Abstract
RecA protein is a crucial and central component of the homologous recombination and DNA repair machinery. Despite numerous studies on the protein, several issues concerning its action, including the allosteric regulation mechanism have remained unclear. Here we report, for the first time, a crystal structure of a complex of Mycobacterium smegmatis RecA (MsRecA) with dATP, which exhibits a fully ordered C-terminal domain, with a second dATP molecule bound to it. ATP binding is an essential step for all activities of RecA, since it triggers the formation of active nucleoprotein filaments. In the crystal filament, dATP at the first site communicates with a dATP of the second site of an adjacent subunit, through conserved residues, suggesting a new route for allosteric regulation. In addition, subtle but definite changes observed in the orientation of the nucleotide at the first site and in the positions of the segment preceding loop L2 as well as in the segment 102–105 situated between the 2 nt, all appear to be concerted and suggestive of a biological role for the second bound nucleotide.
INTRODUCTION
RecA protein is a crucial and central component of the homologous genetic recombination machinery (1). It also plays an important role in DNA repair and SOS response processes. ATP binding is essential for all activities of RecA, since it initiates the formation of active nucleoprotein filaments (2,3). Crystal structures of the protein from Escherichia coli (4–7), Mycobacterium tuberculosis (8,9), Mycobacterium smegmatis (10) and Deinococcus radiodurans (11) have revealed a three-domain [N-terminal (N: 1–30), Major (M: 31–269) and C-terminal (C: 270–349)] architecture. The major M domain harbors the ‘P-loop containing nucleotide triphosphate hydrolases’ fold containing the nucleotide-binding site as well as the two loops L1 and L2 involved in DNA binding. Similar structural organizations are seen in several other proteins, encompassing physiologically diverse groups of proteins such as Rad51 (12), the eukaryotic orthologue of bacterial RecA and small G-proteins such as Rac1 (13). The M domain in RecA, in addition to being associated with catalytic activity, also brings about filament assembly by recruiting the N-terminal domain of an adjacent subunit. The C-terminal domain, for which a regulatory role has been suggested (14,15), is not fully ordered in any of the previously known RecA crystal structures. This has resulted in lack of clarity on the structural basis of any such role. Here, we report, for the first time, a crystal structure of M.smegmatis RecA (MsRecA) complexed with dATP, with a fully ordered C domain. Remarkably, the structure also shows the presence of a second dATP-binding site, presumably caused by the complete ordering of the C-terminal domain.
MATERIALS AND METHODS
Crystallization and data collection
MsRecA was purified (16) and crystallized (10) in the presence of magnesium and dATP, a close structural analogue of ATP, as described before, except for a small variation during the protein crystallization step [0.2 M ammonium acetate was used instead of 0.1 M as in the study of PDB:1UBC (MsRecA apo structure)]. The crystals, space group P61 with a = b = 108.2, c = 73.0 Å, diffracted upto 3.2 Å resolution. This structure will be referred to as MsRecA-II, hereafter, to differentiate from the previously reported MsRecA-dATP structure [PDB:1UBG, (10)], which will be referred to as MsRecA-I. X-Ray diffraction data were collected at room temperature using a 30 cm MAR imaging plate mounted on Rigaku RU200 X-ray generator. The data were processed using DENZO and SCALEPACK (17). Data collection and refinement statistics are listed in Table 1.
Table 1.
Data collection and refinement statistics of the MsRecA+dATP complex
| Space group | P61 |
|---|---|
| Unit cell dimension (Å) | 108.2, 73.0 |
| Resolution limit (Å) | 30–3.2 |
| Last resolution (Å) | 3.31–3.20 |
| Rmerge (%) | 10.3 (53.1) |
| Total number of unique reflections | 7941 (807) |
| Completeness | 97.5 (100.0) |
| Multiplicity | 4.1 |
| Rfactor (%) | 23.7 |
| Rfree (%) | 30.7 |
| Root mean square deviation from ideal | |
| Bond length (Å) | 0.01 |
| Bond angle (degrees) | 2.8 |
| Dihedral angle (degrees) | 24.4 |
| Improper angle (degrees) | 1.5 |
| No. of protein atoms | 2399 |
| No. of ligand atoms | 74 |
| No. of solvent atoms | 108 |
| Ramachandran plot (%) | |
| Allowed region | 91.9 |
| Generously allowed region | 7.5 |
| Disallowed region | 0.7 |
Values in parentheses refer to the lowest resolution shell.
