Abstract
Reverse transcription of retroviral RNA genomes produce a double-stranded linear cDNA molecule. A host degradation system prevents a majority of the cDNA molecules from completing the obligatory genomic integration necessary for pathogenesis. We demonstrate that the human TFIIH complex proteins XPB (ERCC3) and XPD (ERCC2) play a principal role in the degradation of retroviral cDNA. DNA repair-deficient XPB and XPD mutant cell lines exhibited an increase in transduction efficiency by both HIV- and Moloney murine leukemia virus-based retroviral vectors. Replicating Moloney murine leukemia virus viral production was greater in XPB or XPD mutant cells but not XPA mutant cells. Quantitative PCR showed an increase in total cDNA molecules, integrated provirus, and 2LTR circles in XPB and XPD mutant cells. In the presence of a reverse transcription inhibitor, the HIV cDNA appeared more stable in mutant XPB or XPD cells. These studies implicate the nuclear DNA repair proteins XPB and XPD in a cellular defense against retroviral infection.
Keywords: AIDS, ERCC2, ERCC3, HIV, xeroderma pigmentosum
After entry into a host cell, retroviruses must copy their genomic RNA to a linear cDNA. A preintegration complex (PIC) is formed that contains the retroviral cDNA and viral and host proteins. Lentiviral PICs are able to traverse an intact nuclear membrane. Once inside the nucleus, the viral integrase protein catalyzes covalent joining of the cDNA into the host chromosome, yielding a provirus. A functional provirus is necessary to continue the viral life cycle. Alternative fates for the viral cDNA include formation of 1LTR circles, 2LTR circles, or degradation. Circle formation has long been taken as a measure of successful nuclear import of the PIC, because these products are not observed in the cytoplasm. The mechanism of cDNA degradation has not yet been elucidated.
Many recent studies implicate roles for host DNA repair proteins in the retroviral life cycle (1–3). XPB (ERCC3) and XPD (ERCC2) are DNA helicases with opposing polarity that function as integral components of the TFIIH protein complex. TFIIH is required for basal transcription and nucleotide excision repair (NER) (4). The helicase activity of TFIIH is required to separate DNA strands at a promoter during transcription or at a site of DNA damage during NER. Both XPB and XPD are conserved and are essential in eukaryotes, precluding the establishment of null cell lines (5). Hypomorphic mutations of either XPB or XPD may lead to three recessive diseases with varying severity: trichothiodystrophy (TTD), xeroderma pigmentosum (XP), or associated XP and Cockayne syndrome (6–8). TTD-associated mutations appear to affect mainly transcription activity, whereas mutations associated with XP affect NER activity (9, 10). Although >20 mutations of XPD have been described, only three mutations of XPB have been observed in the human population, suggesting that mutations of XPB may be incompatible with survival (11).
Here we demonstrate that transduction by HIV or Moloney murine leukemia virus (MMLV)-based retroviral vectors was substantially greater in XPB or XPD mutant cells compared with isogenic complemented cells. Replicating MMLV viral production was greater in XPB or XPD mutant cells. There was no effect on transduction efficiency or MMLV viral production with other DNA repair-deficient cell lines. Cell death induced by retroviral infection did not account for the difference in transduction efficiency. Total retroviral cDNA, 2LTR circles, and integrated provirus were all increased in XPB or XPD mutant cells. Our results are consistent with the conclusion that XPB and XPD reduce HIV and MMLV integration efficiency by enhancing the degradation of retroviral cDNA and, thereby, reducing the available pool of cDNA molecules for integration.
Results
HIV and MMLV Transduction.
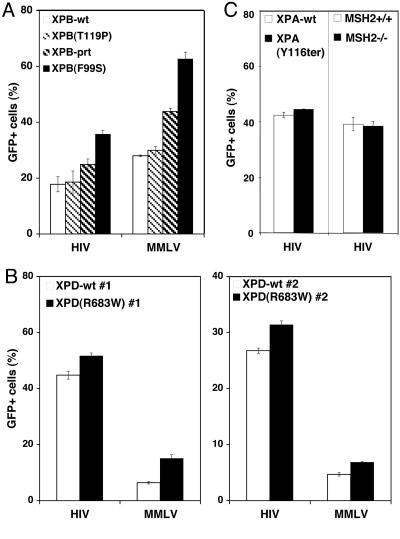
HIV-based retroviral vector transduction efficiency was evaluated in XPB and XPD mutants and complemented cell lines (Table 1, which is published as supporting information on the PNAS web site). XPB mutant cell lines were derived from one patient with the TTD-associated mutation T119P [XPB(T119P)] and a second patient with the XP/CS-associated mutation F99S [XPB(F99S)] (12). The more severely NER-defective XPB(F99S) cell line was complemented with a WT XPB allele (XPB-wt) or the XPB(T199P) allele (XPB-prt) (prt, partially). The XPB-prt cell line exhibits a prt rescued NER phenotype compared to the parent XPB(F99S) cell line and the XPB(T119P) cell line (12). The XPB mutant and complemented cell lines are isogenic with the exception of the XPB(T119P) cell line. The isogenetic backgrounds of these cell lines provide a unique platform for evaluating the singular effect(s) of the complemented gene.
