed by aminoguanidine (1 mg). These data suggest that aminoguanidine suppresses the enzyme activity of iNOS without affecting its protein expression and that allopurinol does not affect the expression or activity of iNOS.
Fig. 6.
Effects of aminoguanidine and allopurinol on iNOS protein expression and nitrite-plus-nitrate concentration in LPS-treated skin. C57BL/6J mice were treated with LPS (0.1 mg), LPS (0.1 mg) plus aminoguanidine (0.1 mg), and LPS (0.1 mg) plus allopurinol (0.1 mg) for 6 h. (Upper) iNOS protein expression in LPS-treated skin at 6 h. Shown are the results of immunoblot analyses using skin tissue homogenate (50 μg per lane). Samples were resolved by SDS/PAGE, transferred to a nitrocellulose membrane, and probed with an anti-iNOS monoclonal antibody. Shown are the results from representative data from an experiment repeated independently three times with equivalent results. RAW 264.7 cells treated with LPS plus IFN-γ were used as a positive control. (Lower) Nitrite-plus-nitrate concentration in skin tissue at 6 h. Values represent the mean ± SE derived from three independent experiments. ∗, P < 0.01 vs. untreated group; ##, P < 0.001 vs. LPS-treated group.
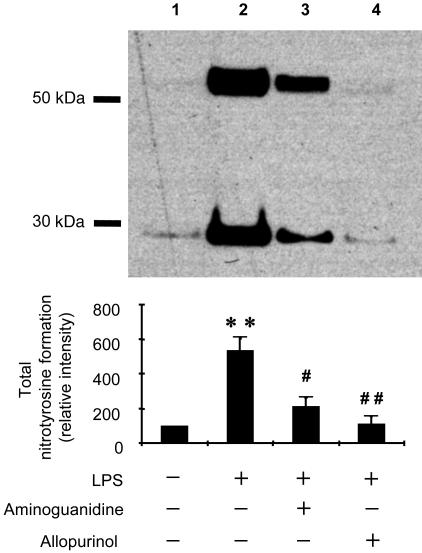
To obtain additional insight into the mechanism of LPS-induced free radical production, we examined nitrotyrosine formation in skin protein using a monoclonal nitrotyrosine antibody. As shown in Fig. 7, we detected nitrotyrosine mainly in the protein bands of ≈30 and 50 kDa in skin. These levels increased dramatically in LPS-treated skin (Fig. 7, lane 2). Both aminoguanidine (1 mg per mouse) and allopurinol (1 mg per mouse) independently suppressed nitrotyrosine formation as can clearly be seen in the densitometric analysis of the bands, suggesting that both iNOS and XO are required for nitrotyrosine formation in LPS-treated skin.
Fig. 7.
Effects of aminoguanidine and allopurinol on nitrotyrosine formation in LPS-treated skin. C57BL/6J mice were treated with LPS (0.1 mg), LPS (0.1 mg) plus aminoguanidine (0.1 mg), and LPS (0.1 mg) plus allopurinol (0.1 mg) for 6 h. (Upper) Results of immunoblot analyses using skin tissue homogenate (50 μg per lane). Samples were resolved by SDS/PAGE, transferred to a nitrocellulose membrane, and probed with an anti-nitrotyrosine monoclonal antibody. Immunoreactive bands of ≈30 and ≈50 kDa were detected. Shown are the results from representative data. (Lower) Results of densitometric analysis from pooled data. Values represent the mean ± SE derived from three independent experiments. ∗∗, P < 0.001 vs. untreated group; #, P < 0.01 vs. LPS-treated group; ##, P < 0.001 vs. LPS-treated group.
To investigate the role of transition metals in the generation of •OH induced by LPS, experiments were carried out in which animals were treated with the FeIII chelating agent Desferal. In earlier work, Desferal was found to inhibit oxidative damage in the lungs (8). However, in the lipid extract of mouse skin, the ESR spectrum was unaffected by the combined treatment of Desferal and LPS or by Desferal injected 5 h after LPS (data not shown).
