Abstract
Mitochondrial transformation of Chlamydomonas reinhardtii has been optimized by using a particle-gun device and cloned mitochondrial DNA or PCR fragments. A respiratory-deficient strain lacking a 1.2-kb mitochondrial DNA region including the left telomere and part of the cob gene could be rescued as well as a double-frameshift mutant in the mitochondrial cox1 and nd1 genes. High transformation efficiency has been achieved (100–250 transformants per microgram of DNA), the best results being obtained with linearized plasmid DNA. Molecular analysis of the transformants suggests that the right telomere sequence can be copied to reconstruct the left telomere by recombination. In addition, both nondeleterious and deleterious mutations could be introduced. Myxothiazol-resistant transformants have been created by introducing a nucleotide substitution into the cob gene. Similarly, an in-frame deletion of 23 codons has been created in the nd4 mitochondrial gene of both the deleted and frameshift recipient strains. These 23 codons are believed to encode the first transmembrane segment of the ND4 protein. This Δnd4 mutation causes a misassembly of complex I, with the accumulation of a subcomplex that is 250-kDa smaller than the wild-type complex I. The availability of efficient mitochondrial transformation in Chlamydomonas provides an invaluable tool for the study of mitochondrial biogenesis and, more specifically, for site-directed mutagenesis of mitochondrially encoded subunits of complex I, of special interest because the yeast Saccharomyces cerevisiae, whose mitochondrial genome can be manipulated virtually at will, is lacking complex I.
Keywords: green alga, mitochondrial DNA mutagenesis, telomere, complex I assembly, respiratory-deficient mutant
Chlamydomonas reinhardtii is a single-cell eukaryotic alga that is amenable to genetic manipulation and, thus, has been extensively used for studies on photosynthesis, respiration, and cross-talk between the nucleus and chloroplast or mitochondria (1). Both the nucleus and the chloroplast compartments can be efficiently transformed (2, 3), and markers have been developed to facilitate the selection of transformants (4, 5). In contrast, modification or replacement of Chlamydomonas mitochondrial genes has not been routinely performed to date. So far, the only organism whose mitochondrial genome can be manipulated virtually at will is the yeast Saccharomyces cerevisiae (6–8). Mitochondrial transformation of other organisms is a current challenge that stimulates extensive research, especially concerning mammalian mitochondrial genomes (9).
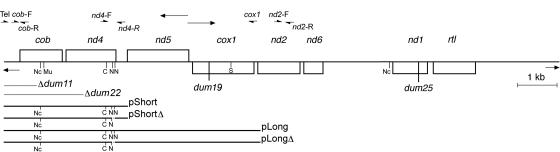
The mitochondrial genome of C. reinhardtii is a 15.8-kb linear molecule containing, at each extremity, telomeres corresponding to inverted repeats of ≈500 bp, with 40-bp single-stranded extensions (10). This genome has been totally sequenced and encodes only eight proteins (Fig. 1). Several mutations altering mitochondrial genes encoding apocytochrome b (cob gene) and subunit 1 of cytochrome c oxidase (cox1 gene) have been characterized (11). Phenotypically, these mutants have lost the capacity to grow under heterotrophic conditions, i.e., in the dark, with acetate as carbon source. In contrast, mutants altered in nd mitochondrial genes, which encode subunits of NADH/ubiquinone oxidoreductase (complex I), are able to grow in the dark, but considerably more slowly than the wild-type strain (12, 13). Most of the mutations located in the cob gene are deletions covering not only the cob gene but also the left telomere, whereas mutations in other genes are usually frameshifts consisting of the deletion or addition of one thymine (14).
Fig. 1.
Partial physical map of the 15.8-kb mitochondrial genome of C. reinhardtii. The rectangles represent protein-coding genes: cob, gene encoding apocytochrome b of complex III; nd1, 2, 4, 5, and 6, genes encoding the corresponding subunits of complex I; cox1, gene encoding the subunit 1 of complex IV, rtl: reverse transcriptase-like protein. The inverted telomeric ends are represented by short arrows, the bidirectional origin of transcription between nd5 and cox1 by longer arrows. Only restriction sites used in this work are presented: Nc, NcoI; C, ClaI; N, NdeI; S, SstI. Mu, resistance to myxothiazol. Positions of the dum11 and dum22 deletions and of the dum19 and dum25 point mutations are indicated. Fragments of mitochondrial genome contained in pShort, pShortΔ, pLong, and pLongΔ are shown as well as the primers used for PCR amplifications. Using the GenBank u03843 numbering for the Chlamydomonas mitochondrial DNA, primers positions are Tel, 1–21; cob-F, 431–450; cob-R, 564–545; nd4-F, 2765–2780; nd4-R, 3301–3282; cox1, 6653–6634; nd2-F, 6636–6655; and nd2-R, 7343–7323.