The NER activity of these cell lines was confirmed by examining UV sensitivity (Fig. 5A, which is published as supporting information on the PNAS web site). The XP/CS cell line XPB(F99S) was most sensitive to UV irradiation, but viability was rescued by complementation with the WT XPB gene. The TTD cell line XPB(T119P) exhibits near WT UV sensitivity, as reported in ref. 12. The prt complemented XPB-prt cell line has an intermediate UV-sensitive phenotype, indicating only partial rescue of the NER defect. The UV sensitivity of the XPB cell lines is XPB(F99S) > XPB-prt > XPB-wt ≈ XPB(T119P).
These XPB mutant and complemented cell lines were transduced with an HIV-based retroviral vector expressing the GFP gene driven by a CMV promoter (HIV-GFP), and transduction was quantified by flow cytometry (Fig. 1; ref. 13). The HIV-GFP vector faithfully recapitulates the early steps of the HIV life cycle from reverse transcription through integration (14, 15). The percentage of cells expressing GFP significantly decreased with increasing XPB activity, suggesting a decrease in transduction efficiency (Fig. 1A, left). The fully rescued XPB-wt cell line and the XPB(T119P) cell line displayed the least transduction efficiency. The XPB(T119P) mutation is the least defective for XPB activity (12), suggesting that transcription-associated mutations of XPB only marginally affect HIV transduction. The more severe XPB(F99S) mutation led to a >100% increase in transduction efficiency (P = 0.01). The XPB-prt yielded an intermediate transduction increase of 39.9% (P = 0.04). The least severe XPB mutation XPB(T119P) had only a 4.5% increase in transduction efficiency, which was not statistically significant (P = 0.4). Infection with an MMLV-based retroviral vector (MMLV-GFP) showed the same trend of decreasing transduction efficiency with increasing XPB activity, suggesting conservation of these effects in retroviruses (Fig. 1A, right). Transduction efficiency of the XPB cell lines was XPB(F99S) > XPB-prt > XPB(T119P) ≈ XPB-wt.
Fig. 1.
Evaluation of transduction efficiency in DNA repair mutant and rescued cell lines. Isogenic mutant and rescued cell lines were transduced with HIV-based and MMLV-based retroviral vectors pseudotyped with VSV-G. The only ORF of the vectors is GFP driven by a CMV promoter leading to expression of GFP after successful integration. The percentage of cells expressing GFP (GFP+) at 48 h was measured by flow cytometry. Each bar represents an individual cell line. (A) From the most NER activity to the least, XPB cell lines are XPB(F99S) fully complemented with the WT XPB allele (XPB-wt), the TTD patient-derived XPB(T119P) mutant, XPB(F99S) complemented with the XPB(T119P) mutant allele (XPB-prt), and the XP/CS patient-derived XPB(F99S) mutant. Paired t test analysis yielded the following two-tailed P values for HIV-GFP infections: XPB-wt and XPB(T119P), P = 0.4; XPB-wt and XPB-prt, P = 0.04; XPB-wt and XPB(F99S), P = 0.01. Analysis of MMLV-GFP infection of XPB-wt and XPB(F99S) yielded P = 0.003. (B) XPD cell lines include two XP patient-derived XPD(R683W) mutant cell lines [XPD(R683W) #1 and XPD(R683W) #2], and each line complemented with the WT XPD allele (XPD-wt #1 and XPD-wt #2). Paired t test analysis yielded the following two-tailed P values: HIV-GFP infections of XPD-wt #1 and XPD(R683W) #1, P = 0.009; XPD-wt #2 and XPD(R683W) #2, P = 0.0002. Analysis of MMLV-GFP infections: XPD-wt #1 and XPD(R683W) #1, P = 0.0006; XPD-wt #2 and XPD(R683W) #2, P = 0.003. (C) XPA cell lines include a patient-derived XPA mutant cell line [XPA(Y116ter)] and complemented with the WT XPA allele (XPA-wt). MSH2 cell lines include WT MEFs (MSH2+/+) and MSH2−/− littermate MEFs. Error bars indicate the standard deviation between duplicate infected wells. An identical trend was observed in at least three separate experiments for all cell lines.
HIV transduction efficiency also was evaluated in XPD cells (Fig. 1B). Two independent XPD cell lines were derived from XP patients expressing an R683W mutation [XPD(R683W)] that accounts for ≈64% of observed XPD patients (10, 16, 17). Both of the XPD(R683W) cell lines were complemented with WT XPD (XPD-wt), yielding two pairs of matched isogenic mutant and complemented cell lines. The NER defects in XPD(R683W) cell lines and rescued in XPD-wt cell lines was confirmed by sensitivity to UV irradiation where XPD(R683W) > XPD-wt (Fig. 5B; data not shown).
Transduction of these cell lines with HIV-GFP revealed fewer XPD-wt cells expressing GFP than XPD(R683W) mutant cells (Fig. 1B). The increase of HIV transduction efficiency in XPD(R683W) cell lines compared to the XPD-wt cell lines was 15.3% (P = 0.009) and 17.2% (P = 0.0002). Transduction of these cell lines with MMLV-GFP also showed a similar decrease in transduction efficiency in XPD-wt cells (Fig. 1B). The observed differences in transduction efficiency between mutant and complemented XPB or XPD cell lines were not affected by varying the multiplicity of infection (MOI293T) below 10 MOI293T (Fig. 6 A and B, which is published as supporting information on the PNAS web site). Both transduction efficiency and UV sensitivity show the same trend in the two pairs of cell lines, XPD(R683W) > XPD-wt.
The Role of Other DNA Repair Factors on Integration.