Finally, we examined histological changes in LPS-treated skin. It has been reported that treatment with LPS induces accumulation of leukocytes in mouse skin at 24 h (18). However, we observed no significant changes in skin treated with LPS after 6 h (data not shown). These data suggest that LPS-induced free radical production in skin is leukocyte-independent at this early stage.
Discussion
In the current study, we found that a 6-h treatment with LPS increased lipid radical formation in the skin of wild-type C57BL/6J mice. The lipid radical formation was also increased in LPS-treated gp91phox−/− mice but not in iNOS−/− mice. Notably, allopurinol and aminoguanidine independently suppressed the increase in the lipid radical formation. •NO oxidation products were increased in LPS-treated skin at 6 h and were inhibited by aminoguanidine but not by allopurinol. In addition, both allopurinol and aminoguanidine independently suppressed nitrotyrosine formation in LPS-treated skin at 6 h.
No significant histological change was observed in LPS-treated skin at 6 h. In particular, no neutrophils had been recruited as occurs in inflammation. In skin treated with LPS, NADPH oxidase in infiltrating neutrophils is well known to produce a large amount of superoxide anion (9) as it does in other organs. Although NADPH oxidase in the nonphagocytic cells has been less well characterized, a physiologically relevant low level of generation of free radical and neutrophil-like expression of NADPH oxidase subunits is also present in skin keratinocytes (10, 11) and fibroblasts (12). However, there was a significant increase in the lipid radicals from the skin of mice treated with LPS for 6 h, even in gp91phox knockout animals. Thus, NADPH oxidase is not a main free radical generator in LPS-treated skin at 6 h. Presumably, NADPH oxidase may be involved after 24 h when many neutrophils have infiltrated into the skin (9).
Another important source of free radicals is XO. This enzyme reduces molecular oxygen, leading to the formation of both O2•− and hydrogen peroxide (19). Studies have shown that in mice, XO in the skin is activated by photosensitization (20), benzoyl peroxide, cumene hydroperoxide (21, 22), and 12-O-tetradecanoylphorbol-13-acetate (23, 13), and in rats it is activated in burn-injured skin (24). Thus, the regulation of XO activity is important during skin inflammation. Although allopurinol inhibits XO activity in vivo and in clinical practice, it is known to scavenge the hydroxyl radical in chemical systems and has been proposed to scavenge the hydroxyl radical in vivo.
Although allopurinol reacts with hydroxyl radical at the near diffusion-limited rate of ≈109 M−1•s−1 (25), this very high rate is typical for reactions of the hydroxyl radical that reacts with chloride ion at the rate of 4.3 × 109 M−1•s−1 (26) and with GSH at the rate of 1.3 × 1010 M−1•s−1 (27). Because both of these compounds as well as many other biochemicals react with the hydroxyl radical at near diffusion-limited rates, reactions of hydroxyl radical with allopurinol will be a very, very small fraction of the total reactions of the hydroxyl radical in vivo; therefore, hydroxyl radical scavenging by allopurinol cannot be the explanation of the effects we observed (Figs. 3, 6, and 7). In view of our observation that allopurinol significantly suppressed the lipid radical and nitrotyrosine formation, XO is most probably increased in LPS-treated skin at 6 h. Because few phagocytic cells infiltrated the skin, XO must be activated in nonphagocytic cells like keratinocytes, fibroblasts, or endothelial cells. The XO activity in these nonphagocytic cells is considered to have a role in the initiation of inflammation (28).
In the skin, iNOS is involved in the regulation of blood flow (29), UV-induced melanogenesis (30), and inflammatory skin diseases such as psoriasis (31), lupus erythematosus (32), and dermatosis (33). This enzyme also affects cutaneous wound healing (34) and protects against pathogens (35). The present study showed that free radical generation induced by LPS in the skin depends on iNOS. Moreover, the increase in iNOS expression levels and nitrite-plus-nitrate accumulation at 6 h support the relevance of iNOS in LPS-treated skin. In addition to the formation of O2•− and H2O2, XO is known to reduce nitrite to •NO, thereby recycling the •NO produced by iNOS that had been oxidized to nitrite (36).