Only two reports of transformation of the mitochondrial genome of Chlamydomonas have been published so far. More than 10 years ago, a mitochondrial mutant deleted for the left telomere and apocytochrome b gene (1.5-kb deletion) was successfully transformed to respiratory competence with partially purified mitochondrial DNA from C. reinhardtii or Chlamydomonas smithii by using a particle-gun device (15). Very recently, the particle gun was also used to transform the same kind of deletion mutant with cloned mitochondrial DNA or PCR constructs (16). In both cases, a wild-type mitochondrial genome replaced the deleted genome after selection of transformants under heterotrophic conditions. Transformation efficiency was low (0.4–5 transformants per microgram of DNA) and did not lead to any genetic manipulation of the mitochondrial genome. Moreover, transformation of point mutants failed, and it was concluded that only mitochondrial genomes deleted for their left end could be transformed (16).
We present here an optimized biolistic transformation procedure of Chlamydomonas mitochondria using cloned mitochondrial DNA or PCR fragments. In contrast to previous reports, both a deletion and a frameshift mutant could be rescued, and high transformation efficiency has been achieved (100–250 transformants per microgram of DNA). Two mutations have been introduced: a nucleotide substitution in the cob gene, conferring resistance to myxothiazol, and an internal deletion in nd4. This deletion probably eliminates the region encoding the first transmembrane segment of the ND4 protein and leads to a misassembly of complex I. These results open the way to reverse genetics with the Chlamydomonas mitochondrial genome and, more specifically, to site-directed mutagenesis of mitochondrially encoded subunits of NADH/ubiquinone oxidoreductase.
Results
Mitochondria of the dum11 Mutant, Lacking the Telomere and cob Gene, Can Be Transformed with High Efficiency.
To set up the mitochondrial transformation technique, we choose as recipient the mutant dum11, which we found to be completely devoid of the left telomere by PCR experiments and Southern blot analysis (data not shown and Fig. 2B). The transforming DNA was either plasmid or PCR DNA carrying the telomere and cob region deleted in the mutant and extending up to the nd4 gene (pShort), or up to the cox1 gene (pLong and PCR fragments) as described in Materials and Methods and shown in Fig. 1. Cells were incubated for one night in the light (4,000 lux) before being transferred to the dark. Microcolonies were first detected with a stereoscopic microscope after 4–6 weeks of incubation in the dark. After 2 months of growth in the dark, plates were transferred to dim light (260 lux) for 1 week. Control plates without bombardment did not present any growth, whereas plates bombarded with DNA exhibited colonies in the periphery of the plate, probably because cells in the center were killed by the impact of the beads.
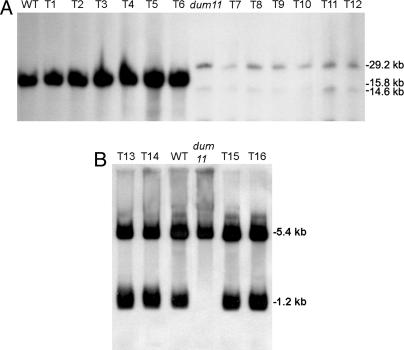
Fig. 2.
Southern blot analysis of mitochondrial transformants using dum11 as recipient strain. (A) TTC+ (T1 to T6) and TTC− (T7 to T12) transformants isolated after transformation by using pShort (T1, T2, T3, T7, T8, and T9) or a Tel/cox1 PCR fragment (T4, T5, T6, T10, T11, and T12). Total undigested DNA was probed with a nd2 PCR fragment amplified with primers nd2-F/R (Fig. 1). Wild type (WT) and dum11 mutant are shown as controls. Equal amounts (10 μg) of each total DNA preparation were loaded per lane. As shown, dum11 and the TTC− transformants repeatedly gave lower signal intensities, reflecting the fact that deletion mutant strains contain less mitochondrial DNA (17). (B) TTC+ transformants (T13 to T16) isolated after transformation by using a cob-F/cox1 PCR fragment (T13, T14) or a Tel/cox1 PCR fragment (T15, T16). Total DNA digested with NcoI was hybridized with a probe covering the telomeric region (primers Tel/cob-R). The WT and the dum11 mutant are shown as controls. In this case, the dum11 lane has been overloaded to ensure the correct detection of the fragments.
Various parameters of transformation were tested by using the dum11 deletion mutant as recipient strain (see below and Materials and Methods). For the same mitochondrial insert, colonies were obtained with both linearized and supercoiled plasmids as well as PCR fragments. Linearized plasmids and PCR fragments were ≈5 and 3 times more efficient than supercoiled plasmids, respectively. Typically, 300–660 transformants were obtained per plate for 3 μg of linearized plasmid (pLongΔ) and 200–400 transformants when 1.5 μg of Tel/cox1 PCR fragment were used.
Restoration of the Cytochrome Pathway in dum11 Mitochondrial Transformants Correlates with Reconstitution of the Telomere and cob Gene.
