Abstract
Shewanella species are renowned for their respiratory versatility, including their ability to respire poorly soluble substrates by using enzymatic machinery that is localized to the outside of the cell. The ability to engage in “extracellular respiration” to date has focused primarily on respiration of minerals. Here, we identify two gene clusters in Shewanella oneidensis strain MR-1 that each contain homologs of genes required for metal reduction and genes that are predicted to encode dimethyl sulfoxide (DMSO) reductase subunits. Molecular and genetic analyses of these clusters indicate that one (SO1427–SO1432) is required for anaerobic respiration of DMSO. We show that DMSO respiration is an extracellular respiratory process through the analysis of mutants defective in type II secretion, which is required for transporting proteins to the outer membrane in Shewanella. Moreover, immunogold labeling of DMSO reductase subunits reveals that they reside on the outer leaflet of the outer membrane under anaerobic conditions. The extracellular localization of the DMSO reductase in S. oneidensis suggests these organisms may perceive DMSO in the environment as an insoluble compound.
Keywords: DMSO, metabolism, cold temperature adaptation
Abundance of niches has long been thought to be a critical limitation to species diversity within an ecosystem (1). Exploitation of unused or under-used niches is likely a driving force for evolution within all scopes of biology, particularly on the microbial scale. The incredible diversity of microbes from hydrothermal vent systems to subglacial Antarctic lake environments is a testament to metabolic innovation. Microbes are found essentially anywhere they can take advantage of chemical gradients to generate energy by using a diverse repertoire of biochemical tools.
Shewanella species have been isolated from many aquatic environments (2). The cosmopolitan nature of this species is likely because of their incredible respiratory versatility. Various Shewanella strains are reported to use ≈20 different terminal electron acceptors for respiration (3). One of these compounds, dimethyl sulfoxide (DMSO), is found in significant concentrations throughout aquatic environments, sometimes representing the most abundant methylated sulfur compound present (4). DMSO can be produced from the photochemical oxidation of dimethyl sulfide (DMS) (5), but the major source appears to be eukaryotic microplankton (6, 7). Another potential source is bacterial oxidation of DMS to DMSO (8), although it is unclear whether this process is significant in marine systems (9, 10). DMS can be produced both from the reduction of DMSO and from the enzymatic breakdown of dimethylsulfoniopropionate (DMSP) (11). DMSP, in turn, is synthesized by a variety of marine microplankton as an osmoregulator (12), cryoprotectant (13), and radical scavenger (14). Globally, the DMSP/DMS/DMSO cycle is important with respect to climate, because DMS is an antigreenhouse gas, directly impacting cloud formation where it is produced in significant quantities (15).
Despite this importance, very little is understood regarding how aquatic bacteria produce DMS through respiration of DMSO. Although commonly thought to be a soluble compound (16), DMSO can be associated with particulate material in marine systems (4, 7). Accessing this source of DMSO for respiration might be difficult using traditional mechanisms. The DMSO reductase enzyme in Escherichia coli (17) and Rhodobacter spp. (18) is periplasmically located. Here we demonstrate that the DMSO reductase in Shewanella oneidensis is localized to the outside of the cell. The localization of this enzyme suggests that DMSO acquisition may be difficult in the environments Shewanella inhabits, perhaps because of its physical inaccessibility or the challenge of transporting it. We discuss how utilization of this compound by Shewanella might influence the geochemical cycling of sulfur in aquatic systems.
Results
Identification of Extracellular Respiration Gene Clusters in S. oneidensis.
Bacteria like S. oneidensis have developed special metabolic strategies to use poorly soluble substrates in anaerobic respiration. For example, in respiring iron (Fe) and manganese (Mn) (hydr)oxides, S. oneidensis faces the challenge that these substrates are effectively insoluble at near-neutral pH conditions, meaning that the bacterium must deliver electrons to an extracellular oxidant. S. oneidensis solves this problem by using a system that can be described in three modules: an extracellular cell-associated terminal reductase, an integral outer membrane anchor, and a periplasmic electron carrier. Although several mechanistic questions regarding the details of this process remain, results from many laboratories are consistent with this model for Fe and Mn (hydr)oxide reduction in S. oneidensis (19). We define the process of directly transferring electrons to an acceptor located outside of the cell as extracellular respiration. This metabolic strategy is clearly useful for respiration of insoluble electron acceptors but could also be beneficial for respiration of compounds toxic to the organism.
