Abstract
Agrobacterium tumefaciens induces crown gall tumors by transferring a piece of its tumor-inducing plasmid into plant cells. This transferred DNA encodes the synthesis of indole acetic acid (IAA) and cytokinin, and their overproduction results in tumor formation. The transfer is initiated by a two-component regulatory system, VirA/G recognizing plant signal molecules in the plant rhizosphere and activating a regulon on the tumor-inducing plasmid, which is required for the processing and transfer of DNA and protein. Although a great deal is known about vir gene activation, nothing is known about whether or how the vir gene regulon is inactivated after plant cell transformation. Presumably, just as a mechanism exists for activating the vir gene regulon only when a plant is in the immediate environment, a mechanism should exist for inactivating the same regulon once it has fulfilled its mission to transferred DNA into plant cells. We now show that IAA inactivates vir gene expression by competing with the inducing phenolic compound acetosyringone for interaction with VirA. IAA does not inhibit the vir genes in cells containing a constitutive sensor virA locus, which does not require any signal molecules to become phosphorylated. At higher concentrations, IAA inhibits the growth of Agrobacterium and many other plant-associated bacteria but not the growth of bacteria that occupy other ecological niches. These observations provide the missing link in the cycle of vir gene activation and inactivation.
The transformation of plant cells by Agrobacterium is initiated by the bacterium-recognizing signal molecules in the rhizosphere of the plant. This recognition by a two-component regulatory system, VirA/G, sets in motion the activation of the genes (vir) required for the processing and transfer of DNA and proteins into the plant cell (1, 2). These plant signal molecules are a phenolic compound, typically acetosyringone (AS), sugars, which are components of the plant cell wall, and acidic conditions (pH 5.5). In addition, low phosphate concentrations, which characterize many soils, are required for maximum vir gene induction (3). All of these conditions are typical of the rhizosphere of a plant. The sensor protein, VirA, is a membrane-spanning histidine kinase and experimentally can be divided into four domains that function independently of one another: periplasmic, linker, kinase, and receiver (4). The periplasmic sugar binding protein, a product of a chromosomal gene, chvE, first binds the sugars and then interacts with the periplasmic domain of VirA (5). The periplasmic module also recognizes acidic conditions (pH 5.5) (6), whereas the phenolic compounds most likely interact directly with the linker domain (7). The kinase domain is the site of phosphorylation (His-474), and the receiver domain inhibits phosphorylation of the kinase domain (8). The response regulator, VirG, is phosphorylated by VirA (Asp-52) and then directly activates a regulon of ≈30 genes on the tumor-inducing (Ti) plasmid (9). Interestingly, only ≈20 of the 30 genes are required for plant cell transformation under laboratory conditions (10).
The vir genes are responsible for the processing and transfer of ≈20 kb of single-stranded transferred DNA (T-DNA), which map to the Ti plasmid. The T-DNA encodes two enzymes that convert tryptophan to indole acetic acid (IAA) via indole acetamide. Another enzyme encoded on the T-DNA is involved in cytokinin synthesis. The overproduction of auxin and cytokinin by the transformed plant cells results in the typical crown gall tumor. Other transferred genes encode enzymes involved with the synthesis of amino acid and sugar derivatives, the opines, which the strain of Agrobacterium that induces the tumor can use as a source of carbon, nitrogen, and energy. In addition, some opines, termed conjugal, induce the transcription of genes involved in the conjugal transfer of the Ti plasmid between bacteria (11).
The sensing of plant signal molecules by the VirA protein and the environmental conditions that activate the vir genes have been studied extensively by a number of laboratories and are reasonably well understood (2). Much less attention has been paid to the possibility that various environmental conditions might serve to down-regulate the vir regulon. Two laboratories have demonstrated that vir gene induction can be down-regulated by a class of compounds, the benzoxazinones, major secondary metabolites exuded only by graminaceous plants. One member of this group, synthesized by maize, 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one (DIMBOA) inhibited both growth and vir gene induction (12). The related compound, 2-hydroxyl-4,7-dimethoxy-benzoxazin-3-one inhibited vir gene induction but not growth (13). It was suggested that both compounds could serve to inhibit transformation of the host plant, maize, a plant long recognized as being notoriously difficult to transform (13).