A final critical question is the reaction between free radicals generated from XO and from iNOS (Fig. 7). Although the superoxide radical (O2•−) produced by XO is unstable and rapidly dismutated to H2O2 and is generally restricted to the cell compartment in which it is produced, in the presence of a large amount of •NO produced from iNOS, O2•− may form the reactive species peroxynitrite (ONOO−). If •NO and O2•− are formed concomitantly by XO, their proximity would facilitate peroxynitrite formation (36). ONOO− is known to decompose to •NO2 and •OH (37). In the presence of carbon dioxide, ONOO− forms an adduct which decomposes to •NO2 and CO3•− (38–40), but CO3•− has not been reported to form •CH3 from DMSO and is not expected to do so. Detection through the ESR spin-trapping technique or detection of nitrotyrosine formation does not allow the conclusive identification of all of the primary radical oxidants. However, in the current study, the simulation of ESR data demonstrated that through the detection of the •CH3 from DMSO, •OH was formed in LPS-treated skin. In addition, all known pathways for nitrotyrosine formation require the formation of •NO and tyrosyl radical (41). Therefore, the formation of both nitrotyrosine and •CH3 formation from DMSO support the formation of ONOO−.
In conclusion, we have demonstrated that hydroxyl- and lipid-derived radicals and nitrotyrosine were formed after 6 h in LPS-treated skin. Our results imply that both iNOS and possibly XO, but neither NADPH oxidase nor ferric iron, work synergistically to form free radical metabolites and nitrotyrosine early in skin inflammation caused by LPS.
Experimental Procedures
Materials.
POBN was obtained from Alexis. LPS from Escherichia coli serotype 055:B5, aminoguanidine, allopurinol, and deferoxamine mesylate (Desferal) were obtained from Sigma. N-(3-aminomethyl)benzylacetamidine (1400W) was from Calbiochem-Novabiochem.
Animals and Treatments.
Adult male mice containing the disrupted gp91phox gene (gp91phox−/−) and iNOS gene (iNOS−/−) were obtained from The Jackson Laboratory. C57BL/6J strain mice were used as control animals. The animals were group-housed in a temperature-controlled room at 23–24°C with a 12/12-h light/dark cycle and allowed free access to food and water. The studies adhered to National Institutes of Health guidelines for the care and handling of experimental animals. All animal studies were approved by the institutional review board. LPS (0.1 mg) was dissolved in 0.1 ml of 0.9% saline water. Allopurinol (0.1 mg, 1 mg), aminoguanidine (0.1 mg, 1 mg), and 1400W (15 mg/kg) were mixed with the LPS solution and injected at the same time. LPS (0.1 mg) was injected s.c. into the dorsal area of a mouse under anesthesia with isoflurane. Six hours later, POBN was injected into the same area (20 mg) under anesthesia with pentobarbital. Where indicated, Desferal (100 mg/kg i.p.) was injected 60 min before POBN. Animals were killed 30 min after POBN administration, and the lipid phase of the skin was extracted for radical adduct detection by ESR.
In Vitro Methyl and Oxygen-Centered Radical Formation.
The mixture of 100 mM phosphate buffer (pH 7.4), 250 mM13C-DMSO, 50 mM POBN, 100 μM H2O2, and FeSO4 was incubated at room temperature for 1 h under N2 or O2 saturation. The reaction mixture was then treated with chloroform plus methanol.
ESR Studies.
Extraction of the lipid components of the skin was carried out as previously described by Sato et al. (7). Briefly, skin tissue was homogenized with a Polytron in a mixture containing 2.5 ml of 2:1 chloroform:methanol, 0.5 ml of 30 mM 2,2′-dipyridyl, and 4 ml of deionized water under cooling conditions. 2,2′-Dipyridyl, a ferrous chelator, was used to inhibit ex vivo ferrous-dependent free radical generation. For extraction, 16 ml of 2:1 chloroform:methanol was added, and the resultant mixture was shaken and then centrifuged at 931 × g for 10 min. The chloroform layer was then isolated and dried by passing through a sodium sulfate column. The sample was evaporated with nitrogen gas, and ESR spectra were recorded immediately at room temperature by using a quartz flat cell in a Bruker EMX EPR spectrometer equipped with a super high-Q cavity. Spectra were recorded on an IBM-compatible computer interfaced with the spectrometer by using instrument settings of 9.79 GHz, 20.2 mW microwave power, 100 kHz modulation frequency, 1,300 ms conversion time, and 655 ms time constant. Simulations of ESR spectra were performed by using a program (winsim) developed in our laboratory (42).