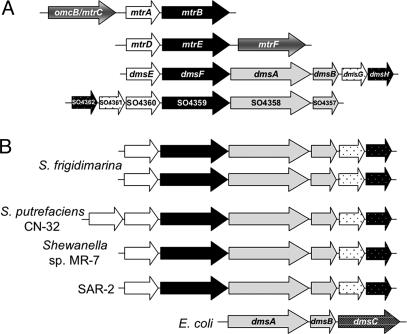
To further analyze the role of extracellular respiration in S. oneidensis, we began by analyzing its genome for homologs of proteins involved in this process. We identified four gene clusters that could encode for extracellular respiratory processes (Fig. 1A). The mtrDEF gene cluster is similar to the metal reduction gene cluster (mtrC/omcB, mtrAB); however, mutations constructed in this locus do not affect metal reduction (20). The other two gene clusters were similar to the mtr gene cluster in that they contain genes predicted to encode proteins with significant similarity to MtrA and MtrB (Fig. 1A; proteins encoded by SO1427 and SO4360 are 73% and 65% similar to MtrA, and SO1428 and SO4359 are 54% and 43% similar to MtrB). Located downstream of these homologs are genes predicted to encode DMSO reductase subunits (DmsA, SO1429 and SO4358; DmsB, SO1430 and SO4359), suggesting that reduction of DMSO in S. oneidensis is an extracellular respiratory process. Several additional sequenced Shewanella genomes contain similarly constructed gene clusters, implying a similar mechanism for DMSO respiration (Fig. 1B). Shewanella putrefaciens has two homologs of the periplasmic electron carrier module (i.e., MtrA). Both S. oneidensis and Shewanella frigidimarina contain two clusters that potentially encode DMSO respiratory proteins. Genes encoding homologs of DmsA or DmsB were not detected in the genomes of Shewanella amazonensis, Shewanella baltica, Shewanella denitrificans, or Shewanella sp. PV-4, and none of these strains are capable of growing on DMSO (data not shown). This correlation suggests that, if Shewanella species have the capacity to respire DMSO, the genes encoding their enzymatic machinery are organized in “extracellular” modules.
Fig. 1.
Alignments of potential extracellular respiratory gene clusters. Genes are color-coded for putative function in terms of extracellular respiratory modules (white, periplasmic electron carrier; black, outer membrane anchor; gray, extracellular cell-associated terminal reductase). (A) Alignment of gene clusters predicted to encode extracellular respiration pathways in S. oneidensis. (B) Alignment of gene clusters containing homologues of DMSO reductases from sequenced Shewanella sp. and comparison to dmsABC from E. coli. dms G and SO4361 and homologs are predicted to encode TorD-like molybdenum cofactor insertion chaperones, whereas dms H and SO4360 and homologs encode hypothetical proteins. All identified clusters contain genes encoding homologues of MtrA and MtrB. S. frigidimarina, S. putrefaciens, and Shewanella sp. MR-7 sequences were made available through the Integrated Microbial Genomes database at the Joint Genome Institute (http://img.jgi.doe.gov), and sequence for SAR-2 was from by Venter et al. (51).
Analysis of Genes Encoding DMSO Reductase in S. oneidensis.
Respiration of DMSO by bacteria has been studied predominantly in two model systems, E. coli and Rhodobacter. In E. coli, DMSO respiration is catalyzed by an enzyme complex, consisting of a molybdenum cofactor-containing subunit (DmsA) and an iron-sulfur cluster containing subunit (DmsB) (21, 22). This complex is anchored to the periplasmic side of the inner membrane by a membrane-spanning subunit (DmsC) (17). In Rhodobacter sphaeroides, a soluble periplasmic enzyme, lacking the iron-sulfur subunit, catalyzes the reduction of DMSO (18, 22). The genes found in S. oneidensis are similar to those used by E. coli for DMSO respiration (the proteins encoded by SO1429 and SO4358 are 59% and 52% similar to DmsA, and SO1430 and SO4357 are 67% and 72% similar to DmsB). Both homologs of DmsA in S. oneidensis (SO1429 and SO4358) contain twin arginine leader sequences at their N termini, suggesting export into the periplasm through the twin arginine translocation protein secretion pathway (23). No homolog of dmsC from E. coli is present either in the gene clusters encoding putative DMSO reductases or in the genome itself (24). Unlike the E. coli DmsA protein, SO1429 and SO4358 are predicted to be lipoproteins (www.tigr.org). The outer membrane metal reductase proteins (OmcA and OmcB/MtrC) have also been shown to be lipoproteins (25). This analysis is consistent with the hypothesis that respiration of DMSO occurs extracellularly, with the DMSO reductase being tethered to the outer membrane by a lipid modification and possibly associated with an MtrB-like outer membrane protein.
We recently identified a locus in S. oneidensis that is required for DMSO respiration (26). To further test the requirement for the SO1427–SO1432 gene cluster for DMSO respiration and to eliminate the involvement of the SO4357–SO4362 gene cluster, we monitored anaerobic expression of genes present in both clusters using quantitative RT-PCR and generated mutations in SO1427 and SO1430. Gene expression was greatly induced under anaerobic growth conditions for genes tested in the SO1427–SO1432 gene cluster (7- to 25-fold) compared with a slight induction for genes tested in the SO4357–SO4362 gene cluster (1.2- to 1.4-fold) (Fig. 2A). These results suggest that only the SO1427–SO1432 gene cluster is required for anaerobic DMSO respiration. To test this hypothesis, we analyzed three mutations: nonpolar in-frame deletions of SO1427 and SO1430, and a polar gene replacement of SO1427 with a gene conferring kanamycin resistance. All three mutant strains displayed a defect using DMSO anaerobically (Fig. 2B) but no defect using fumarate (Fig. 6, which is published as supporting information on the PNAS web site). As suggested in ref. 26, the SO1427–SO1432 gene cluster should be referred to dmsEFABGH. Reverse transcriptase PCR experiments verified that adjacent genes in this cluster are transcribed together (Fig. 7, which is published as supporting information on the PNAS web site). The severe defects of the dmsB (SO1430) deletion and the dmsE(SO1427)::Km gene replacement strains demonstrate that this gene cluster is required for DMSO respiration in S. oneidensis. The partial defect observed in the dmsE (SO1427) deletion strain indicates that this gene is required for maximal utilization of DMSO. Moreover, this strain showed ≈3.5-fold increase in DMSO reductase activity (with an artificial electron donor; see Methods) and ≈2.5- to 4.5-fold increase in transcription of dmsFAB when compared with wild type (Table 2, which is published as supporting information on the PNAS web site). The correlation of increased reductase activity with the increased expression of dmsFAB indicates that there is likely significantly more DMSO reductase (DmsA and DmsB) protein present in the dmsE deletion strain compared with wild type. We interpret the growth defect of this strain to be a consequence of disrupting electron flow from CymA (24) to the terminal DMSO reductase (see Discussion). Demonstrating that a predicted periplasmic c-type cytochrome (DmsE) is required for maximal DMSO reduction supports the hypothesis that this metabolism is an extracellular respiratory process in S. oneidensis.