Bacteria have highly sophisticated mechanisms for regulating the synthesis of metabolites only when they are needed for specific physiological processes. Agrobacterium provides an excellent example. Growing in the soil, in the absence of a plant, the bacterial genes necessary to bring about plant cell transformation are not expressed. However, in the rhizosphere of a plant, the bacteria recognize several plant signal molecules via a two-component regulatory system, which activates the 30 vir gene regulon. The expression of many other genes are likely to be affected indirectly by the activation of the VirA/G regulatory system. Because the vir genes of the Ti plasmid are dedicated to plant cell transformation, it seems wasteful for the bacteria to continue to synthesize at least 30 proteins whose function is no longer necessary. A recent paper reported genetic evidence that VirA can dephosphorylate VirG in the absence of inducing plan signal molecules, thereby inhibiting vir gene induction (14). The data in this report demonstrate that Agrobacterium shuts down vir gene expression by recognizing the plant hormone IAA, which is overproduced by the transformed plant and, thereby, acts as a signature molecule of plant cell transformation.
Results
IAA Inhibits vir Gene Induction and Growth of A. tumefaciens.
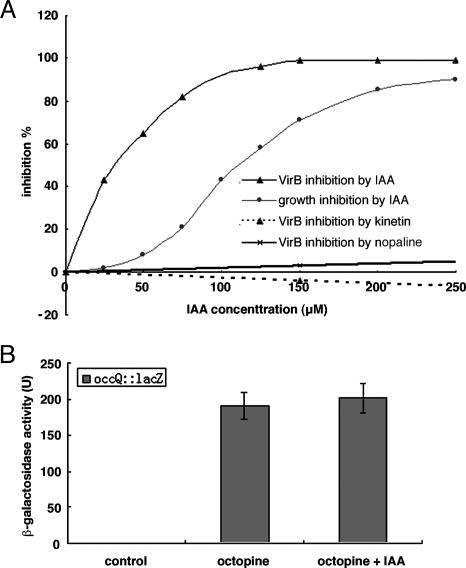
Because Agrobacterium intercepts plant signal molecules to activate genes required for T-DNA processing and transfer, it would not be surprising if this organism could recognize a signature molecule of transformed plant cells. If true, candidate molecules for plant cell transformation are the gene products of the introduced T-DNA. Accordingly, we tested the ability of the three tumor metabolites, IAA, cytokinin, and nopaline, for their ability to inhibit vir gene induction as measured by expression of a β-gal reporter gene fusion in the virB gene (15). Only IAA had a significant inhibitory effect (Fig. 1A). Thus, at concentrations as low as 25 μM, vir gene induction was inhibited significantly. The IC50, the concentration of IAA that inhibits vir gene induction by 50%, in the presence of 100 μM AS, is ≈32 μM. Further, IAA at a concentration of 150 μM, had no effect on the induction of octopine dehydrogenase, which is an enzyme also on the Ti plasmid and induced by octopine (Fig. 1B; ref. 16). Thus, we conclude that the IAA is specifically inhibiting expression of the vir genes and is not a nonspecific general inhibitor of induction of Ti plasmid genes.
Fig. 1.
The effect of IAA on vir gene expression and growth of A. tumefaciens. (A) Effect of IAA, kinetin, and nopaline on virB gene expression. Only one concentration of kinetin and nopaline was used. (B) Effect of IAA on octopine dehydrogenase gene expression. A. tumefaciens A348 with pSM102 (16) was grown in AB minimal medium overnight and inoculated into induction medium with 0.2% arabinose as carbon source. The cells were assayed for β-gal activity by following the method of Miller (17). Control, induction medium without octopine; octoptine, induction medium with 100 μg/ml octopine; octoptine plus IAA, induction medium with 100 μg/ml octoptine and 150 μM IAA.