Structure refinement
The structure was solved by molecular replacement using AMoRe (18) with 1UBC as the template and refined using CNS (19). The structure was first refined by treating the three domains as rigid groups followed by cycles of positional refinement of all atoms and simulated annealing. The ligands were identified using 2Fo–Fc and Fo–Fc maps. Finally grouped B-factor refinement was carried out. The model building was carried out using FRODO (20). The geometric parameters for dATP and citrate ion were obtained from the HICCUP data base (21). Water molecules were built into the electron density map where peaks were visible at contour levels of at least 2.5 σ in a Fo–Fc map and 0.8 σ in 2Fo–Fc map. Electron density was clear for the entire N, M and C domains, excepting the two DNA-binding loops L1 (residues 158–166) and L2 (196–211) in the M domain. Electron density was also clear for the dATP molecule at the expected P-loop-binding site on the M domain, the second dATP and a citrate-like molecule. The co-ordinates have been deposited in Protein Data Bank with accession code 2G88.
Structure analysis
The stereochemical quality of the structures were validated using PROCHECK (22). Superposition of structures was carried out using ALIGN. A distance of <3.6 Å between donor and acceptor atoms and an angle >90° at the hydrogen atoms were used as criterion for determining hydrogen bonds. Cavities indicating putative binding sites were also separately identified theoretically using the α spheres algorithm as implemented in CASTP (23). The volumes, molecular and solvent accessible surfaces of the cavities were also computed using the same software package.
Isothermal titration calorimetry
The protein solution was prepared in 100 mM sodium citrate buffer, pH 7.0, containing 100 mM NaCl, 12 mM MgCl2 and 2.5 mM DTT. The solution of the inhibitor ATPγS (Roche Diagnostics) was prepared in the same buffer. The concentration of the protein and the nucleotide were determined spectrophotometrically. Calorimetric measurements were performed using VP-ITC (MicroCal LIC Northhampton, MA) at 27°C with a reference power of 15 µcal/s, injection volume of 10 µl and a speed of rotation of 280 r.p.m. The data were analysed using Origin 7.0 software.
RESULTS AND DISCUSSION
While the overall structure of MsRecA-II has been, as expected, similar to that of MsRecA-I and all other RecA structures known, it differed significantly from the other structures by exhibiting an ordered C-terminal domain concurrent with the binding of a second nucleotide molecule. Several other significant differences in detail in the binding site regions, are also observed, which are discussed below.
Ordering of C domain and generation of the second nucleotide site
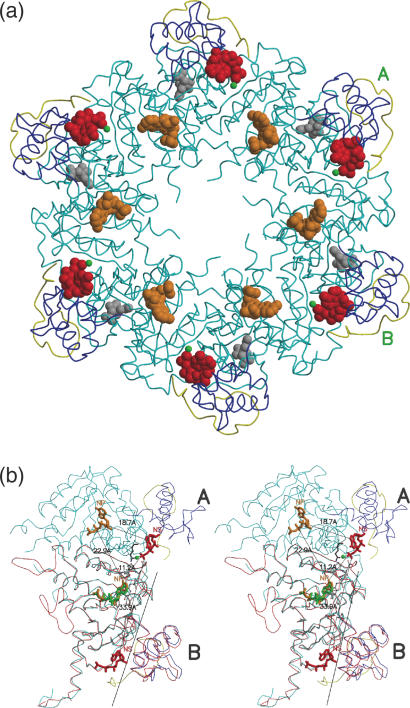
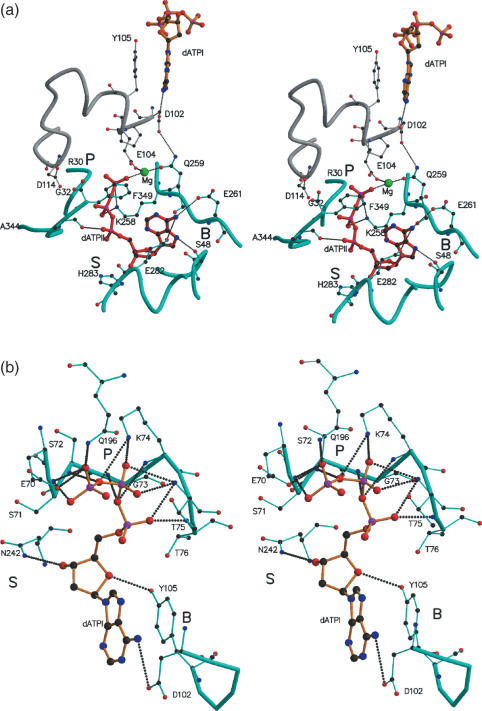
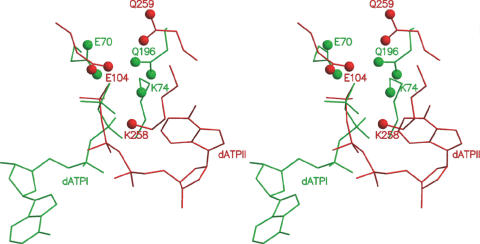