Fig. 2.
Genes located in the SO1427–SO1432 gene cluster are anaerobically induced and specifically required for anaerobic growth by using DMSO. (A) Quantitative RT-PCR expression analysis of representative genes within gene clusters containing homologues of dmsAB in S. oneidensis. Expression of each gene was normalized to recA. (B) Anaerobic growth of dmsB (SO1430, ♦) and dmsE (SO1427, ▴) deletion strains were compared with a mutant containing a polar insertion in SO1427 (▾) and to wild-type MR-1 (■) using DMSO as the sole anaerobic electron acceptor. Error bars represent data range of duplicate cultures in all experiments.
DMSO Reductase Is Localized Extracellularly in S. oneidensis.
To test whether DMSO utilization is an extracellular respiratory process, we took two approaches. First, we tested anaerobic growth of mutants defective in type II secretion, which is the mechanism Gram-negative bacteria use to translocate proteins across the outer membrane from the periplasm (27). This system is required for proper localization of the metal reductase complex in S. putrefaciens strain 200 (28). All S. oneidensis strains defective in type II secretion tested were able to use fumarate (Fig. 8, which is published as supporting information on the PNAS web site); however, they were defective in respiring DMSO anaerobically (Fig. 3). This result suggests that extracellular localization of the terminal DMSO reductase is required for maximal utilization of DMSO.
Fig. 3.
Type II secretion is required for maximal DMSO reduction. Anaerobic growth of S. oneidensis strains with DMSO as the sole electron acceptor. Strains are MR-1 (■), JG91 (gspE, ▴), JG92 (gspJ, •), and JG94 (pilD, ▾). Error bars represent data range for duplicate cultures.
To directly test whether the DMSO reductase is localized to the outer membrane, we visualized the location of the DmsB subunit. DmsB was followed instead of DmsA because its localization should be DmsA dependent because only DmsA has both a twin arginine translocation signal sequence and a lipoprotein attachment motif. Therefore, if we were to find evidence for DmsB on the outside of the cell, DmsA should be localized there as well. We constructed a vector [pDMSB-hemagglutinin (HA)] to constitutively express a modified version of DmsB containing a 3x epitope tag from the hemagglutinin of the human influenza A virus (HA-tag) on the C terminus of the protein (DmsB-HA). Western blots using cell extracts verified that no endogenous S. oneidensis protein reacted with anti-HA antibodies and that similar amounts of DmsB-HA were detected under aerobic and anaerobic conditions (data not shown). This construct complements the defect in DMSO respiration of the dmsB deletion strain, although not to wild-type capacity, indicating that the HA-tag is interfering with some aspect (subunit association, localization, cofactor assembly, etc.) of DMSO reductase activity (Fig. 9, which is published as supporting information on the PNAS web site). In support of this idea, complementation of the dmsB deletion strain by a wild-type copy of dmsB was comparable with the wild type (data not shown). Normal growth profiles with fumarate and oxygen eliminated the possibility that pDMSB-HA simply caused a general growth defect (data not shown).
As discussed above, if the localization of DmsB-HA is dependent on DmsA, anaerobic conditions will be required for proper localization because expression of dmsA is anaerobically induced (Fig. 2A). To test this hypothesis and determine the localization of DmsB-HA, we used immunogold labeled anti-HA antibodies to probe both whole cells and cryo-ultrathin sections of bacteria, which were then visualized with transmission electron microscopy. In examining ≈1,000 aerobically grown cells of a dmsB deletion strain containing pDMSB-HA, significant labeling was never observed on intact cells (Fig. 4A). Conversely, when the same strain was grown anaerobically, the majority of cells (≈50%) were strongly labeled as shown in Fig. 4B. The remaining anaerobic cells showed either more or less labeling than Fig. 4B. These observations were verified by examining cryo-ultrathin sections of aerobic (Fig. 4C) and anaerobic (Fig. 4D) cells. The following two significant observations can be made: (i) cytoplasmic DmsB-HA content is similar between both aerobic and anaerobic samples (because of constitutive expression of dmsB-HA) and (ii) the cryo-ultrathin sections provided substantially less outer membrane surface area for the HA antibody to react with compared with whole cell labeling. No labeling was detected in strains lacking the pDMSB-HA construct under aerobic or anaerobic conditions (data not shown). These results confirm the localization of the DMSO reductase to the outer leaflet of the outer membrane, given that the antibody used is unlikely to cross the outer membrane of fixed cells.