Not only does IAA inhibit vir gene induction, but at slightly higher concentrations, it also inhibits bacterial growth (Fig. 1A). The cells are not killed by IAA because they resume growth once the IAA is removed (data not shown). This inhibition is similar to the situation observed with IAA and several fungi (18). In that report, IAA, at concentrations approximating those we used for inhibition of vir gene expression, resulted in developmental changes in Saccharomyces, which precedes fungal plant infection (18). At slightly higher concentrations of IAA, the growth of Saccharomyces and Ustilago maydis, a fungal pathogen of maize, was inhibited. As in our experiments, the fungi were not killed by the IAA.
IAA Inhibition Is Rescued by Constitutive virA and Constitutive virG.
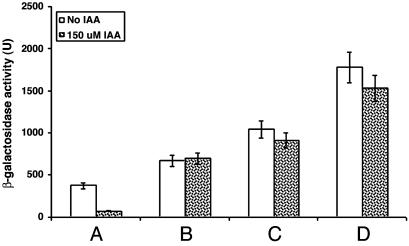
The inhibition of vir gene induction by IAA might involve the VirA/G system, which regulates the vir genes. To confirm this likelihood and gain some insight into the precise site of action, we tested whether strains carrying either a constitutive virA or virG locus were also inhibited. The virA constitutive mutant on a low copy number plasmid does not require a phenolic compound, sugar, or acid for phosphorylation and subsequent activation of VirG, and the VirG constitutive protein does not require phosphorylation to activate the vir gene regulon (19, 20). The constitutive virG locus only functions as a constitutive mutation when it is on a high copy number plasmid (19). vir gene induction was not inhibited if the strain contains either the single-copy constitutive virA or the multicopy constitutive virG locus (Fig. 2). The WT virG locus on the same multicopy plasmid also resulted in the vir genes not being inhibited. The higher level of vir gene induction in this strain can be explained by the multiple copies of virG (21, 22). We conclude that the inhibition by IAA involves the VirA/G system. The fact that a strain containing a single-copy constitutive virA locus is not inhibited suggests that the inhibition involves VirA. However, vir gene inhibition and growth inhibition in Agrobacterium can be disassociated from one another. Although the bacteria carrying the constitutive virA and virG loci were not inhibited by IAA in vir gene induction, they were still inhibited in their growth. Thus, the two types of inhibition have some, but not all, of the same requirements.
Fig. 2.
Effect of constitutive virA and constitutive virG loci on virB gene expression. Lanes: A, C58 with pSM243cd and pVirA (19); B, C58 with pSM243cd and pMutA (19); C, C58 with pSM243cd and pSY203 (20); D, C58 with pSM243cd and pSY204 (20).
The Competition Between IAA and AS.
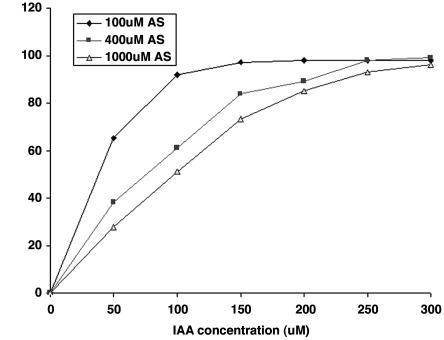
To test the possibility that IAA competitively inhibits the interaction of AS with VirA, we varied the concentration of AS from 100 to 1,000 μM and varied the concentration of IAA from 50 to 300 μM. Increasing the concentration of AS reduced the inhibitory effect at each of three concentrations of IAA. We conclude that IAA is interfering with the sensing of the phenolic compound by VirA (Fig. 3). To determine whether arabinose, which is required for maximum vir gene induction, is involved in the inhibition, we carried out the same experiment with the concentration of AS at 100 μM and the IAA concentration at 150 μM and increased the level of the sugar arabinose from 0.2% to 2.0%. No effect on the inhibition by IAA was observed, suggesting that the sugar plays no competitive role in the inhibition (data not shown).