The additional 21 residues (329–349; referred to as C*) in the C domain seen only in the present structure, though not containing any definable secondary structural elements, folded into a compact structure, making 19 hydrogen bonds with the rest of the molecule. They occur on the surface of the filament formed by RecA molecules around the crystallographic 61 screw axis (Figure 1a). As compared with other MsRecA structures, the C domain was observed to be rotated by about 7.3°, moving it closer to the M domain (Figure 1b). The ordering of C* appears to be concurrent with the generation of a second ATP-binding site between M and C domains. Clear density for a dATP molecule was observed at this site (Figure 2b). This second dATP has a conformation, different from that of the first dATP molecule. A magnesium ion in the vicinity, is co-ordinated by a γ-phosphate oxygen, Gln259OE1 and the Glu104OE2 from a neighbouring subunit. The interactions of dATP and the magnesium ion are listed in Table 2. The phosphate group interacts with Lys258 and Ala348; the adenine base with Glu261, Ser48 and Glu282, and the sugar with His283 (Figure 3a). Residues Lys258, Glu104 and Gln259, at the second site are in fact broadly superposable on Lys74 and Glu70 (of the Walker-A motif) (24) and Gln196 of the first binding site (Figures 3b and 4). The lysine and glutamine residues positioned about 9 Å apart, around the γ-phosphate of ATP, has also been identified previously, as part of the NTP-binding signature in the P-loop NTPase structural family of proteins (9). Further, Asp114 from a neighbouring subunit, positioned appropriately to form a water mediated interaction with the terminal phosphate, is similar to Asp144 in the first site forming the Walker-B motif. The similarities in the residues and their disposition at the second binding site with those of the first and well-characterized sites in other proteins, suggest that the second site in RecA could indeed be biologically relevant. A theoretical exploration of binding site pockets (23) predicts three significant sites, corresponding to (i) the first dATP site, (ii) second dATP site and (iii) a smaller site at the N-M interface in each protein subunit. The same calculation when carried out in the absence of C*, predicted only sites (i) and (iii), thus indicating the requirement of the ordering of the whole C domain for the second dATP site.
Figure 1.
Filament formation in MsRecA. C* is coloured in yellow. The rest of the C domain is in blue. The nucleoside triphosphates dATPI, dATPII and citrate molecules are coloured in orange, red and grey. The magnesium ion is shown in green. (a) Cross sectional view. (b) Stereo view of a blow up of subunits A and B. The subunit B of this structure is superposed with a subunit of MsRecA-I (red). Residues connecting dATPII in A and dATPI in B are shown in ball-and-stick representation. The rotation axis about which the C domain moves, is also shown. The primary and the secondary nucleotide-binding sides are labelled NP and NS, respectively. The distances among the sites in the two subunits are indicated.