Fig. 4.
Localization of DmsB-HA under aerobic and anaerobic conditions. Localization of DmsB-HA was detected by using immunogold labeled anti-HA antibody on whole cells grown aerobically (A) and anaerobically (B) and on cryo-ultrathin sections prepared from cells grown aerobically (C) or anaerobically (D). (Scale bar: 200 nm.) Anaerobically grown cells used DMSO as the terminal electron acceptor. Arrows point to examples of where DmsB-HA was located.
DMSO Reductase of S. oneidensis Is Protease Resistant.
In an attempt to further demonstrate the extracellular localization of the DMSO reductase in S. oneidensis, we treated whole cells with proteases. If the enzyme were indeed localized on the outside of the cell, the protease should decrease DMSO reductase activity when compared with untreated controls. To our surprise, we found that DMSO reductase activity in S. oneidensis was resistant to both proteinase K (Table 1) and pronase (data not shown). This resistance was exhibited both in whole cells and in cell lysates. As a control, fumarate reductase activity [a periplasmic enzyme in S. oneidensis (29)] was monitored with and without protease with both whole cells and lysates (Table 1). As expected, 40% of the fumarate reductase activity was eliminated in cell lysates after 1 h, yet DMSO reductase activity was unchanged (Table 1), even after 5 h of protease treatment (data not shown). DMSO reductase from E. coli lysates showed a 25% decrease in enzymatic activity after 1 h (Table 1), suggesting either the DMSO enzyme complex from S. oneidensis is protease resistant or is protected, possibly by the putative integral outer membrane protein DmsF. Type II secretion mutations did not influence protease degradation of the DMSO reductase, suggesting that the complex is inherently resistant (data not shown). In the environment, resistance to proteases would be a beneficial characteristic of an externally localized enzyme.
Table 1.
DMSO reductase from S. oneidensis is protease resistant
| Protease addition | DMSO reductase activity |
Fumarate reductase activity |
||
|---|---|---|---|---|
| − | + | − | + | |
| MR-1 cells | 1.21 ± 0.11 | 1.24 ± 0.02 | 9.29 ± 0.58 | 7.69 ± 1.85 |
| MR-1 lysate | 1.00 ± 0.18 | 1.06 ± 0.17 | 17.9 ± 0.47 | 10.5 ± 2.02 |
| E. coli lysate | 0.46 ± 0.02 | 0.35 ± 0.00 | 3.16 ± 0.08 | 0.22 ± 0.03 |
Reductase activity reported as ΔA600 per min per OD600. Average values and standard deviations were generated from at least duplicate samples. MG1655 was the strain of E. coli used.
Extracellular Respiration of DMSO.
It is logical to orient a reductase on the outer leaflet of the outer membrane when the oxidant is insoluble, as in the case of Fe and Mn (hydr)oxides. DMSO is generally thought to be a soluble organic molecule that is highly permeable to biological membranes (16), therefore, the periplasmic localization of the DMSO reductases in E. coli and Rhodobacter is not surprising. Why would S. oneidensis, and likely other Shewanella spp. (Fig. 1), localize their DMSO reductase to the outside of the cell? Insight into this question comes from considering where Shewanella typically grows in the environment. S. oneidensis was isolated from Oneida Lake in upstate New York (30), which completely freezes over during the winter months (31). Moreover, S. oneidensis has recently been shown to have the capacity to grow aerobically at low temperatures (32), similar to many other Shewanella species (2, 33, 34). We determined that S. oneidensis could grow anaerobically at 4°C with DMSO, showing only residual growth with no electron acceptor added (Fig. 10, which is published as supporting information on the PNAS web site). Although not directly testing the hypothesis that extracellular respiration of DMSO allows S. oneidensis to respire this compound at low temperatures, it does demonstrate the capacity of the organism to grow under these conditions. Given the prevalence of DMSO in marine ecosystems (4, 6), and that most of the marine environment exists at low temperatures, the ability of S. oneidensis to respire DMSO extracellularly may confer a selective advantage for growth in cold anoxic microenvironments, for reasons that will be discussed below.
Discussion
Based on multiple lines of evidence, we have demonstrated that DMSO utilization is an extracellular respiratory process in S. oneidensis. A model for this process is presented in Fig. 5. We propose that the path of electron flow from the menaquinone pool (MQ) to the membrane-bound c-type cytochrome CymA (24) continues through the putative periplasmic decaheme cytochrome DmsE. The integral outer membrane protein DmsF could facilitate electron transfer across the outer membrane by providing a channel to mediate direct interaction between the extracellular DMSO reductase and DmsE, similar to a model for metal reduction put forward by Beliaev et al.‖ Analysis of type II secretion mutants and localization studies of DmsB in S. oneidensis provide evidence that DMSO utilization is an extracellular respiratory process. It should be noted that type II secretion mutants were not completely defective in respiring DMSO, suggesting either an alternative mechanism for protein translocation to the outside of the cell, or that the reductase retains some capacity to function in the periplasm. The latter case would require the DMSO reductase to interact directly with DmsE (consistent with our model for electron transfer) or potentially with CymA itself. The growth defect caused by the dmsE deletion (Fig. 2B) clearly demonstrates a requirement for the periplasmic electron carrier component of the system. Additionally, this strain exhibits >3-fold more DMSO reductase activity over wild type (Table 2), likely masking a more severe growth defect.