XPB and XPD proteins do not exist as singular proteins in the cell: They are part of the TFIIH complex, which plays a role in both transcription and NER. The XPA protein is required for NER but has no role in transcription because it is not part of TFIIH (18). To determine whether other NER DNA repair pathway genes affect transduction efficiency, isogenic mutant and complemented XPA cell lines were examined (19). The mutant cell line encodes an XPA mutation at the splice site of exon 3 resulting in early termination of the protein [XPA(Y116ter)]. The NER defect of the XPA(Y116ter) cell line was confirmed by sensitivity to UV (Fig. 5C). We observed no difference in the HIV-GFP transduction efficiency between isogenic XPA mutant and rescued cell lines (Fig. 1C, left). We conclude that the effect on HIV transduction efficiency observed in XPB and XPD cells is associated with TFIIH but not necessarily other NER proteins.
The MSH2 protein is a necessary component of the mismatch repair (MMR) pathway. MEFs derived from MSH2 WT and null littermates were transduced with HIV-GFP (20). We observed no difference in the transduction efficiency between MSH2 WT and null cell lines, suggesting that the MMR pathway has no effect on retroviral transduction (Fig. 1C, right).
Replicating Retroviral Infection.
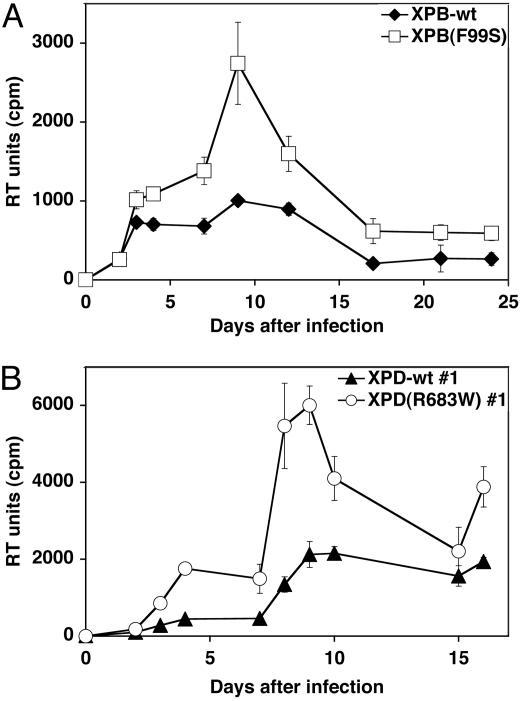
The effects of NER-associated mutations were evaluated by replicating retroviral infection. XPB, XPD, and XPA mutant and complemented cell lines were infected with an amphotropic MMLV (21). Unlike HIV, MMLV requires cellular division for successful infection. Reverse transcription (RT) activity in the supernatant of infected cultures indicated that replicating MMLV viral production was higher in XPB(F99S) and XPD(R683W) cell lines compared with their complemented counterparts (Fig. 2). RT activity increased 3-fold 9 days after infections in XPB cells and 4-fold 8 days after infections in XPD cells. The greater difference observed in the XPD cell lines contrasts transduction experiments where the difference was more subtle. This effect may be due, in part, to the decreased doubling time of the XPD-wt cells compared with XPD(R683W), resulting in fewer cell divisions necessary for MMLV infection. In contrast to XPB and XPD cell lines, MMLV replication in XPA(Y116ter) and XPA-wt cell lines was equivalent (Fig. 7, which is published as supporting information on the PNAS web site). We conclude that the TFIIH/NER components XPB and XPD, and not the basal NER recognition factor XPA, affect retroviral replication.
Fig. 2.
Retroviral replication in XP cell lines. Isogenic mutant and complemented XPB (A) and XPD (B) cell lines were infected with an amphotropic murine leukemia virus. Replicating viral production was monitored by RT activity (RT units). Error bars indicate the SD between triplicate infections.
Reduction of HIV Transduction Is Not Related to Apoptosis.
Several studies with cell lines defective for DNA repair have proposed apoptosis as a response to retroviral infection (1, 2, 22–24). To test this possibility, the isogenic XPB and XPD mutant and rescued cell lines were transduced with the HIV-GFP vector, and apoptosis was examined by trypan blue exclusion and/or activation of caspase 3 (Fig. 8, which is published as supporting information on the PNAS web site; data not shown). Comparison of nontransduced cells to transduced cells suggested that there is little, if any, apoptotic response to transduction (Fig. 8). We conclude that the differences in integration efficiency are not due to apoptosis.
Kinetics of HIV cDNA.