Fig. 3.
Competition between IAA and AS.
Specificity of IAA Inhibition.
We next studied the specificity of the inhibition by IAA. We assayed the inhibition of vir gene induction by several compounds that had aromatic rings and/or acetate moieties. The compounds were tested at a concentration of 150 μM IAA, which maximally inhibited both vir gene induction and growth. They included naphthalene acetic acid (NAA), phenylacetic acid, indole, indole-3-pyruvic acid, tryptophan, and 2,4-dichlorophenoxyacetic acid. Inside the plant, IAA is often found in conjugated forms, which are not biologically active (23). Therefore, several conjugates including N-(indole-3-acetyl)-l-alanine and N-(indole-3-acetyl)-l-leucine were also tested. Of all these compounds, only NAA and phenylacetic acid showed any inhibition (Table 1). We assayed inhibition at various concentrations of NAA and phenylacetic acid to compare their IC50 with IAA. Neither compound was as effective an inhibitor as IAA, indicating that an acetate moiety and an aromatic ring may be necessary for the compound to inhibit vir gene induction (Table 1).
Table 1.
The inhibition of vir gene expression by various compounds
| Compounds | IC50 of vir gene inhibition |
|---|---|
| IAA | 32 |
| NAA | 135 |
| Phenylacetic acid | 140 |
| Indole | — |
| Indole-3-pyruvic acid | — |
| Trytophan | — |
| 2,4-dichlorophenoxyacetic acid | — |
| N-(indole-3-acetyl)-l-alanie | — |
| N-(indole-3-acetyl)-l-leucine | — |
—, no inhibition.
NAA and phenylacetic acid also inhibited growth at higher concentrations. The compounds that did not inhibit vir gene induction also did not inhibit growth (data not shown).
The Inhibition of the Growth of Other Plant-Associated Bacteria by IAA.
To determine whether Agrobacterium is unusual, the ability of IAA to inhibit the growth of other bacteria, both plant-associated and non-plant-associated, was assessed. Bacteria were inoculated into induction broth (pH 5.5) with and without 200 μM IAA. The OD at 600 nm was measured before and after incubation at 28°C with shaking. Because the growth rate of the different bacteria varied, the time points of the final measurements were taken after the control cells showed significant growth. All of the bacteria grew in the absence of IAA under the conditions of incubation. Eight of the 10 plant-associated bacteria tested were inhibited significantly in their growth by 200 μM IAA, a concentration that inhibited growth of Agrobacterium C58 (Table 2). At the same time, none of the eight non-plant-associated bacteria tested, which included both Gram-positive and Gram-negative bacteria, were inhibited. Although the number of organisms tested is relatively small, it does suggest a bias toward inhibition of plant-associated bacteria. Thus, this observation suggests an underlying genetic basis for this sensitivity in plant-associated bacteria. After treatment with IAA, the cells were plated on mannitol glutamate/Luria salts medium, which demonstrated that, as in the case with Agrobacterium, IAA inhibited the growth but did not kill the bacteria (data not shown). This observation suggests that, in addition to shutting down vir gene activation at an appropriate concentration, IAA can serve as a chemical agent in plant defense under the conditions that exist in the rhizosphere.
Table 2.
Growth inhibition of various plant-associated and nonplant-associated bacteria by IAA
| Strains | Growth inhibition by 200 uM IAA in induction medium (pH 5.5), % |
|---|---|
| Plant-associated bacteria | |
| Agrobacterium tumefaciens | 85 |
| Pseudomonas putida | — |
| Pseudomonas fluorescens | 54 |
| Xanthomonas campestris pv. Campestris* | 85 |
| Xanthomonas campestris pv. Vesicatoria* | 64 |
| Pseudomonas syringae | 90 |
| Sinorhizobium meliloti | 92 |
| Erwinia carotovora susp atroseptica ATCC 33260 | 82 |
| Acidovorax avenae subsp avenae† | 21 |
| Erwinia herbicola | — |
| Non-plant-associated bacteria | |
| Acidovorax temperans† | — |
| Bacillus subtilis | — |
| Enterococcus faecalis | — |
| Staphylococcus epidermidis | — |
| Pseudomonas aeruginosa | — |
| Enterobacter aerogenes | — |
| Serratia marcescens | — |
| Salmonella typhi | — |
—, no inhibition.