To check whether the cytochrome pathway of respiration was restored in the transformants, an in vivo test was performed on colonies by using 2,3,5-triphenyltetrazolium chloride (TTC). Generally, 95–100% of the transformants were found TTC+ and should thus possess a functional cob gene. TTC+ clones grew like the wild type in the dark (data not shown). Thirty TTC+ clones from a transformation experiment using pShort and thirty TTC+ clones from a transformation with a Tel-cox1 PCR fragment were subcloned and analyzed by PCR. All clones contained the cob gene and the left telomere end (data not shown). Undigested DNA of 20 of these TTC+ clones was analyzed by Southern blotting using as a probe an nd2 fragment. A unique signal at 15.8 kb was detected in each TTC+ transformant as well as in the wild type and corresponded to the wild-type mitochondrial genome (Fig. 2A, clones T1 to T6). In addition, the mitochondrial genome of the TTC+ transformants was linear like in the wild type, because digestion with an enzyme cutting once in the genome (SstI) gave two fragments (data not shown). To the contrary, the mitochondrial genome of six TTC− clones analyzed in parallel (Fig. 2A, T7 to T12) presented the same complex pattern as the deleted dum11 recipient strain: monomers with the deletion of 1.2 kb and dimers arising from head-to-head fusions between monomers (17). Hybridization signals were weak compared with wild type and TTC+ transformants, as already observed in deletion mutants (17), suggesting that the mitochondrial DNA is present in reduced amount in these strains. These TTC− clones had probably initially been transformed to be able to grow in the dark but might have remained heteroplasmic, allowing the recombinant mitochondrial DNA copies to be diluted and eventually lost when the selection pressure was removed by transferring plates into the light. We found that the proportion of these TTC− clones could reach up to 20% if the plates were kept in the dark for shorter periods of time, e.g., 5–6 weeks instead of 8 weeks.
Total DNA of the TTC+ clones was then digested by using restriction enzymes (NcoI and ClaI, Fig. 1) that cut in the left part of the genome, and Southern blot analysis was performed by using a probe that covered cob. TTC+ clones presented the same restriction profile as the wild-type strain (data not shown). To confirm this observation, the nd4-cob-telomere region of one clone issued from transformation with pShort was sequenced and found to be identical to wild type and donor plasmid DNA. As proposed in refs. 15 and 16, these results are consistent with the interpretation that defective left ends of the recipient mitochondrial genomes were replaced by the corresponding wild-type sequences from the donor DNA.
The Telomeric Region Enhances the Rate of Transformation of the dum11 cob Deletion Mutant but Is Not Essential.
To analyze the role of the telomeric region in the recombination process, transformation experiments of dum11 were also performed by using two different PCR fragments, carrying either the entire 500-bp left telomere (Tel/cox1 primers) or only 28 bp (cob-F/cox1 primers).
We found that the presence of the entire left telomere in the donor DNA is essential to reach a high level of transformation (Table 1). However, a few colonies were reproducibly recovered after transformation with the cob-F/cox1 PCR product. These clones were TTC+ and grew like wild type in the dark. Two of them (T13 and T14) were analyzed by Southern blotting, using as control two TTC+ transformants coming from a transformation experiment with the Tel/cox1 fragment (T15 and T16), the wild type, and the dum11 deletion mutant. After restriction of total DNA by NcoI and hybridization with a probe covering the telomeric region, a fragment of 5.2 kb was detected in all strains and corresponded to the NcoI fragment of the right telomere. Contrary to dum11 and like the wild-type strain, the four transformants displayed the 1.2-kb fragment corresponding to the left extremity of the genome (Fig. 2B).
Table 1.
Numbers of TTC+ transformants obtained after two independent shootings of dum11 with PCR fragments containing the entire left telomere (primers Tel/cox1) or a very reduced portion of it (cob-F/cox1)
| Plate/experiment | Transforming PCR fragment |
|||||
|---|---|---|---|---|---|---|
| Tel/cox1 |
cob-F/cox1 |
|||||
| Plate no. | 1 | 2 | 3 | 1 | 2 | 3 |
| No. of transformants (experiment 1) | 226 | 172 | 180 | 16 | 10 | 2 |
| No. of transformants (experiment 2) | 372 | 254 | 317 | 5 | 8 | 0 |
Thus, although the presence of the telomeric sequence in the transforming DNA enhances the transformation rate, our data suggest that the information contained in the right telomere can be copied to reconstruct the left telomere.
A Myxothiazol-Resistance Mutation Can Be Stably Introduced by Transformation of the dum11 cob Deletion Mutant.
To test whether the transformation technique could be used to modify or replace mitochondrial genes in Chlamydomonas, we first decided to introduce a nondeleterious mutation into the cob gene. We choose a point mutation conferring resistance to mucidine and myxothiazol (18) located ≈60-bp upstream of the putative end of the deletion in the recipient dum11 strain (Fig. 1).
We used the DNA from a myxothiazol-resistant strain as a template to amplify the Tel-cox1 PCR product and bombard the dum11 recipient. Transformants were selected by growth in the dark and screened by TTC staining. Among 78 TTC+ clones picked up from two independent transformation experiments, 69 clones were resistant to myxothiazol in the dark, whereas the other 9 were sensitive like the wild type. The apocytochrome b gene of one sensitive and four resistant clones was sequenced. The four resistant clones showed the substitution responsible for myxothiazol resistance, whereas the sensitive clone exhibited the wild-type sequence. In this latter case, DNA exchange had thus presumably occurred in the 60 bp downstream of the Mu-resistance mutation (Fig. 1).
Together, these data show that a nondeleterious mutation can be efficiently integrated into mitochondrial DNA, without direct selection.