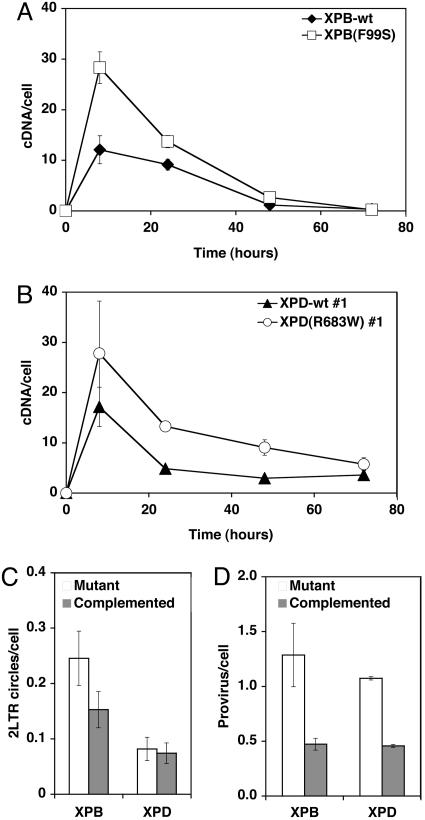
We examined the kinetic production of HIV cDNA during transduction by real-time quantitative fluorescent PCR (qPCR) (25). The PCR primers and probe detect late RT products and are a measure of full-length, double-stranded cDNA molecules. This value was compared to the host genomic DNA marker PBGD to calculate the number of viral cDNA molecules per cell (26). The XPB-wt and XPB(F99S) mutant cell lines were transduced with the HIV-GFP retroviral vector at 20 MOI293T (Fig. 3A) and 3 MOI293T (Fig. 6C), and the kinetics of viral cDNA were monitored after infection. We found that the number of cDNA molecules was greater in the XPB(F99S) mutant cells compared with the XPB-wt cells (20 MOI293T, P = 0.0005; 3 MOI293T, P = 0.004). Quantitation also revealed more 2LTR circles (P = 0.005) and integrated provirus (P = 0.017) in XPB(F99S) cells compared with XPB-wt cells (Fig. 3 C and D).
Fig. 3.
Quantitative PCR analysis of HIV cDNA in XPB and XPD cell lines. Isogenic mutant and rescued XPB and XPD cell lines were transduced with an HIV-based retroviral vector pseudotyped with VSV-G. (A and B) Cells were transduced at 20 MOI293T based on 293T titers. Total DNA extracts were purified at 8, 24, 48, and 72 h. Late reverse transcripts and a cellular genomic marker gene were quantified by qPCR, yielding the number of HIV cDNA molecules per cell. (A) Transductions of XPB(F99S) cells and XPB-wt cells are compared. Paired t test analysis yielded the two-tailed P value for 20 MOI293T P = 0.0005. (B) Transductions of XPD(R683W) #1 cells and XPD-wt #1 cells are compared. Paired t test analysis yielded the two-tailed P value for 20 MOI293T, P < 0.0001. The number of 2LTR circles (C) and integrated proviruses (D) per cell were determined by qPCR. Error bars indicate the SD between duplicate infected wells. An identical trend was observed in at least three separate experiments for all cell lines.
HIV cDNA accumulation also was evaluated by qPCR in isogenic XPD(R683W) mutant and XPD-wt cell lines (Figs. 3B and 6D). The number of full-length cDNA molecules was greater in the XPD(R683W) mutant cells compared with the XPD-wt cells (20 MOI293T, P < 0.0001; 3 MOI293T, P = 0.002). As the cDNA is degraded, the relative difference between the cDNA in paired cell lines remains nearly constant at each time point. We also observed more integrated proviruses (P = 0.005) in the XPD(R683W) mutant cells compared with XPD-wt (Fig. 3D). The number of 2LTR circles was slightly decreased in XPD(R683W) mutant cells, but the difference was not statistically significant (P = 0.3; Fig. 3C). These results are consistent with the conclusion that XPB and XPD affect transduction efficiency by reducing the relative number of linear cDNA molecules.
The effect of XPB and XPD mutations on the accumulation of HIV cDNA also was tested with an integrase mutant virus [HIV(IN-D116N)-GFP; Fig. 9, which is published as supporting information on the PNAS web site]. The HIV(IN-D116N)-GFP virus encodes the integrase mutation D116N in the catalytic site DDE motif necessary for integration (27). The D116N mutation allows the expression of integrase protein, but it is unable to complete integration. Equivalent RT units of mutant or WT integrase virus particles were used to transduce XPB and XPD isogenic mutant and rescued cell lines. The accumulation of viral cDNA appeared the same for both the mutant HIV(IN-D116N)-GFP and WT HIV-GFP vectors (Fig. 9). We conclude that integrase catalytic activity does not affect the accumulation of HIV cDNA. These results suggest that XPB and XPD act on the HIV cDNA before integration. Moreover, XPB and XPD would not appear to be recognizing any form of integration intermediate associated with the host chromosome.
HIV cDNA Stability.
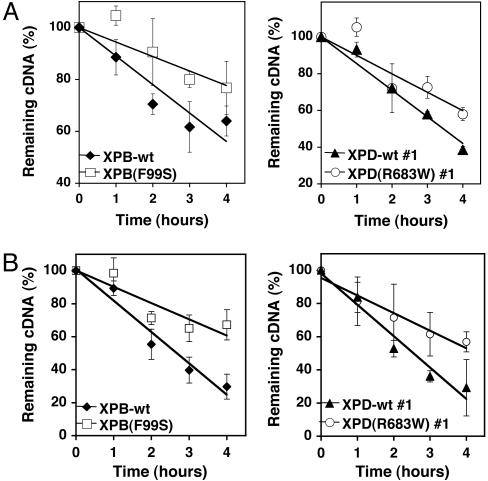
The reduced accumulation of HIV cDNA in XPB-wt and XPD-wt cell lines could be a result of reduced RT-dependent cDNA synthesis or increased cDNA degradation after RT. To distinguish these two possibilities, we examined the stability of existing HIV cDNA in cells treated with the nonnucleoside reverse transcriptase inhibitors (NNRTI) efavirenz or foscarnet. If XPB and XPD affect cDNA synthesis, then the rate of cDNA degradation should be similar for mutant and rescued (wt) cell lines. We compared the total HIV cDNA of infected cells treated with an NNRTI to infected untreated cells (Fig. 4). The XPB(F99S) mutant cells showed a significantly slower decline in the number of cDNA molecules per cell than the isogenic XPB-wt cells (Fig. 4A Left; P = 0.002). The slope suggests that the HIV cDNA half life in XPB(F99S) cells was ≈7.7 h compared with 4.6 h in XPB-wt cells. Furthermore, the XPD(R683W) mutant cells also displayed a slower degradation of cDNA per cell compared with the isogenic XPD-wt cells (Fig. 4A Right, P = 0.01). The slope suggests that the HIV cDNA half life was ≈5.0 h in XPD(R683W) cells and 3.5 h in XPD-wt cells. Efavirenz had no effect on transductions with the HIV-RT(K103N) resistance mutant (Fig. 10, which is published as supporting information on the PNAS web site), suggesting that efavirenz is specifically inhibiting RT during the transduction. We observed a similar effect on the half life of HIV cDNA after treatment with the unrelated NNRTI foscarnet (Fig. 4B). These results are consistent with the conclusion that the TFIIH component proteins XPB and XPD enhance the degradation of HIV cDNA, thereby decreasing the pool of available cDNA for integration and, ultimately, decreasing the integration efficiency.