Fig. 5.
Model for DMSO respiration in S. oneidensis. Arrows represent pathway of electron flow. See text for discussion.
There are several reasons why an organism might localize a terminal reductase on the outside of the cell. A simple explanation is that the product, the substrate, or both, are toxic to the organism. Localizing the reductase extracellularly would enable the cell to use the substrate with minimal toxic exposure. Another possible advantage may be to optimize substrate utilization. For example, algae and other eukaryotic microplankton are known to produce DMSO in aquatic environments (6, 7), and strains of Shewanella have been isolated from the surface of algae (35, 36). If positioning the DMSO reductase externally allows Shewanella to more efficiently capture DMSO, Shewanella might have a selective advantage for using this substrate over other organisms with an internal DMSO reductase.
Positioning a terminal reductase on the outer leaflet of the outer membrane is logical if the substrate is insoluble, as is the case for Mn and Fe (hydr)oxides. The chemical properties of DMSO suggest that it might be difficult to access under certain environmental conditions. First, at high concentrations (>80%) and at low temperatures (<15°C), the phase diagram of DMSO predicts that it will be a solid (37). It is possible, albeit unlikely, that sources produce DMSO in high enough local concentrations that it could exist in the solid state. A more likely scenario is that DMSO could exist in pseudoparticulate form by adsorbing to the silicate and calcite surfaces of marine diatoms and coccolithophorids, given its strong capacity to adsorb to positively charged surface (7, 16). Finally, cold temperatures may inhibit diffusion of DMSO into the cell. Precedent for this exists in eukaryotic cell-culture lines, where it has been shown that the permeability of DMSO across membranes strongly decreases with decreasing temperature (38, 39). If the same holds for bacterial membranes, extracellular localization of the DMSO reductase could allow Shewanella species to use DMSO under conditions where diffusion-dependent mechanisms would be inefficient.
The capacity to respire DMSO at low temperatures could enable Shewanella species to directly contribute to the production of DMS in aquatic environments. DMS production is a significant environmental concern because DMS is an antigreenhouse gas, directly impacting cloud formation and potentially influencing global climate (15). DMSO is predominant in aquatic systems in both soluble and particulate (algae-associated) phases (4, 7, 9, 10). Anaerobic respiration of DMSO by bacteria could be an important input into the marine DMS budget in oxic waters, given that anoxic microenvironments exist in marine snow aggregates (40, 41). It remains to be determined whether the innovative strategy used by S. oneidensis to respire DMSO in the laboratory extends to diverse bacteria in low-temperature aquatic environments.
Methods
Media and Growth Conditions.
Growth of S. oneidensis aerobically and anaerobically was as described in ref. 26 or as indicated.
Generation of Mutations in S. oneidensis.
Random transposon mutagenesis was performed as described in ref. 42. Type II secretion transposon mutants were isolated in a screen for mutants defective in Fe-mineral [amporphous iron oxy(hydr)oxide] reduction (J.A.G. and D.K.N., unpublished work). Genes disrupted by transposons were identified as described in ref. 42 and PCR verified. Directed mutagenesis was carried out as described in ref. 43. The construction of the SO1427::Km strain was described in ref. 26. Primer sequences used can be found in Table 3, which is published as supporting information on the PNAS web site.
Quantitative RT-PCR.
Quantitative RT-PCR analysis of RNA extracted from mid-logarithmic growing cells (OD600, 0.4–0.5) was as described in ref. 26. Primer pairs used for this experiment are provided in Table 3. Relative expression was standardized to both recA and envZ expression, yielding similar fold differences.
Methyl Viologen-Dependent Terminal Reductase Assays.
Oxygen-limited S. oneidensis cultures were grown in 10 ml of LB medium in capped 18 × 150 mm test tubes. The cultures were inoculated from colonies and were incubated aerobically at 250 rpm (Innova 4430 incubator-shaker; New Brunswick Scientific) at 30°C for 8–12 h to an OD600 of 3.0–4.5. E. coli cultures were grown to an OD600 of 0.5 in a Coy anaerobic chamber at 30°C with 30 mM glycerol, 25 mM DMSO, and 5 mM fumarate in LM medium (44) supplemented with 20% LB. S. oneidensis grown in this way (lactate instead of glycerol) gave similar levels of activity to oxygen-limited S. oneidensis cultures (data not shown).