Complex I Mutants Bearing a 69-bp Deletion in nd4 Can Be Isolated After Transformation of the dum11 cob Deletion Mutant.
Because a myxothiazol-resistance mutation could be successfully introduced in the cob gene, we tried to integrate a deleterious mutation in a complex I gene. In contrast to a cob mutant, such as dum11, complex I mutants can grow in the dark but much more slowly than the wild type (12). Thus, we used the dum11 mutant as recipient to shoot the plasmid pLongΔ, which misses codons 2–24 of the nd4 gene (Fig. 1).
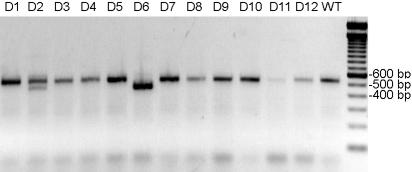
Plates were incubated for 8 weeks in the dark, as usual. After a week in dim light, Chlamydomonas TTC+ colonies of medium to small size were picked up and tested by PCR using primers nd4-F/nd4-R that encompass the deletion (Fig. 1). In two independent experiments, we obtained three clones bearing the deletion among 90 tested. The PCR amplification showed that two clones were homoplasmic (e.g., Fig. 3, clone D6), whereas the last one was heteroplasmic (e.g., Fig. 3, clone D2) but could be purified to homogeneity by subcloning. These three clones grew slowly in the dark like complex I mutants, whereas the others grew like wild type. To recover Δnd4 clones more efficiently, we found, in subsequent experiments, that it was beneficial to first confirm the slow growth in the dark before characterizing the clones by PCR.
Fig. 3.
Detection of the Δnd4 mutation by PCR amplification. PCR on 12 transformants (D1 to D12) was performed by using primers nd4-F and nd4-R. D2 and D6 present the 69-bp NdeI deletion in nd4 at the heteroplasmic (D2) and homoplasmic (D6) state. The 100-bp ladder (Eurogentec, Liège, Belgium) is shown at the right.
We also used pShortΔ for transformation. pShortΔ bears the same deletion on a shorter mitochondrial insert (Fig. 1). Of 31 clones analyzed, 1 was found to be heteroplasmic for the deletion and could not be purified by subcloning, probably because the Δnd4-deleted genome was present in too low an amount in this clone (data not shown). This plasmid was not used in further experiments.
The nd4 Deletion Can also Be Introduced in a cox1 nd1 Double Mutant.
Because complex I is composed of several dozens of subunits, it is interesting to study the genetic interactions among them. To set up a strategy allowing us to create a double mutant in different mitochondrial nd genes, we took advantage of a double mutant (dum19 dum25) harboring a deletion of one T in cox1 and an in-frame deletion of two codons in nd1. Using pLongΔ, we tested whether Δnd4-Δnd1 mutants could be isolated, while restoring the wild-type cox1 and, thus, TTC staining at the same time because of a recombination event upstream of the frameshift mutation in cox1.
Fifty TTC+ clones were analyzed for their growth in the dark. As expected, they all grew slowly, because the nd1 mutation should be present in all of them. PCR with primers nd4-F/nd4-R that encompass the deletion showed that one clone (D34) carried the Δnd4 deletion. In addition, sequencing results showed that the wild-type cox1 gene sequence was restored, whereas the nd1 mutation was still present in this transformant.
Thus, this experiment shows that a mutant affected in two different nd genes can be created and that a strain still containing the left telomeric region can be used as a recipient for transformation.
The Δnd4 Transformants Assemble a Subcomplex Instead of the Full-Size Complex I.
The Δnd4 (D6) and Δnd4-Δnd1 (D34) mutants were analyzed molecularly. Southern blot analysis revealed that, like the wild-type strain, they contain a linear mitochondrial genome (data not shown).
In Chlamydomonas, the origin of transcription of the mitochondrial genome is bidirectional and would be located between nd5 and cox1 (Fig. 1). Long cotranscripts are produced, and mature transcripts are generated by precise endonucleolytic cleavage, implying secondary structures made by direct and inverted repeats (19). Because the nd4 deletion is close to the intergenic region between nd4 and nd5, we investigated its possible impact on the amount of nd4 or nd5 transcripts. Northern blot analysis of the D6 and D34 strains using a nd4 and a nd5 probe did not reveal any difference in either mutant (data not shown).