*Grown in induction medium for 3 days.
†Grown in induction medium for 2 days.
Discussion
As a general rule, bacteria conserve carbon and energy by synthesizing only those molecules that they require in any particular environment. In the case of Agrobacterium, once T-DNA becomes integrated and expressed in the plant genome, vir gene expression is no longer required. Second, the recognition of IAA by Agrobacterium as the signature molecule for the transformed cell makes good sense in that the other two potential indicators of transformation have some serious deficiencies. Cytokinin is synthesized as a product of the vir genes in many strains of Agrobacterium (24). Because its synthesis is under the control of VirA/G, it is synthesized before plant cell transformation in these strains. Strain C58, on the other hand, does not have genes for IAA synthesis as indicated by the annotation of its genome (25). The problem with the opines being signature molecules is that there are ≈20 different opines synthesized by different strains (2). Their structures vary enough that it is unlikely that the VirA protein, which is highly conserved, would be capable of sensing all of these various structures. Thus, IAA seems the logical choice for Agrobacterium to use as a signature molecule for plant cell transformation. The fact that IAA has the chemical structure to compete with the phenolic inducers for interaction with VirA also makes this molecule the one of choice.
IAA is not the only natural inhibitor of vir gene induction that competes with phenolic compounds for interaction with VirA. The benzoxazinone 2-hydroxyl-4,7-dimethoxy-benzoxazin-3-one was also shown to inhibit vir gene induction by competing with acetosyringone (13). Further, it seems likely that a related benzoxazinone, 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one, which also inhibits vir gene induction, acts by the same mechanism. Certainly competition with the phenolic inducer is probably the simplest way to shut down vir gene induction. However, the benzoxazinones appear to be strictly involved in defense against Agrobacterium infection in a limited number of plants: members of the family graminae. Members of this family are notoriously resistant to transformation by Agrobacterium, and this observation may help explain their resistance (13).
One question that we have not directly answered in this study relates to the concentration of IAA in the environment of the tumor to which Agrobacterium would be exposed. Freshly isolated tumor tissue consists primarily of untransformed cells, presumably because the transformed cells are exuding plant hormones into the environment, which support the growth of nontransformants (26). It is in this environment in which Agrobacterium resides and is exposed to IAA. Further, Fink and colleagues (18) reported that levels of IAA, which approximate those that inhibit vir gene induction in the present study, enhance filamentation and surface adhesion of cells of S. cerevisiae, early stages in the invasion of plant tissue by bona fide fungal plant pathogens. Presumably these concentrations of IAA are present on the plant surfaces on which the fungi alight. If anything, one might expect the levels of IAA to be higher in the environment of tumor cells overproducing and exuding IAA. We emphasize that all of our observations were made under conditions that mimic those found in the rhizosphere.
IAA appears to play several roles. Not only does it inhibit vir gene induction of Agrobacterium, but it also inhibits growth in Agrobacterium and a wide variety of plant-associated bacteria. Thus, at slightly higher concentrations than is required for inhibition of vir gene induction, growth is inhibited. Therefore, IAA seems to be a molecule that can serve in plant defense against a variety of bacteria. Whether this inhibition is a natural phenomenon that occurs in the rhizosphere of a tumor is not known. It is surprising, and perhaps meaningful, that this inhibition seems to extend primarily to plant-associated bacteria. What feature(s) is/are shared by these bacteria, which is not found in most of the non-plant-associated bacteria, is not clear.