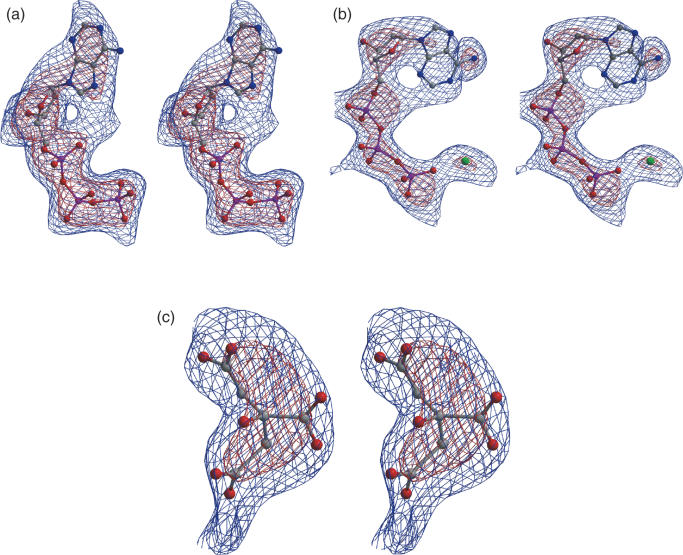
Figure 2.
Stereo view of the electron density from Fo–Fc simulated annealed omit maps for (a) First dATP molecule (b) second dATP molecule and (c) citrate-like molecule. Contours at 2 σ (blue) and 3.5 σ (red) are shown.
Table 2.
Interactions (distance <3.7 Å) involving dATP-II and the associated magnesium ion
| Ligand atoms | Protein atoms |
|---|---|
| O1G | Lys 258 NZ |
| O1B | Lys 258 NZ |
| O1A | Ala 348 O |
| O3* | His 283 ND1 His 283 NE2 |
| N7 | Ser 48 OG |
| N6 | Glu 282 OE1 Glu 261 OE2 |
| Mg2+ | Phe 257 O |
| Gln 259 OE1 | |
| Glu 104 OE2 | |
| HOH 934 O | |
| DTP 833 O3G |
O1A, O1B and O1G belong to α-, β- and γ- phosphates, respectively. O3* belongs to the sugar and N6 and N7 to the base.
Figure 3.
Stereo views of the two nucleotide binding sites of (a) the second dATP (dATPII) molecule and (b) the first dATP (dATPI) molecule. P, B and S refer to the phosphate, base and sugar binding regions. dATPI and dATPII are shown as large ball-and-stick models in orange and red respectively. In (a) the first dATP molecule (labelled dATPI) from an adjacent subunit along with an associated stretch of the polypeptide chain (grey) are also shown.
Figure 4.
Stereo view of the structurally comparable residues around dATPI (green) and dATPII (red). Phosphorous atom from the terminal phosphate, NZ of the lysines and the corresponding Cδ atoms of the glutamines and glutamic acids in the two structures were used for superposition.
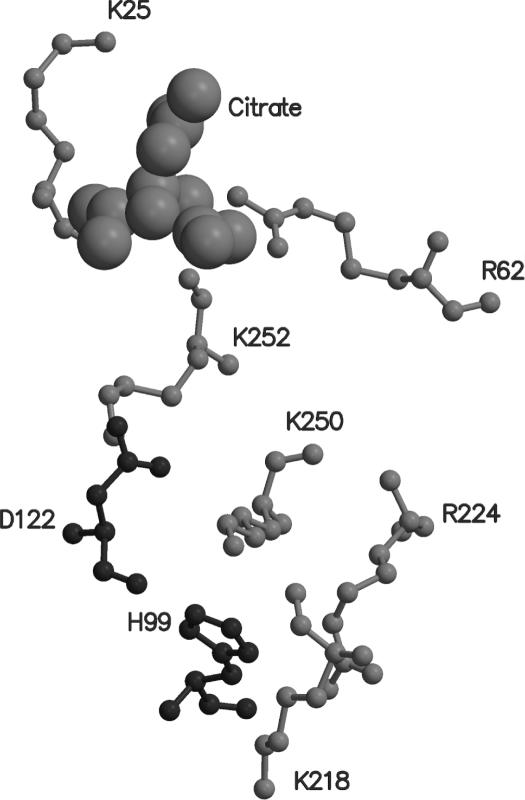
Clear electron density appears at the third smaller site referred to above (Figure 2c), which has been interpreted as a citrate ion. Lys25 and Arg62 directly interacts with the citrate molecule (Figure 5). Lys252 is in its close vicinity. All the three are conserved in the known RecA sequences. This conserved regions extends to include Lys218, Arg224, Lys250 from the same monomer and Asp122 and His99 from neighbouring molecule. Interestingly, the residues in the region exhibit rec− phenotypes when mutated. A glycerol has been refined at this site in a recent EcRecA structure (6). Further exploration of the functional role of this site is warranted.