Fig. 4.
Evaluation of HIV cDNA degradation after addition of an RT inhibitor. XPB and XPD cell lines were transduced with an HIV-based retroviral vector. Three hours after the addition of the virus, the media was replaced and the RT inhibitor efavirenz (A) or foscarnet (B) was added. The number of cDNA molecules per cell was quantified by qPCR. At each time point, the remaining cDNA (%) was calculated from the number of cDNA molecules per cell treated with efavirenz/foscarnate divided by the number of cDNA molecules per untreated cell. Time indicates the number of hours after the addition of efavirenz. P values associated with efavirenz treatment: XPB-wt and XPB(F99S), P = 0.002; XPD-wt #1 and XPD(R683W) #1, P = 0.01. P values associated with foscarnet treatment: XPB-wt and XPB(F99S), P = 0.001; XPD-wt #1 and XPD(R683W) #1, P = 0.01. An identical trend was observed in at least three separate experiments. Error bars indicate the standard deviation between duplicate infected wells.
Discussion
The TFIIH proteins XPB and XPD appear to play a principal role in the degradation of retroviral cDNA. Moreover, the XPB/XPD-dependent reduction of HIV cDNA results in decreased successful integration events. Although TFIIH has been shown to interact with the HIV protein Tat and enhance transcription from the LTR promoter (28, 29), this effect appears unrelated to our results because Tat is not present in these retroviral vectors or is not replicating MMLV.
Because XPB and XPD are essential components of the basal transcription machinery, it is not possible to generate null cells. The mutant XPB and XPD cell lines tested in these studies are functional hypomorphs with reduced XPB and XPD transcription or NER activity. The HIV and MMLV transduction efficiencies uniquely correlated with decreased NER function (XPB(F99S) > XPD(R683W) > XPB(T119P) ≈ WT). However, an XPA NER mutant did not affect the transduction efficiency. These results suggest that the function of TFIIH (XPB/XPD) in HIV cDNA degradation is discrete from both the transcription or NER apparatus. Other novel activities of TFIIH have been described that include ubiquitin ligase, interaction with the molecular chaperone Hsp90, and inhibition of short sequence recombination (30–32). Because TFIIH is not known to exhibit an exonuclease activity, it is likely that host factors in addition to XPB and XPD are required to mediate the degradation of LTR retroelement cDNA.
Other host defense mechanisms against HIV infection have been described that appear to be cytoplasmic (33–37). The XPB/XPD defense pathway appears to represent a previously undescribed example of a nuclear host defense against retroviral infection, because these proteins are reportedly exclusive nuclear residents (10).
Similar studies have been performed with the yeast LTR retrotransposon Ty1. Mutation of the S. cerevisiae XPB homolog, Ssl2, or XPD homolog, Rad3, led to an increase in Ty1 cDNA and a subsequent increase in transposition frequency (38, 39). Mutations of other NER genes had no effect on Ty1 transposition frequency. The mutated residues of Ssl2(E556K) and Rad3(G595R) identified in the yeast screen differ from the human mutations of XPB(F99S) and XPD(R683W) studied here. Both the yeast studies and our results suggest a greater effect of XPB (Ssl2) compared with XPD (Rad3) on LTR retroelement integration. This difference may reflect a greater significance of XPB protein activity in the cDNA degradation pathway. The similarity between the human and yeast studies suggests an evolutionarily conserved defense against invading LTR retroelements.
XPB or XPD patients are very rare and typically do not advance to sexual maturity. The rarity and genetics suggest that any possible increased risk for HIV infection might not be detected. However, a number of polymorphisms of the XPB and XPD genes exist in the human population with unknown significance to HIV pathogenesis. Because several of these alterations have been associated with increased cancer risk, their role in HIV pathogenesis deserves investigation (40).
Materials and Methods
Cell Lines.
All media reagents were obtained from Mediatech. Cell lines are described in Table 1 and are available upon request. Media was supplemented with 10% heat-inactivated FCS, penicillin, streptomycin, and l-glutamine.