Measurement of Nitrite-Plus-Nitrate.
The accumulation of nitrite-plus-nitrate in tissue was measured as described in ref. 43. The supernatant of skin tissue homogenate was centrifuged at 7,740 × g with a micropore filter (Ultrafree-MC microcentrifuge device, UFC3; Millipore) to remove substances larger than 10 kDa. The micropore filter was washed with distilled water before applying the sample. The filtrates were analyzed by using a NO2−/NO3− assay kit-C II (Dojindo, Kumamoto, Japan). After nitrate was reduced to nitrite by nitrate reductase, the absorbance at 540 nm was measured after reaction with the Griess reagent. This value was expressed as the total of NO end products, nitrite plus nitrate. Nitrite-plus-nitrate was expressed in micromolar concentrations based on tissue weight, assuming a density of 1.
Western Blotting.
The degree of iNOS protein expression in mouse skin was determined by Western blot analysis in the following procedure. Skin tissue was homogenized with the Polytron in RIPA buffer containing 1% vol/vol Nonidet P-40, 20 mM Tris (pH 7.7), 150 mM NaCl, 1 mM EDTA, and a mixture of protease inhibitors (Calbiochem). After incubation at 4°C for 20 min, samples were centrifuged at 23,700 × g for 10 min. The supernatant was analyzed for protein concentration, and an equal amount of cellular protein (50 μg) was separated on reducing NuPAGE 4–12% Bis-Tris or 7% Tris-acetate gel (Invitrogen) and transferred to a nitrocellulose membrane. The membrane was then probed with a monoclonal iNOS primary antibody (Transduction Laboratories). Membrane-bound primary antibodies were visualized by using secondary antibodies conjugated with horseradish peroxidase and chemiluminescent substrate (Pierce). For the analysis of nitrotyrosine formation, the membrane was probed with monoclonal nitrotyrosine primary antibody (Cayman Chemical). The other staining procedures were the same as for the iNOS antibody. The images were subjected to densitometry analysis by using nih image software (National Institutes of Health, Bethesda).
Statistical Analysis.
Data for ESR hyperfine couplings were given as mean ± SD. Others were given as mean ± SE. A statistical analysis was performed with a commercially available statistical package for Macintosh (statview-j, Version 5; Abacus Concepts, Berkeley, CA) using an analysis of variance followed by the Scheffé test. P < 0.05 was considered statistically significant.
Acknowledgments
We thank Jean B. Corbett for technical support and Dr. Ann Motten and Mary Mason for editorial help in the preparation of this manuscript.