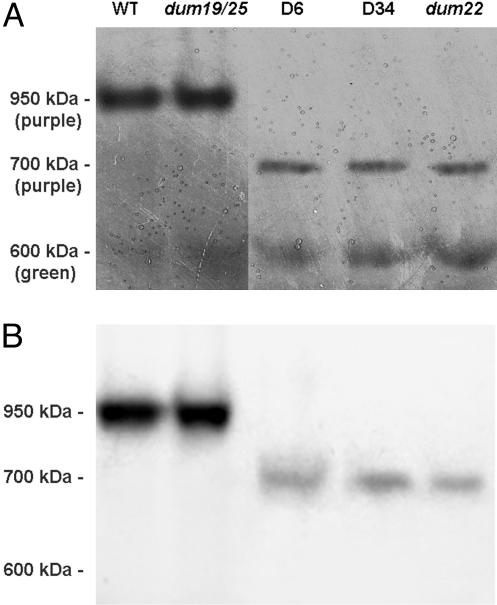
The rotenone-sensitive NADH/duroquinone activity of complex I was measured in membrane fractions and was shown to be null in both mutants. Assembly of complex I was then investigated by Blue Native (BN)/PAGE. Complex I was identified by an NADH/NBT (nitroblue tetrazolium) staining reaction involving the NADH dehydrogenase activity associated with the peripheral arm of the enzyme (Fig. 4A). A purple-stained band corresponding to whole complex I was found at 950 kDa for the wild type and the dum19 dum25 mutant. It has been shown that the in-frame deletion in nd1 present in the dum19 dum25 mutant does not affect the assembly and the NADH dehydrogenase activity of complex I (13). In contrast, Δnd4 (D6) and Δnd4-Δnd1 (D34) transformants contained low amounts of a subcomplex that was stained in purple and, thus, had retained NADH dehydrogenase activity. This subcomplex migrated ≈700 kDa, i.e., was ≈250-kDa smaller than the wild-type complex I (Fig. 4A). Another complex appearing green on the BN gel was also seen at 600 kDa: This complex was not stained by the NADH/NBT reaction and could correspond to Photosystem I associated to light-harvesting complex (20). These results were confirmed by Western blotting using a polyclonal antibody directed against the whole complex I from Neurospora crassa (Fig. 4B). A similar subcomplex was also observed in the dum22 mutant carrying a large deletion of the left part of the genome, including cob and 300 bp of the 3′ end of nd4 (13).
Fig. 4.
Analysis of mitochondrial NADH dehydrogenases from Δnd4 transformants after BN/PAGE. (A) Membrane extracts (120 μg of protein) of wild type, dum19 dum25, dum22, and two transformants. D6 carries the Δnd4 mutation, and D34 harbors both the Δnd4 and dum25 mutations. The NADH/NBT staining revealed purple bands corresponding to complexes at 950 and 700 kDa. The green 600-kDa complex, which is not detected by the staining, is shown as a loading control and probably corresponds to Photosystem I associated to light-harvesting complex (20). (B) The gel was blotted and probed with an antiserum against the N. crassa whole complex I.
Discussion
The ability to genetically transform an organism by introducing DNA into its nuclear and/or organellar genome greatly enhances its utility as a model system for experimental research. In this study, we describe an efficient method to transform Chlamydomonas mitochondria. In contrast to reports in refs. 15 and 16, both point mutations and deletions >1 kb could be rescued, and several hundreds of colonies were recovered in transformation assays, allowing a detailed phenotypical and molecular analysis of transformants. Chlamydomonas represents, therefore, the first photosynthetic organism whose mitochondrial genome can be easily manipulated.
An interesting molecular feature of the recombination after the transformation is the fact that a strain lacking both the cob gene and the left telomere could be rescued, even when the transforming DNA is nearly completely devoid of the left telomere, suggesting that this end could be reconstructed through a recombination event involving the very short portion of the left telomere (28 bp) present on the transforming DNA and the right telomere of the endogenous mitochondrial DNA.
Whatever the construct or the recipient strain used, the selection process was generally extremely slow, and, even after 2 months in the dark, heteroplasmic clones could still be found. Because microcolonies transferred from darkness to light contain only a small number of cells (200 at most), the number of divisions was probably not always sufficient to allow the complete segregation of recombinant and recipient mitochondrial genomes. To speed up the selection process, we made several attempts to develop a cotransformation strategy allowing a primary selection in the light by bombarding the recipient strain simultaneously with two different plasmids, one carrying a nuclear marker and the other containing the mitochondrial DNA of interest (data not shown). Nuclear transformants were first selected in the light and then tested for growth in the dark or TTC staining to detect concomitant mitochondrial transformation events. This type of selection procedure is commonly used for yeast mitochondrial transformation (8). However, these experiments were not successful in our hands, and direct selection for the mitochondrial transformants, although lengthy appears to be the most efficient strategy.
In addition, using this direct-selection scheme, we demonstrate that slowly growing complex I mutants can be recovered after transformation of cob or cox1-Δnd1 mutants. The Δnd4 deletion that we have constructed this way removes a region that probably codes part of the first transmembrane domain of ND4 according to the topology identified in prokaryotic NUOM corresponding to mitochondrial ND4 (21). Both Δnd4 and Δnd4-Δnd1 transformants showed a subcomplex that is 250-kDa smaller than the full-size complex I of the wild type. A similar situation was found in strains with larger deletions in nd4, such as dum22 (13), suggesting that the deleted part of ND4 plays an important role in the assembly of a module of 250 kDa or its attachment to the 700-kDa subcomplex. However, one cannot rule out a synthesis defect of ND4 that could be due to a modified nucleotide context downstream of the initiation codon, even though the nd4 RNA level is unaffected. Unfortunately, antibodies recognizing ND4 are not available to test whether the protein is produced and stable. Moreover, because ND1 is thought to be part of the 700-kDa subcomplex (13, 22), our data imply that the 6-bp in-frame deletion in nd1, which does not destabilize the whole complex I (13), also has no effect on the stability of the 700-kDa subcomplex when present simultaneously with the Δnd4 mutation. More generally, our data demonstrate the feasibility of site-directed mutagenesis of the very hydrophobic mitochondrially encoded subunits located in the membrane domain of complex I.