XPB cell lines were originally described by Riou et al. (12). XPB cell lines include the XP/CS patient-derived cell line expressing the mutant allele XPB(F99S), a TTD patient-derived cell line expressing the mutant allele XPB(T119P), the XPB(F99S) cell line complemented with the XPB(T119P) allele (yielding a partial rescue of XPB activity and referred to here as XPB-prt), the XPB(F99S) cell line complemented with the WT XPB allele (referred to here as XPB-wt) (12). XPB(F99S) and XPB-wt cell lines were grown in DMEM/F10. The XPB-prt growth media was supplemented with 400 μg/ml G418. The XPB(T119P) cell line was grown in MEM.
XPD cell lines were originally described by Gozukara et al. and are available through the Coriell Cell Repository (16) as well as by Marionnet et al. (17). XPD cell lines include one XP patient-derived cell line [XPD(R683W) #1] encoding two XPD mutations, one allele encoding a 78-nt deletion that is not expressed, and the second allele encoding the R683W mutation (16). This XPD(R683W) mutant cell line was complemented with the WT XPD allele (16). The XPD(R683W) #1-derived cell lines were grown in DMEM. The XPD-wt #1 growth media was supplemented with 600 μg/ml G418. A second XP patient-derived XPD mutant cell line [XPD(R683W) #2] encodes the R683W mutation on both alleles (17). This XPD mutant cell line also was complemented with the WT XPD allele (17). The XPD(R683W) #2-derived cell lines were grown in MEM.
XPA cell lines have been described by Levy et al. (19) and are available through the Coriell Cell Repositories (Camden, NJ) (19). The patient-derived XPA cell line [XPA(Y116ter)] was complemented with the WT XPA allele (referred to in the text as XPA-wt). Both complemented and mutant XPA cell lines were grown in DMEM.
MSH2 mice were described by Cranston et al. (20). MSH2 WT (MSH2+/+)and MSH2−/− littermate murine embryonic fibroblasts (MEFs) were at passage 3 during transduction. MEFs were grown in DMEM (20).
UV Survival.
Cells were assayed for UV sensitivity as described with the following modifications (41). Incubation of 105 cells was performed in triplicate dishes for 2 days in media with 0.2% FCS, washed with PBS, and irradiated at 254 nm (Stratalinker; Stratagene). PBS was replaced with media containing 0.5% FCS. Ten days later, cellular viability was determined by trypan blue exclusion.
Retrovirus and Retroviral Vector Particles.
The construct pAMS encodes a provirus of amphotropic MMLV (21). 293T cells were transiently transfected with the pAMS plasmid by calcium phosphate precipitation. Viral supernatants were collected, filtered to remove producer cells, and added to 3T3 cells to test for infectivity. Viral replication was monitored by RT activity in the media.
Target cells for pAMS infections were plated at 105 cells per well in triplicate in six-well dishes. The RT activity from infected 3T3 cells was measured, and 105 cpm was added to the target cells in triplicate and in the presence of 10 μg/ml DEAE Dextran (Sigma). RT activity of supernatants was monitored for at least 2 weeks. Cells were split to 2 × 105 per well after becoming confluent throughout the course of infection.
HIV-based retroviral vector particles were generated by transfection of 293T cells. The three plasmids include an envelope construct-encoding VSV-G, the HIV-packaging construct ΔR9 (14), and the genomic construct p156RRLsinPPTCMVGFPPRE-expressing GFP from a CMV promoter (13). The integrase D116N catalytic site mutant and RT K103N efavirenz-resistant mutant were engineered into ΔR9 and confirmed by sequencing (27, 42). Similar to HIV vector particles, MMLV vector particles were generated by transfection of 293T cells. The three MMLV vector plasmids include the envelope construct encoding VSV-G, the MMLV packaging construct pHIT60, and the genomic pLEGFP-CI expressing GFP from a CMV promoter (Clontech). Transfections were by the calcium phosphate method (43). Vector supernatants were collected and filtered to remove vector producer cells. Vector supernatants were treated with 10–20 units of DNaseI (Roche) for 1 h at ambient temperature to degrade producer plasmids before transductions (25, 44). Titers of vector particles were determined by transduction of 293T cells and quantified by expression of GFP by flow cytometry (Coulter XL-MCL or Becton Dickinson FACSCalibur). Thus, the functional quantification of vector particles expressed as “MOI” (MOI293T) is determined by transduction of 293T cells.
Mutant and rescued cell lines were plated in six-well dishes at 4 × 105 cells per well at the beginning of infections. HIV-based vector particles were added to mutant and rescued cell lines in the presence of 10 μg/ml DEAE dextran at 1 MOI293T for flow cytometry studies and at 3 MOI293T and 20 MOI293T for qPCR experiments. MMLV-based vector particles were added to mutant and rescued cell lines in the presence of 10 μg/ml DEAE dextran at 2 MOI293T. Cells were fixed with 4% paraformaldehyde at 48 h and analyzed by flow cytometry.
RT Assays.
MMLV replication was monitored by RT activity in the culture supernatants. MMLV RT activity was measured by the Quan-T-RT assay (Amersham Pharmacia Biosciences) with the following modification, 1× assay buffer was 12.5 mM Hepes, pH 7.6/30 mM KCl/4 mM MnCl2/12 mM spermidine/0.05% Nonidet P-40/3 mM 2-mercaptoethanol (45).
Because HIV integrase D116N catalytic site mutants do not integrate, these mutant vector particles could not be titered by GFP expression (27). HIV RT activity of WT integrase vector particles and D116N integrase mutant vector particles was measured by the Quant-T-RT assay as per the manufacturer’s instructions. Equivalent HIV RT units of WT integrase and D116N mutant integrase vector particles were added to cells during infections.