Abbreviations
- ESR
electron spin resonance
- iNOS
inducible nitric oxide synthase
- POBN
α-(4-pyridyl-1-oxide)-N-tert-butylnitrone
- XO
xanthine oxidase.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Aderem A., Ulevitch R. J. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 3.Chow J. C., Young D. W., Golenbock D. T., Christ W. J., Gusovsky F. J. Biol. Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 4.Beutler B. Curr. Opin. Microbiol. 2000;3:23–28. doi: 10.1016/s1369-5274(99)00046-6. [DOI] [PubMed] [Google Scholar]
- 5.Kadiiska M. B., Mason R. P., Dreher K. L., Costa D. L., Ghio A. J. Chem. Res. Toxicol. 1997;10:1104–1108. doi: 10.1021/tx970049r. [DOI] [PubMed] [Google Scholar]
- 6.Kadiiska M. B., Ghio A. J., Mason R. P. Spectrochim. Acta. A. 2004;60:1371–1377. doi: 10.1016/j.saa.2003.10.035. [DOI] [PubMed] [Google Scholar]
- 7.Sato K., Kadiiska M. B., Ghio A. J., Corbett J., Fann Y. C., Holland S. M., Thurman R. G., Mason R. P. FASEB J. 2002;16:1713–1720. doi: 10.1096/fj.02-0331com. [DOI] [PubMed] [Google Scholar]
- 8.Arimoto T., Kadiiska M. B., Sato K., Corbett J., Mason R. P. Am. J. Respir. Crit. Care Med. 2005;171:379–387. doi: 10.1164/rccm.200402-248OC. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura Y., Murakami A., Ohto Y., Torikai K., Tanaka T., Ohigashi H. Cancer Res. 1998;58:4832–4839. [PubMed] [Google Scholar]
- 10.Beak S. M., Lee Y. S., Kim J. A. Biochimie. 2004;86:425–429. doi: 10.1016/j.biochi.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Chamulitrat W., Stremmel W., Kawahara T., Rokutan K., Fujii H., Wingler K., Schmidt H. H. H. W., Schmidt R. J. Invest. Dermatol. 2004;122:1000–1009. doi: 10.1111/j.0022-202X.2004.22410.x. [DOI] [PubMed] [Google Scholar]
- 12.Ceolotto G., Bevilacqua M., Papparella I., Baritono E., Franco L., Corvaja C., Mazzoni M., Semplicini A., Avogaro A. Diabetes. 2004;53:1344–1351. doi: 10.2337/diabetes.53.5.1344. [DOI] [PubMed] [Google Scholar]
- 13.Alam A., Khan N., Sharma S., Saleem M., Sultana S. Pharmacol. Res. 2002;46:557–564. doi: 10.1016/s1043661802002268. [DOI] [PubMed] [Google Scholar]
- 14.Deliconstantinos G., Villiotou V., Stavrides J. C. Biochem. Pharmacol. 1996;51:1727–1738. doi: 10.1016/0006-2952(96)00110-4. [DOI] [PubMed] [Google Scholar]
- 15.Fujii E., Yoshioka T., Ishida H., Irie K., Muraki T. Br. J. Pharmacol. 2000;130:90–94. doi: 10.1038/sj.bjp.0703277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iuvone T., D’Acquisto F., Van Osselaer N., Di Rosa M., Carnuccio R., Herman A. G. Br. J. Pharmacol. 1998;123:1325–1330. doi: 10.1038/sj.bjp.0701730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue Qian S., Kadiiska M. B., Guo Q., Mason R. P. Free Radical Biol. Med. 2005;38:125–135. doi: 10.1016/j.freeradbiomed.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 18.Bochkov V. N., Kadl A., Huber J., Gruber F., Binder B. R., Leitinger N. Nature. 2002;419:77–81. doi: 10.1038/nature01023. [DOI] [PubMed] [Google Scholar]
- 19.Porras A. G., Olson J. S., Palmer G. J. Biol. Chem. 1981;256:9096–9103. [Google Scholar]
- 20.Athar M., Elmets C. A., Bickers D. R., Mukhtar H. J. Clin. Invest. 1989;83:1137–1143. doi: 10.1172/JCI113993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sultana S., Alam A., Khan N., Sharma S. Redox Rep. 2003;8:105–112. doi: 10.1179/135100003125001422. [DOI] [PubMed] [Google Scholar]