Methyl viologen (MV)-dependent terminal reductase assays were set up in the anaerobic chamber in screw-capped quartz cuvettes. The assay contained 50 mM Tris·HCl (pH 7.5), 150 mM MV, 25 mM DMSO or 10 mM fumarate, and 100 mM dithionite in a final volume of 3 ml. The assays were started by adding 30 µl of sample, capped and mixed by inversion, then followed spectrophotometrically with absorbance readings taken every 10 s at 600 nm at room temperature outside the chamber. Cultures or cell lysates were used undiluted or diluted with the anaerobic Tris buffer to yield an assay that lasted for 150–300 s. Rates were calculated from the slope of the decrease in absorbance over time, normalized to cell density.
Protease Treatments.
Protease treatments were performed on whole cells washed once with anaerobic Tris buffer or with cell lysates prepared by centrifuging 500 µl of culture and resuspending the pellet in 250 µl of CelLytic Express solution (Sigma; prepared at 50 mg/ml in anaerobic Tris buffer) followed by incubation for 15 min at room temperature. Samples of 100 µl of washed cells or cell lysates were incubated with 10 µl of 10 mg/ml proteinase K or pronase (prepared in anaerobic Tris buffer) at room temperature in the anaerobic chamber for the desired time. A control sample contained 10 µl of the anaerobic Tris buffer instead of protease.
Construction of pDMSB-HA.
dmsB (SO1430) was PCR amplified from S. oneidensis genomic DNA by using the primers dmsB-EcoRI-N terminal (ATGAATTCATGACTCAACAAACACAATATG) and dmsB-BamHI-C terminal (ATGGATCCCACTTCTGCAGGGTTTAATAAT). The product was digested with EcoRI and BamHI, gel purified and ligated into a similarly digested plasmid derived from pBBR1MCS2 (45), pLARS (L. E. P. Dietrich and D.K.N., unpublished work), yielding pDMSB-HA. Expression of the insert is driven by the tac promoter from pAK20 (46), and the product is fused to a 3x HA epitope tag.
Preparation of Cells for Immunogold Labeling and Cryo-EM.
Strains were grown aerobically or anaerobically to an OD600 between 0.4 and 0.5. One-milliliter samples were spun down and washed once in 1× PBS, then resuspended in 1 ml of 1× PBS containing 0.5% gluteraldehyde and 4% paraformaldehyde. Anaerobically grown samples were prepared identically, except anoxic solutions were used throughout, and manipulations were carried out in an anaerobic glove box (Coy Laboratory Products, Ann Arbor, MI). After preparation, anoxic samples were kept in closed microfuge tubes until processed.
Cryo-Ultramicrotomy.
Cryofixation and cryo-ultramicrotomy were used to investigate the internal structure of the bacteria in their near-natural state. Immunogold labeling was used to ascertain the location of the DmsB-HA in transmission electron microscopy (TEM). Cryo-ultrathin sections of bacteria were obtained following a modified procedure of Tokuyasu (47) and Zierold (48). A small pellet of bacteria was dispersed in a solution of 2.3 M sucrose in PBS for 1 h at room temperature. Five milliliters of the solution was transferred onto a metal stub, rapidly frozen by plunging into liquid propane cooled by liquid nitrogen, and stored in liquid nitrogen before cryo-ultramicrotomy. The stub was mounted on the specimen holder of a Ultracut-E ultramicrotome (Reichert) equipped with an FCS cryosystem. Cryo-ultrathin sections (70- to 100-nm thick) were cut from the frozen pellet in a cryo chamber at −120°C with a dry glass knife. The sections were picked up with a gold loop filled with a 2.3 M sucrose solution, transferred onto 200-mesh Cu TEM grids, and kept on ice in a drop of PBS and 2.5% BSA/2% casein/0.5% ovalbumin (BCO).
Immunogold Labeling.
For immunogold labeling, the sections on the grids were immersed in a PBS solution containing 0.02 M glycine to block nonspecific labeling. Monoclonal anti-HA antibodies (primary) were purchased from Sigma-Aldrich. The sections were incubated with the primary antibody for 1 h (1:10 and 1:5 dilutions), washed six times in PBS, incubated with the secondary antibody conjugated with 10-nm gold particles, and washed six times with PBS. To increase contrast, the cryo-ultrathin sections were stained with a 2.5% solution of uranyl acetate for 2 min, rinsed with deionized water, and transferred to a drop of methylcellulose containing a 4% solution of uranyl acetate.
A similar procedure was applied for immunogold labeling of the whole bacteria. Approximately 10 µl of the original culture containing 0.5% gluteraldehyde and 4% paraformaldehyde was transferred to a formvar coated 300-mesh Cu TEM grid and washed three times with PBS. Bacteria adsorbed on the formvar film were exposed to 0.02 M glycine for 5 minutes to block nonspecific labeling. The grids were washed with PBS and exposed to the primary antibody for 5 minutes (1:1 dilutions), washed six times in PBS, exposed to the secondary antibody conjugated with 10-nm gold particles, and washed six times with PBS. The bacteria were not stained with uranyl acetate to enable observation of the gold particle on the whole surface of the bacteria in TEM.
All samples were imaged with TEM [FEI Tecnai (Eindhoven, The Netherlands) 12 at 120 kV and JEOL JEM-2011 at 200 kV]. For additional details of sample preparation, see Griffiths et al. (49, 50).