The RT activities of WT HIV and HIV-RT(K103N) were tested in the presence or absence of efavirenz. WT RT was sensitive to efavirenz, and RT(K103N) was resistant (data not shown).
Cellular Viability Assays.
Cell lines were treated with 0, 0.5, or 2 MOI293T HIV vector particles in the presence of 10 μg/ml DEAE dextran for 2 h and then refed with fresh media. Viable cells were counted by trypan blue exclusion at 0, 24, and 48 h. Apoptotic cells were measured with the phycoerythrin-conjugated polyclonal active caspase-3 antibody apoptosis kit (BD Pharmingen) and flow cytometry.
Quantitative PCR.
Cells were transduced with HIV vector particles in the presence of 10 μg/ml DEAE dextran for 2 h. Genomic DNA from duplicate wells was harvested at 8, 24, 48, and 72 h by the DNeasy Tissue Kit (Qiagen, Valencia, CA). DNA samples were precipitated and resuspended in 10 mM Tris/1 mM EDTA, pH 8. Late reverse transcripts and 2LTR circles were quantified as described in ref. 25. Standards were generated by subcloning the amplicons into pGEM-T Easy (Promega). Integrated provirus was quantified as described in ref. 46. The genomic marker porphobilinogen deaminase (PBGD) was quantified as described in ref. 26; standards were generated from the genomic DNA of matched uninfected cells. Late RT values were divided by PBGD values to yield the number of cDNA molecules per cell. All reactions were amplified with Taqman Universal PCR Master Mix (Applied Biosystems) in an ABI Prism 7700 or 7900HT Sequence Detection System. Thermal cycling conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min.
HIV cDNA Degradation Rate.
The rate of HIV vector particle cDNA degradation was measured by qPCR. Cells were transduced for 3 h and refed with fresh media in the presence or absence of efavirenz (National Institutes of Health AIDS Research and Reference Reagent Program, Germantown, MD) or foscarnet (Sigma). Genomic DNA from duplicate wells of treated and untreated cells was harvested at 0, 1, 2, 3, and 4 h after efavirenz treatment, corresponding to 3, 4, 5, 6, and 7 h after infection. The number of cDNA molecules per cell treated with efavirenz or foscarnet was divided by the number of cDNA molecules per untreated cell (% remaining cDNA).
Statistical Analysis.
Data presented in Figs. 1 and 3 were analyzed by paired t test to generate two-tail P values [GraphPad (San Diego) prism 4]. Linear trendlines in Fig. 4 were calculated by Microsoft excel. Data in Fig. 4 was also analyzed by two-way ANOVA to generated P values (GraphPad prism). P values were rounded to one significant figure.
Supplementary Material
Acknowledgments
We thank Vicki Whitehall, Samir Acharya, and David Garfinkel for critical reading of the manuscript and Carsten Muenk and Stephen Hughes for helpful suggestions. Efavirenz was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. This work was supported by National Institutes of Health Grants T32 DK07705 (to K.Y.) and CA56542 (to R.F.).
Abbreviations
- MMLV
Moloney murine leukemia virus
- MOI
multiplicity of infection
- NER
nucleotide excision repair
- prt
partially
- qPCR
quantitative fluorescent PCR
- RT
reverse transcriptase
- TTD
trichothiodystrophy
- XP
xeroderma pigmentosum.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Daniel R., Katz R. A., Skalka A. M. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 2.Kilzer J. M., Stracker T., Beitzel B., Meek K., Weitzman M., Bushman F. D. Virology. 2003;314:460–467. doi: 10.1016/s0042-6822(03)00455-0. [DOI] [PubMed] [Google Scholar]
- 3.Lau A., Kanaar R., Jackson S. P., O’Connor M. J. EMBO J. 2004;23:3421–3429. doi: 10.1038/sj.emboj.7600348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egly J. M. FEBS Lett. 2001;498:124–128. doi: 10.1016/s0014-5793(01)02458-9. [DOI] [PubMed] [Google Scholar]
- 5.van Brabant A. J., Stan R., Ellis N. A. Annu. Rev. Genomics Hum. Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- 6.Cleaver J. E., Thompson L. H., Richardson A. S., States J. C. Hum. Mutat. 1999;14:9–22. doi: 10.1002/(SICI)1098-1004(1999)14:1<9::AID-HUMU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann A. R. Genes Dev. 2001;15:15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann A. R. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Taylor E. M., Broughton B. C., Botta E., Stefanini M., Sarasin A., Jaspers N. G. J., Fawcett H., Harcourt S. A., Arlett C. F., Lehmann A. R. Proc. Natl. Acad. Sci. USA. 1997;94:8658–8663. doi: 10.1073/pnas.94.16.8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubaele S., Proietti De Santis L., Bienstock R. J., Keriel A., Stefanini M., Van Houten B., Egly J. M. Mol. Cell. 2003;11:1635–1646. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 11.Garfinkel D. J., Bailis A. M. J. Biomed. Biotechnol. 2002;2:55–60. doi: 10.1155/S1110724302201023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riou L., Zeng L., Chevallier-Lagente O., Stary A., Nikaido O., Taieb A., Weeda G., Mezzina M., Sarasin A. Hum. Mol. Genet. 1999;8:1125–1133. doi: 10.1093/hmg/8.6.1125. [DOI] [PubMed] [Google Scholar]