- 22.Sultana S., Khan N., Sharma S., Alam A. J. Ethnopharmacol. 2003;85:33–41. doi: 10.1016/s0378-8741(02)00360-4. [DOI] [PubMed] [Google Scholar]
- 23.Reiners J. J., Jr., Pence B. C., Barcus M. C., Cantu A. R. Cancer Res. 1987;47:1775–1779. [PubMed] [Google Scholar]
- 24.Shimizu S., Tanaka H., Sakaki S., Yukioka T., Matsuda H., Shimazaki S. J. Trauma. 2002;52:683–687. doi: 10.1097/00005373-200204000-00011. [DOI] [PubMed] [Google Scholar]
- 25.Moorhouse P. C., Grootveld M., Halliwell B., Quinlan J. G., Gutteridge J. M. C. FEBS Lett. 1987;213:23–28. doi: 10.1016/0014-5793(87)81458-8. [DOI] [PubMed] [Google Scholar]
- 26.Jayson G. G., Parsons B. J., Swallow A. J. J. Chem. Soc. Faraday Trans. 1973;69:1597–1607. [Google Scholar]
- 27.Quintiliani M., Badiello R., Tamba M., Esfandi A., Gorin G. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1977;32:195–202. doi: 10.1080/09553007714550891. [DOI] [PubMed] [Google Scholar]
- 28.Martin H. M., Hancock J. T., Salisbury V., Harrison R. Infect. Immun. 2004;72:4933–4939. doi: 10.1128/IAI.72.9.4933-4939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldsmith P. C., Leslie T. A., Hayes N. A., Levell N. J., Dowd P. M., Foreman J. C. J. Invest. Dermatol. 1996;106:113–118. doi: 10.1111/1523-1747.ep12328204. [DOI] [PubMed] [Google Scholar]
- 30.Roméro-Graillet C., Aberdam E., Clément M., Ortonne J. P., Ballotti R. J. Clin. Invest. 1997;99:635–642. doi: 10.1172/JCI119206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruch-Gerharz D., Fehsel K., Suschek C., Michel G., Ruzicka T., Kolb-Bachofen V. J. Exp. Med. 1996;184:2007–2012. doi: 10.1084/jem.184.5.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn A., Fehsel K., Lehmann P., Krutmann J., Ruzicka T., Kolb-Bachofen V. J. Invest. Dermatol. 1998;111:149–153. doi: 10.1046/j.1523-1747.1998.00253.x. [DOI] [PubMed] [Google Scholar]
- 33.Rowe A., Farrell A. M., Bunker C. B. Br. J. Dermatol. 1997;136:18–23. [PubMed] [Google Scholar]
- 34.Yamasaki K., Edington H. D. J., McClosky C., Tzeng E., Lizonova A., Kovesdi I., Steed D. L., Billiar T. R. J. Clin. Invest. 1998;101:967–971. doi: 10.1172/JCI2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei X.-Q., Charles I. G., Smith A., Ure J., Feng G.-J., Huang F.-P., Xu D., Muller W., Moncada S., Liew F. Y. Nature. 1995;375:408–411. doi: 10.1038/375408a0. [DOI] [PubMed] [Google Scholar]
- 36.Millar T. M. FEBS Lett. 2004;562:129–133. doi: 10.1016/S0014-5793(04)00218-2. [DOI] [PubMed] [Google Scholar]
- 37.Augusto O., Gatti R. M., Radi R. Arch. Biochem. Biophys. 1994;310:118–125. doi: 10.1006/abbi.1994.1147. [DOI] [PubMed] [Google Scholar]
- 38.Bonini M. G., Radi R., Ferrer-Sueta G., Ferreira A. M. D. C., Augusto O. J. Biol. Chem. 1999;274:10802–10806. doi: 10.1074/jbc.274.16.10802. [DOI] [PubMed] [Google Scholar]
- 39.Goldstein S., Czapski G., Lind J., Merényi G. Chem. Res. Toxicol. 2001;14:1273–1276. doi: 10.1021/tx0100845. [DOI] [PubMed] [Google Scholar]
- 40.Lymar S. V., Hurst J. K. Chem. Res. Toxicol. 1996;9:845–850. doi: 10.1021/tx960046z. [DOI] [PubMed] [Google Scholar]
- 41.Nakai K., Mason R. P. Free Radical Biol. Med. 2005;39:1050–1058. doi: 10.1016/j.freeradbiomed.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 42.Duling D. R. J. Magn. Reson. B. 1994;104:105–110. doi: 10.1006/jmrb.1994.1062. [DOI] [PubMed] [Google Scholar]
- 43.Kosaka H., Yoneyama H., Zhang L., Fujii S., Yamamoto A., Igarashi J. FASEB J. 2003;17:636–643. doi: 10.1096/fj.02-0585com. [DOI] [PubMed] [Google Scholar]