In addition to its role in vir gene induction, IAA may serve as a signal to Agrobacterium that the plant environment is changing, from a pretumorous to a posttumorous state, and that the bacterium should modify its gene expression. This possibility should be explored through the use of microarrays under conditions in which IAA inhibits vir gene induction but not cell growth, as well as under conditions in which IAA inhibits cell growth.
These observations point out a fact that may be underappreciated. IAA does not inhibit growth at pH 7. However, the acidic conditions of the rhizosphere, which are necessary for vir gene activation, on one hand provide an environment that may put Agrobacterium in a vulnerable state with regard to inhibition by other molecules on the other hand. This finding emphasizes the need to measure biological activities under conditions as close to the natural environment as possible.
Materials and Methods
Strains and Growth Conditions.
The list of strains and plasmids are shown in Table 3. A. tumefaciens C58 was grown in either MG/L or AB minimal media (29) with arabinose as a carbon source at 28°C with shaking.
Table 3.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Characteristics | Ref. |
|---|---|---|
| Strains | ||
| C58 | A136 (pTiC58) (nopaline-type) | 27 |
| A348 | A136 (pTiA6NC) (octopine-type) | 28 |
| Plasmids | ||
| pSM243cd | virB::lacZ fusion | 15 |
| pSM102 | occQ::lacZ, IncP | 16 |
| pSY203 | Wild-type virG | 20 |
| pSY204 | Constitutive virG | 20 |
| pVirA | virA in pUCD2, pBR322ori, IncW | 19 |
| pMutA | virA(G665D) in pUCD2, pBR322ori, IncW | 19 |
vir Gene Expression Assays.
A. tumefaciens C58 cells with pSM243cd (virB::lacZ) (15) were grown overnight in AB minimal medium supplemented with kanamycin (100 μg/ml) and carbenicillin (100 μg/ml). Cells were washed with sterile water and inoculated into induction medium (pH 5.5) supplemented with 0.2% arabinose/100 μM AS at an initial OD600 of ≈0.1. The various compounds being tested for inhibition were added to the induction medium (29) at the indicated concentrations. The bacteria were incubated for 16 h and then assayed for β-gal activity by following the method of Miller (17). The concentrations of 50% inhibition (IC50) were determined from the dose–response analyses. Data presented represent the average of three separate experiments.
Growth Inhibition Assays.
Cells were grown in MG/L medium overnight at 28°C, washed twice with sterile water, and inoculated into induction medium (pH 5.5) with 0.2% arabinose and with or without 200 μM IAA. The OD at 600 nm was measured before and after 16 h of incubation, except for the indicated time of incubation. Data presented represent the average of three separate experiments.
Acknowledgments
We thank Alan Collmer (Cornell University, Ithaca, NY), Frank White (Kansas State University, Manhattan, KS), Dean Gabriel (University of Florida, Gainesville), Nic Pinel, and Mark Chandler (University of Washington) for providing plant-associated and non-plant-associated bacteria; and Robert Cleland, Mark-Eugene Duban, and Nic Pinel for valuable discussions and insights. This work was supported by National Institutes of Health Grant GM 32618 (to E.W.N).