Deficiencies in complex I activity due to mutations in mitochondrially or nuclear-encoded subunits have been implicated in the development of several human diseases, including Parkinson's disease (23, 24). The relationship between gene alterations and clinical disease symptoms often remains obscure. Because direct studies of human material are subject to strong restrictions and, in particular, many complex I patients die young, the development of nonhuman models of diseases is desirable. Whereas site-directed mutagenesis of nuclear-encoded subunits is possible in mammals (25), Neurospora crassa (26), and Yarrowia lipolytica (27), models in mitochondrially encoded subunits are not yet available because of specific difficulties associated with gene replacement in mitochondria. Mitochondrial gene mutagenesis is routinely performed in S. cerevisiae, but this organism lacks a respiratory NADH:ubiquinone oxidoreductase. Reconstructing pathogenic mutations in bacteria is unsatisfactory, because the membrane domain of eukaryotic complex I contains many more subunits than the prokaryotic domain (23 versus 7) (28), and it has not yet been demonstrated that the seven common subunits that are mitochondrially encoded in eukaryotes are located exactly at the same position in the membrane domain. Chlamydomonas NADH-dehydrogenase has a complexity and organization that is similar to its human counterpart, the mitochondrially encoded subunits being well conserved (29). Reconstruction of pathogenic mutations in Chlamydomonas would help to promote basic research on complex I that will eventually provide a better understanding of how these alterations interfere with enzyme function at a molecular level. More generally, the transformation procedure presented above will allow a systematic analysis of mitochondrial gene function and expression in Chlamydomonas.
Materials and Methods
Strains and Growth Conditions.
Strains used in this work are derived from strain 137c of C. reinhardtii. Strain Mu, whose DNA was used as template for PCR amplifications, contains, in codon 129 of the cob gene, a T-to-C substitution, conferring resistance to myxothiazol and mucidine (18). Dum22 is a deletion mutant lacking the left telomere, the cob gene, and part of the nd4 gene (14) and was used as a control. The two strains described below were used as recipients for the biolistic transformation. The mutant dum11 (17, 30) exhibits a 1.2-kb deletion extending beyond codon 147 of cob, responsible for loss of complex III activity. The double mutant dum19 dum25 bears a one-T deletion at codon 152 of cox1 (dum19 mutation), responsible for loss of complex IV activity, and an in-frame deletion of two codons at positions 199–200 of nd1 (dum25 mutation), responsible for loss of NADH/duroquinone oxidoreductase activity (12). Cells were routinely grown under mixotrophic conditions, i.e., under light (4,000 lux) on Tris-acetate phosphate (TAP) medium (31).
Phenotypical Analyses.
Under anaerobic conditions, the electrons of the respiratory chain can be transferred by cytochrome c oxidase to TTC, which is then reduced to red formazan (32). Contrary to the deleted recipient strain dum11, which remains green in the dark when overlaid with TTC, strains that possess an intact cytochrome pathway of respiration (functional complex III and complex IV) become brown. The TTC test was performed, essentially as described in ref. 17, by pouring 15 ml of a 0.05% TTC and 0.5% agar solution onto TAP medium plates containing 60 replica-plated transformants. Plates were incubated in the dark overnight.
Growth of colonies in the dark was tested by spotting 10 μl of cell suspension (106 to 107 cells per ml) on TAP agar plates and incubating the plates for 10 days in the dark.
Resistance to myxothiazol was tested in the dark as described above on TAP agar plates containing 2.4 μM myxothiazol (18).
PCR, DNA, and RNA Analyses.
C. reinhardtii total nucleic acids were prepared according to ref. 33. PCR fragments were amplified either from total DNA or directly on Chlamydomonas colonies according to a protocol derived from ref. 15 using Pfu or Taq polymerase. Sequencing was performed directly on amplified products by Genome Express (Grenoble, France).
Southern and Northern blots were performed according to standard protocols (34). PCR fragments labeled with digoxigenin were used as probes and detected as recommended by the manufacturer (Roche Molecular Biology).
DNA Constructs Used for Chlamydomonas Transformation.