- 13.Follenzi A., Ailles L. E., Bakovic S., Geuna M., Naldini L. Nat. Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 14.Naldini L., Blomer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 15.Butler S. L., Johnson E. P., Bushman F. D. J. Virol. 2002;76:3739–3747. doi: 10.1128/JVI.76.8.3739-3747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gozukara E. M., Parris C. N., Weber C. A., Salazar E. P., Seidman M. M., Watkins J. F., Prakash L., Kraemer K. H. Cancer Res. 1994;54:3837–3844. [PubMed] [Google Scholar]
- 17.Marionnet C., Quilliet X., Benoit A., Armier J., Sarasin A., Stary A. Cancer Res. 1996;56:5450–5456. [PubMed] [Google Scholar]
- 18.de Laat W. L., Jaspers N. G., Hoeijmakers J. H. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 19.Levy D. D., Saijo M., Tanaka K., Kraemer K. H. Carcinogenesis. 1995;16:1557–1563. doi: 10.1093/carcin/16.7.1557. [DOI] [PubMed] [Google Scholar]
- 20.Cranston A., Bocker T., Reitmair A., Palazzo J., Wilson T., Mak T., Fishel R. Nat. Genet. 1997;17:114–118. doi: 10.1038/ng0997-114. [DOI] [PubMed] [Google Scholar]
- 21.Miller A. D., Law M. F., Verma I. M. Mol. Cell. Biol. 1985;5:431–437. doi: 10.1128/mcb.5.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baekelandt V., Claeys A., Cherepanov P., De Clercq E., De Strooper B., Nuttin B., Debyser Z. J. Virol. 2000;74:11278–11285. doi: 10.1128/jvi.74.23.11278-11285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Olvera J. M., Yoder K. E., Mitchell R. S., Butler S. L., Lieber M., Martin S. L., Bushman F. D. EMBO J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daniel R., Greger J. G., Katz R. A., Taganov K. D., Wu X., Kappes J. C., Skalka A. M. J. Virol. 2004;78:8573–8581. doi: 10.1128/JVI.78.16.8573-8581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butler S. L., Hansen M. S., Bushman F. D. Nat. Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 26.Buckman J. S., Bosche W. J., Gorelick R. J. J. Virol. 2003;77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engelman A., Craigie R. J. Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Martinez L. F., Mavankal G., Neveu J. M., Lane W. S., Ivanov D., Gaynor R. B. EMBO J. 1997;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou M., Nekhai S., Bharucha D. C., Kumar A., Ge H., Price D. H., Egly J. M., Brady J. N. J. Biol. Chem. 2001;276:44633–44640. doi: 10.1074/jbc.M107466200. [DOI] [PubMed] [Google Scholar]
- 30.Takagi Y., Masuda C. A., Chang W. H., Komori H., Wang D., Hunter T., Joazeiro C. A., Kornberg R. D. Mol. Cell. 2005;18:237–243. doi: 10.1016/j.molcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Flom G., Weekes J., Johnson J. L. Curr. Genet. 2005;47:368–380. doi: 10.1007/s00294-005-0580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maines S., Negritto M. C., Wu X., Manthey G. M., Bailis A. M. Genetics. 1998;150:963–976. doi: 10.1093/genetics/150.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 34.Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 35.Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 36.Hatziioannou T., Perez-Caballero D., Yang A., Cowan S., Bieniasz P. D. Proc. Natl. Acad. Sci. USA. 2004;101:10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keckesova Z., Ylinen L. M. J., Towers G. J. Proc. Natl. Acad. Sci. USA. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee B. S., Lichtenstein C. P., Faiola B., Rinckel L. A., Wysock W., Curcio M. J., Garfinkel D. J. Genetics. 1998;148:1743–1761. doi: 10.1093/genetics/148.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee B. S., Bi L., Garfinkel D. J., Bailis A. M. Mol. Cell. Biol. 2000;20:2436–2445. doi: 10.1128/mcb.20.7.2436-2445.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benhamou S., Sarasin A. Mutagenesis. 2002;17:463–469. doi: 10.1093/mutage/17.6.463. [DOI] [PubMed] [Google Scholar]
- 41.Eveno E., Quilliet X., Chevallier-Lagente O., Daya-Grosjean L., Stary A., Zeng L., Benoit A., Savini E., Ciarrocchi G., Kannouche P., et al. Biochimie. 1995;77:906–912. doi: 10.1016/0300-9084(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 42.Bacheler L., Jeffrey S., Hanna G., D’Aquila R., Wallace L., Logue K., Cordova B., Hertogs K., Larder B., Buckery R., et al. J. Virol. 2001;75:4999–5008. doi: 10.1128/JVI.75.11.4999-5008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCurrach M. E., Lowe S. W. Methods Cell. Biol. 2001;66:197–227. doi: 10.1016/s0091-679x(01)66010-2. [DOI] [PubMed] [Google Scholar]
- 44.Lu R., Limon A., Devroe E., Silver P. A., Cherepanov P., Engelman A. J. Virol. 2004;78:12735–12746. doi: 10.1128/JVI.78.23.12735-12746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malmsten A., Ekstrand D. H., Akerblom L., Gronowitz J. S., Kallander C. F., Bendinelli M., Matteucci D. J. Virol. Methods. 1998;75:9–20. doi: 10.1016/s0166-0934(98)00091-3. [DOI] [PubMed] [Google Scholar]
- 46.Brussel A., Sonigo P. J. Virol. 2003;77:10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.