Abbreviations
- AS
acetosyringone
- IAA
indole acetic acid
- NAA
naphthalene acetic acid
- T-DNA
transferred DNA.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Gelvin S. B. Microbiol. Mol. Biol. Rev. 2003;67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brencic A., Winans S. C. Microbiol. Mol. Biol. Rev. 2005;69:155–194. doi: 10.1128/MMBR.69.1.155-194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winans S. C. J. Bacteriol. 1990;172:2433–2438. doi: 10.1128/jb.172.5.2433-2438.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang C. H., Winans S. C. J. Bacteriol. 1992;174:7033–7039. doi: 10.1128/jb.174.21.7033-7039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cangelosi G. A., Ankenbauer R. G., Nester E. W. Proc. Natl. Acad. Sci. USA. 1990;87:6708–6712. doi: 10.1073/pnas.87.17.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao R., Lynn D. G. J. Bacteriol. 2005;187:2182–2189. doi: 10.1128/JB.187.6.2182-2189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y. W., Jin S., Sim W. S., Nester E. W. Proc. Natl. Acad. Sci. USA. 1995;92:12245–12249. doi: 10.1073/pnas.92.26.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin S., Roitsch T., Ankenbauer R. G., Gordon M. P., Nester E. W. J. Bacteriol. 1990;172:525–530. doi: 10.1128/jb.172.2.525-530.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin S. G., Prusti R. K., Roitsch T., Ankenbauer R. G., Nester E. W. J. Bacteriol. 1990;172:4945–4950. doi: 10.1128/jb.172.9.4945-4950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalogeraki V. S., Zhu J., Stryker J. L., Winans S. C. J. Bacteriol. 2000;182:1774–1778. doi: 10.1128/jb.182.6.1774-1778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dessaux Y., Petit A., Farrand S., Murphy P. J. Opine and Opine-like Molecules Involved in Plant/Rhizobiaceae Interactions. Dordrecht, The Netherlands: Kluwer; 1998. [Google Scholar]
- 12.Sahi S. V., Chilton M. D., Chilton W. S. Proc. Natl. Acad. Sci. USA. 1990;87:3879–3883. doi: 10.1073/pnas.87.10.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Boone L., Kocz R., Zhang C., Binns A. N., Lynn D. G. Chem. Biol. 2000;7:611–621. doi: 10.1016/s1074-5521(00)00007-7. [DOI] [PubMed] [Google Scholar]
- 14.Brencic A., Angert E. R., Winans S. C. Mol. Microbiol. 2005;57:1522–1531. doi: 10.1111/j.1365-2958.2005.04763.x. [DOI] [PubMed] [Google Scholar]
- 15.Stachel S. E., Nester E. W. EMBO J. 1986;5:1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stachel S. E., An G., Flores C., Nester E. W. EMBO J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J. H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 18.Prusty R., Grisafi P., Fink G. R. Proc. Natl. Acad. Sci. USA. 2004;101:4153–4157. doi: 10.1073/pnas.0400659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLean B. G., Greene E. A., Zambryski P. C. J. Biol. Chem. 1994;269:2645–2651. [PubMed] [Google Scholar]
- 20.Jin S., Song Y., Pan S. Q., Nester E. W. Mol. Microbiol. 1993;7:555–562. doi: 10.1111/j.1365-2958.1993.tb01146.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu C. N., Steck T. R., Habeck L. L., Meyer J. A., Gelvin S. B. Mol. Plant–Microbe Interact. 1993;6:144–156. [Google Scholar]
- 22.Rogowsky P. M., Close T. J., Chimera J. A., Shaw J. J., Kado C. I. J. Bacteriol. 1987;169:5101–5112. doi: 10.1128/jb.169.11.5101-5112.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kowalczyk M., Sandberg G. Plant. Physiol. 2001;127:1845–1853. [PMC free article] [PubMed] [Google Scholar]
- 24.Alt-Moerbe J., Neddermann P., Von Lintig J., Weiler E. W., Schroder J. Mol. Gen. Genet. 1988;213:1–8. [Google Scholar]
- 25.Wood D. W., Setubal J. C., Kaul R., Monks D. E., Kitajima J. P., Okura V. K., Zhou Y., Chen L., Wood G. E., Almeida N. F., Jr., et al. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 26.Binns A. N., Sciaky D., Wood H. N. Cell. 1982;31:605–612. doi: 10.1016/0092-8674(82)90316-6. [DOI] [PubMed] [Google Scholar]
- 27.Hamilton R. H., Fall M. Z. Experientia. 1971;27:229–230. doi: 10.1007/BF02145913. [DOI] [PubMed] [Google Scholar]
- 28.Garfinkel D. J., Simpson R. B., Ream L. W., White F. F., Gordon M. P., Nester E. W. Cell. 1981;27:143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- 29.Cangelosi G. A., Best E. A., Martinetti G., Nester E. W. Methods Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]