A mitochondrial PCR fragment was amplified from total C. reinhardtii DNA by using the Pfu polymerase (Promega) and primers Tel and nd4-R (Fig. 1), containing 5′-end extensions, allowing for cloning by recombination in the vector pDNR-Dual (4.9 kb, BD Biosciences). These extensions were acgaagttatcacccggg for Tel and cgaatggtctagcccggg (SmaI restriction site) for nd4-R. The resulting plasmid, called pShort, contains the first 3,301 bp of the mitochondrial genome (GenBank accession no. U03843), including the left telomere, the cob and nd4 genes, and 257 bp of the intergenic nd4–nd5 region (Fig. 1). A PCR fragment was amplified by using primers Tel and cox1 containing 5′-end extensions and inserted in the same vector to obtain pLong. The 5′-end extension of the cox1 primer was identical to the one of nd4-R. This plasmid contains the first 6,653 bp of the mitochondrial genome, including the left telomere, the cob, nd4, nd5, and cox1 genes and 30 bp of the intergenic cox1-nd2 region. To obtain plasmids pShortΔ and pLongΔ, we took advantage of two NdeI restriction sites (ca/tatg) located at positions 2,974 and 3,043. Digestion by NdeI followed by religation removed the first 23 codons of nd4. Because the 24th codon is an ATG, this resulted in an in-frame deletion eliminating codons 2–24 while leaving an intact ATG initiation codon at the beginning of the shortened nd4 sequence (Fig. 1). The sequencing of these mitochondrial inserts did not reveal any differences with the published sequences (GenBank accession no. U03843). These plasmids were difficult to clone and to amplify in E. coli, perhaps because of the presence of the telomeric region. For transformation experiments, they were used at a concentration of 1–2 μg/μl and, if desired, linearized with BglI.
PCR fragments used for transformation were amplified with primers Tel/cox1 or cob-F/cox1, precipitated, and used at a concentration of 1–2 μg/μl for the bombardments.
Transformation Procedure.
Cells were grown in liquid TAP medium up to exponential phase (2–3 ×106 cells) and spread at high density on TAP (1-cm thick, 1.5% wt/vol agar) plates (108 cells per plate). The thickness of the agar avoided its damage by the combined action of the vacuum and the helium burst. Plates were bombarded with tungsten beads coated with DNA by using a Bio-Rad PDS-1000He apparatus under a pressure of 1,100 psi and a partial vacuum in the chamber corresponding to a reading of at least 29 inches Hg, according to ref. 8. The highest transformation rates were obtained with rupture disks of 1,100 psi rather than 1,350 psi and with plates positioned at 7 cm from the macrocarrier assembly (second level from the bottom). Generally, we found that the use of stopping screens mildly enhances the rate of transformation.
Enzyme Activity and Analysis of Protein Complexes.
Rotenone-sensitive NADH/duroquinone oxidoreductase activity of complex I was measured in membrane fractions from cells disrupted by sonication (12).
For detection of active complex I in gels, protein complexes were solubilized from membrane fractions by 0.9% (wt/vol) N-dodecylmaltoside, separated by BN/PAGE and stained for NADH dehydrogenase activity by using nitroblue tetrazolium as an electron acceptor (13). BN gels were electroblotted according to ref. 35. Polyclonal antibodies against the whole complex I from N. crassa were used for immunodetection by using the ECL kit (Roche Molecular Biology) with anti-rabbit POD-conjugated antibodies.
Acknowledgments
We thank Michèle Radoux for expert technical assistance, the team of Dr. Hélène Barbier-Brygoo for access to their plant culture facility, Dr. U. Schulte and H. Weiss (University of Düsseldorf, Germany) for the kind gift of polyclonal antibodies against whole complex I from Neurospora crassa, René Matagne and Christopher J. Herbert for critical reading of the manuscript, and Christopher J. Herbert for looking over the English. This work was supported by Belgian Fonds de la Recherche Fondamentale Collective Grants 2.4587.04 and 2.4582.05 (to C.R.) and grants from the Fonds Spéciaux pour la Recherche Universitaire (to C.R.), the Association Française contre les Myopathies (to N.B.), and a joint France-Wallonie/Bruxelles Tournesol Grant (to N.B. and C.R.). P.C. is a postdoctoral researcher from the Belgian Fonds National de la Recherche Scientifique.
Abbreviations
- BN
Blue Native
- TAP
Tris-acetate phosphate
- TTC
2,3,5-triphenyltetrazolium chloride.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rochaix J.-D., Goldschmidt-Clermont M., Merchant S., editors. The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Dordrecht, The Netherlands: Kluwer; 1998. [Google Scholar]