Supplementary Material
Acknowledgments
We thank the University of Southern California/Agouron Institute 2003 International Geobiology class for technical assistance in isolating type II secretion mutants and Drs. A. Komeili and L. Dietrich for helpful discussions and suggestions. We also thank J. Mui and Dr. S. K. Sears (both of the Facility for Electron Microscopy Research, McGill University) for assistance. This work was supported by grants from the Office of Naval Research, Henry Luce Foundation, Packard Foundation, and the Howard Hughes Medical Institute (to D.K.N.). H.V. acknowledges financial support from the Natural Sciences and Engineering Research Council (NSERC) of Canada.
Abbreviations
- DMS
dimethyl sulfide
- HA
hemagglutinin
- TEM
transmission electron microscopy
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Beliaev, A. S., Marshall, M. J., Kennedy, D. W., Dohnalkova, A., Saffarini, D. A., Culley, D. E., Reed, S. B., Gorby, Y. A., Romine, M. F., Shi, L., et al. The Joint International Symposia for Subsurface Microbiology and Environmental Biogeochemistry, 2005, Jackson Hole, WY, poster.
References
- 1.Begon M., Harper J. L., Townsend C. R. Ecology. Oxford: Blackwell Scientific; 1996. [Google Scholar]
- 2.Venkateswaran K., Moser D. P., Dollhopf M. E., Lies D. P., Saffarini D. A., MacGregor B. J., Ringelberg D. B., White D. C., Nishijima M., Sano H., et al. Int. J. Syst. Bacteriol. 1999;49:705–724. doi: 10.1099/00207713-49-2-705. [DOI] [PubMed] [Google Scholar]
- 3.Nealson K. H., Scott J. In: The Prokaryotes. Dworkin M., editor. Vol. 2004. New York: Springer; 2003. [Google Scholar]
- 4.Lee P. A., de Mora S. J. Journal of Phycol. 1999;35:8–18. [Google Scholar]
- 5.Brimblecombe P., Shooter D. Mar. Chem. 1986;19:343–353. [Google Scholar]
- 6.Andreae M. O. Limnol. Oceanogr. 1980;25:1054–1063. [Google Scholar]
- 7.Simo R., Hatton A. D., Malin G., Liss P. S. Mar. Ecol. Prog. Ser. 1998;167:291–296. [Google Scholar]
- 8.Hanlon S. P., Holt R. A., Moore G. R., McEwan A. G. Microbiology. 1994;140:1953–1958. [Google Scholar]
- 9.Simo R., Pedros-Alio C., Malin G., Grimalt J. O. Mar. Ecol. Prog. Ser. 2000;203:1–11. [Google Scholar]
- 10.Hatton A. D. Deep-Sea Res. 2002;49:3053–3065. [Google Scholar]
- 11.Turner S. M., Malin G., Liss P. S., Harbour D. S., Holligan P. M. Limnol. Oceanogr. 1988;33:364–375. [Google Scholar]
- 12.Vairavamurthy A., Andreae M. O., Iverson R. L. Limnol. Oceanogr. 1985;30:59–70. [Google Scholar]
- 13.Kirst G. O., Thiel C., Wolff H., Nothnagel J., Wanzek M., Ulmke R. Mar. Chem. 1991;35:381–388. [Google Scholar]
- 14.Sunda W., Kieber D. J., Kiene R. P., Huntsman S. Nature. 2002;418:317–320. doi: 10.1038/nature00851. [DOI] [PubMed] [Google Scholar]
- 15.Charlson R. J., Lovelock J. E., Andreae M. O., Warren S. G. Nature. 1987;326:655–661. [Google Scholar]
- 16.David N. A. Annu. Rev. Pharmacol. 1972;12:353–374. doi: 10.1146/annurev.pa.12.040172.002033. [DOI] [PubMed] [Google Scholar]
- 17.Stanley N. R., Sargent F., Buchanan G., Shi J., Stewart V., Palmer T., Berks B. C. Mol. Microbiol. 2002;43:1005–1021. doi: 10.1046/j.1365-2958.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- 18.McEwan A. G., Wetzstein H. G., Ferguson S. J., Jackson J. B. Biochim. Biophys. Acta. 1985;806:410–417. [Google Scholar]
- 19.Croal L. R., Gralnick J. A., Malasarn D., Newman D. K. Annu. Rev. Genet. 2004;38:175–202. doi: 10.1146/annurev.genet.38.072902.091138. [DOI] [PubMed] [Google Scholar]
- 20.Myers C. R., Myers J. M. Appl. Environ. Microbiol. 2002;68:5585–5594. doi: 10.1128/AEM.68.11.5585-5594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiner J. H., Macisaac D. P., Bishop R. E., Bilous P. T. J. Bacteriol. 1988;170:1505–1510. doi: 10.1128/jb.170.4.1505-1510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hille R., Retey J., Bartlewski-Hof U., Reichenbecher W., Schink B. FEMS Microbiol. Rev. 1998;22:489–501. doi: 10.1111/j.1574-6976.1998.tb00383.x. [DOI] [PubMed] [Google Scholar]
- 23.Bronstein P., Marrichi M., DeLisa M. P. Res. Microbiol. 2004;155:803–810. doi: 10.1016/j.resmic.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Schwalb C., Chapman S. K., Reid G. A. Biochemistry. 2003;42:9491–9497. doi: 10.1021/bi034456f. [DOI] [PubMed] [Google Scholar]