- 2.Kindle K. L. Proc. Natl. Acad. Sci. USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boynton J. E., Gillham N. W., Harris E. H., Hosler J. P., Johnson A. M., Jones A. R., Randolph-Anderson B. L., Robertson D., Klein T. M., Shark K. B., et al. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- 4.Goldschmidt-Clermont M. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris E. H. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- 6.Johnston S. A., Anziano P. Q., Shark K., Sanford J. C., Butow R. A. Science. 1988;240:1538–1541. doi: 10.1126/science.2836954. [DOI] [PubMed] [Google Scholar]
- 7.Fox T. D., Sanford J. C., McMullin T. W. Proc. Natl. Acad. Sci. USA. 1988;85:7288–7292. doi: 10.1073/pnas.85.19.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnefoy N., Fox T. D. In: Methods in Molecular Biology: Mitochondria. Leister D., Herrmann J. M., editors. Totowa, NJ: Humana; 2006. in press. [Google Scholar]
- 9.Yoon G. Y., Koob M. D. Nucleic Acids Res. 2005;33:e139. doi: 10.1093/nar/gni140. 10.1093/nar/gni140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vahrenholz C., Riemen G., Pratje E., Dujon B., Michaelis G. Curr. Genet. 1993;24:241–247. doi: 10.1007/BF00351798. [DOI] [PubMed] [Google Scholar]
- 11.Remacle C., Matagne R. F. In: The Molecular Biology of Chloroplasts and Mitochondria. Rochaix J.-D., Goldschmidt-Clermont M., Merchant S., editors. Dordrecht, The Netherlands: Kluwer; 1998. pp. 661–674. [Google Scholar]
- 12.Remacle C., Baurain D., Cardol P., Matagne R. F. Genetics. 2001;158:1051–1060. doi: 10.1093/genetics/158.3.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardol P., Matagne R. F., Remacle C. J. Mol. Biol. 2002;319:1211–1221. doi: 10.1016/S0022-2836(02)00407-2. [DOI] [PubMed] [Google Scholar]
- 14.Remacle C., Duby F., Cardol P., Matagne R. F. Biochem. Soc. Trans. 2001;29:442–446. doi: 10.1042/bst0290442. [DOI] [PubMed] [Google Scholar]
- 15.Randolph-Anderson B. L., Boynton J. E., Gillham N. W., Harris E. H., Johnson A. M., Dorthu M.-P., Matagne R. F. Mol. Gen. Genet. 1993;236:235–244. doi: 10.1007/BF00277118. [DOI] [PubMed] [Google Scholar]
- 16.Yamasaki T., Kurokama S., Watanabe K. I., Ikuta K., Ohama T. Plant Mol. Biol. 2005;58:515–527. doi: 10.1007/s11103-005-7081-3. [DOI] [PubMed] [Google Scholar]
- 17.Dorthu M.-P, Remy S., Michel-Wolwertz M.-R., Colleaux L., Breyer D., Beckers M.-C., Englebert S., Duyckaerts C., Sluse F. E., Matagne R. F. Plant Mol. Biol. 1992;18:759–772. doi: 10.1007/BF00020017. [DOI] [PubMed] [Google Scholar]
- 18.Bennoun P., Delosme M., Kück U. Genetics. 1991;127:335–343. doi: 10.1093/genetics/127.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray M. W., Boer P. H. Philos. Trans. R. Soc. London B. 1988;319:135–147. doi: 10.1098/rstb.1988.0038. [DOI] [PubMed] [Google Scholar]
- 20.Rexroth S., Meyer zu Tittingdorf J. M., Krause F., Dencher N. A., Seelert H. Electrophoresis. 2003;24:2814–2823. doi: 10.1002/elps.200305543. [DOI] [PubMed] [Google Scholar]
- 21.Mathiesen C., Hagerhall C. Biochim. Biophys. Acta. 2002;1556:121–132. doi: 10.1016/s0005-2728(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 22.Sazanov L. A., Peak-Chew S. Y., Fearnley I. M., Walker J. E. Biochemistry. 2000;39:7229–7235. doi: 10.1021/bi000335t. [DOI] [PubMed] [Google Scholar]
- 23.Smigrodzki R., Parks J., Parker D. W. Neurobiol. Aging. 2004;25:1273–1281. doi: 10.1016/j.neurobiolaging.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 24.DiMauro S., Hirano M. Neuromuscular Disorders. 2005;15:276–286. doi: 10.1016/j.nmd.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Scheffler I. E., Yadava N., Potluri P. Biochim. Biophys. Acta. 2004;1659:160–171. doi: 10.1016/j.bbabio.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Duarte M., Schulte U., Ushakova A. V., Videira A. Genetics. 2005;171:91–99. doi: 10.1534/genetics.105.041517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerscher S., Grgic L., Garofano A., Brandt U. Biochim. Biophys. Acta. 2004;1659:197–205. doi: 10.1016/j.bbabio.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Gabaldon T., Rainey D., Huynen M. A. J. Mol. Biol. 2005;348:857–870. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- 29.Cardol P., Vanrobaeys F., Devreese B., Van Beumen J., Matagne R. F., Remacle C. Biochim. Biophys. Acta. 2004;1658:212–224. doi: 10.1016/j.bbabio.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Colin M., Dorthu M.-P., Duby F., Remacle C., Dinant M., Wolwertz M.-R., Duyckaerts C., Sluse F., Matagne R. F. Mol. Gen. Genet. 1995;249:179–184. doi: 10.1007/BF00290364. [DOI] [PubMed] [Google Scholar]
- 31.Harris E. H. The Chlamydomonas Sourcebook. San Diego: Academic; 1989. p. 25. [Google Scholar]
- 32.Slater T. F., Sawyer B., Sträuly U. Biochim. Biophys. Acta. 1963;77:383–393. doi: 10.1016/0006-3002(63)90513-4. [DOI] [PubMed] [Google Scholar]
- 33.Newman S. M., Boynton J. E., Gillham N. W., Randolph-Anderson B. L., Johnson A. M., Harris E. H. Genetics. 1990;126:875–888. doi: 10.1093/genetics/126.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 35.Duby F., Cardol P., Matagne R. F., Remacle C. Mol. Genet. Genom. 2001;266:109–114. doi: 10.1007/s004380100529. [DOI] [PubMed] [Google Scholar]