- 25.Myers C. R., Myers J. M. Lett. Appl. Microbiol. 2004;39:466–470. doi: 10.1111/j.1472-765X.2004.01611.x. [DOI] [PubMed] [Google Scholar]
- 26.Gralnick J. A., Brown C. T., Newman D. K. Mol. Microbiol. 2005;56:1347–1357. doi: 10.1111/j.1365-2958.2005.04628.x. [DOI] [PubMed] [Google Scholar]
- 27.Sandkvist M. Mol. Microbiol. 2001;40:271–283. doi: 10.1046/j.1365-2958.2001.02403.x. [DOI] [PubMed] [Google Scholar]
- 28.DiChristina T. J., Moore C. M., Haller C. A. J. Bacteriol. 2002;184:142–151. doi: 10.1128/JB.184.1.142-151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris C. J., Black A. C., Pealing S. L., Manson F. D., Chapman S. K., Reid G. A., Gibson D. M., Ward F. B. Biochem. J. 1994;302:587–593. doi: 10.1042/bj3020587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers C. R., Nealson K. H. Science. 1988;240:1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- 31.Mills E. L., Holeck K. T. Clearwaters. 2001;31:22–25. [Google Scholar]
- 32.Abboud R., Popa R., Souza-Egipsy V., Giometti C. S., Tollaksen S., Mosher J. J., Findlay R. H., Nealson K. H. Appl. Environ. Microbiol. 2005;71:811–816. doi: 10.1128/AEM.71.2.811-816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowman J. P., McCammon S. A., Nichols D. S., Skerratt J. H., Rea S. M., Nichols P. D., McMeekin T. A. Int. J. Syst. Bacteriol. 1997;47:1040–1047. doi: 10.1099/00207713-47-4-1040. [DOI] [PubMed] [Google Scholar]
- 34.Stapleton R. D., Sabree Z. L., Palumbo A. V., Moyer C. L., Devol A. H., Roh Y., Zhou J. Z. Aquat. Microb. Ecol. 2005;38:81–91. [Google Scholar]
- 35.Simidu U., Kita-Tsukamoto K., Yasumoto T., Yotsu M. Int. J. Syst. Bacteriol. 1990;40:331–336. doi: 10.1099/00207713-40-4-331. [DOI] [PubMed] [Google Scholar]
- 36.Patel P., Callow M. E., Joint I., Callow J. A. Environ. Microbiol. 2003;5:338–349. doi: 10.1046/j.1462-2920.2003.00407.x. [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen D. H., Mackenzie A. P. Nature. 1968;220:1315–1317. doi: 10.1038/2201315a0. [DOI] [PubMed] [Google Scholar]
- 38.Liu J., Zieger M. A., Lakey J. R., Woods E., Critser J. K. Transplant. Proc. 1997;29:1987. doi: 10.1016/s0041-1345(97)00198-x. [DOI] [PubMed] [Google Scholar]
- 39.Paynter S. J., Cooper A., Gregory L., Fuller B. J., Shaw R. W. Hum. Reprod. 1999;14:2338–2342. doi: 10.1093/humrep/14.9.2338. [DOI] [PubMed] [Google Scholar]
- 40.Ploug H. Limnol. Oceanogr. 2001;46:1624–1631. [Google Scholar]
- 41.Kuypers M. M. M., Lavik G., Woebken D., Schmid M., Fuchs B. M., Amann R., Jorgensen B. B., Jetten M. S. M. Proc. Natl. Acad. Sci. USA. 2005;102:6478–6483. doi: 10.1073/pnas.0502088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shyu J. B., Lies D. P., Newman D. K. J. Bacteriol. 2002;184:1806–1810. doi: 10.1128/JB.184.6.1806-1810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saltikov C. W., Newman D. K. Proc. Natl. Acad. Sci. USA. 2003;100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myers C. R., Myers J. M. J. Appl. Bacteriol. 1994;76:253–258. doi: 10.1111/j.1365-2672.1994.tb01624.x. [DOI] [PubMed] [Google Scholar]
- 45.Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., II, Peterson K. M. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 46.Komeili A., Vali H., Beveridge T. J., Newman D. K. Proc. Natl. Acad. Sci. USA. 2004;101:3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokuyasu K. T. Histochem. J. 1980;12:381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- 48.Zierold K. Scan. Electron Microsc. 1986;2:713–724. [PubMed] [Google Scholar]
- 49.Griffiths G., Brands R., Burke B., Louvard D., Warren G. J. Cell Biol. 1982;95:781–792. doi: 10.1083/jcb.95.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griffiths G., Simons K., Warren G., Tokuyasu K. T. Methods Enzymol. 1983;96:466–485. doi: 10.1016/s0076-6879(83)96041-x. [DOI] [PubMed] [Google Scholar]
- 51.Venter J. C., Remington K., Heidelberg J. F., Halpern A. L., Rusch D., Eisen J. A., Wu D. Y., Paulsen I., Nelson K. E., Nelson W., et al. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.