Figure 5.
The binding mode of a citrate-like molecule (cpk representation) along with the conserved region associated with it. His99 and Asp122 belonging to an adjacent molecule in the filament.
Communication between nucleotide-binding sites in the filament
In the crystal filament the position of the second dATP molecule (dATPII) in each subunit, is close to the first dATP molecule (dATPI) of an adjacent subunit (Figure 1a). Further, residues 102–105 belonging to the first site interact with Gln259, γ-phosphate and the magnesium at the second site, thus establishing a possible communication channel between the two ATP molecules (Figure 3a). Asp102 hydrogen bonds with the adenine base of the first molecule and also with the side chain of Gln259 of the second site, both well-conserved residues in bacterial RecA. Gln259 (structurally equivalent to Gln196 of the first site) interacts with the γ-phosphate at the second site. Besides, Tyr105 stacks with the adenine at the first site, while Glu104 co-ordinates the magnesium at the second site. Though the latter residue is highly variable among RecA sequences, examination of the structural neighbourhood of this region suggests the possibility of another acidic side chain (e.g. Asp352 in E.coli) from the extreme C-terminal stretch, being available for co-ordination with the magnesium ion.
Concurrent with the ordering of the C* region and the binding of the second nucleotide, several other subtle but significant changes were observed in this structure, as compared with MsRecA-I, as discussed below.
A change was observed in the orientation of the dATP molecule in the first site, resulting in the first dATP of one subunit moving closer to the second dATP of the adjacent subunit (Figure 1b). Superposition of all Cα atoms of one subunit in the two structures indicate the largest shift to be between the N6 atoms of the base in the two structures, reaching 1.5 Å. This shift appears to be even more pronounced when one turn of the filaments from the two structures are superposed. This change reflects the possibility of subtle structural changes among different states during the ATP hydrolytic cycle.
Conformational changes are observed in the segments preceding (192–196) and succeeding (212–216) loop L2 (Figure 6). Changes in the order of 1–4 Å, more prominent in the 212–216 segment, are suggestive of loop L2 (disordered in this structure) being in a different conformation as compared to that in MsRecA-I. A shift is also observed in the position of Gln-196 (1.4 Å in its Cα atom), yet it makes a strong hydrogen bond with the terminal phosphate of the first nucleotide.
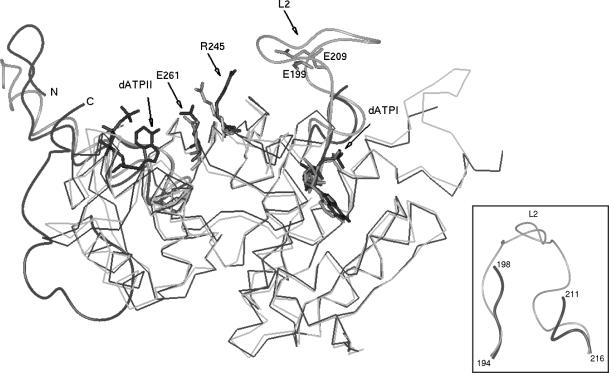
A shift of 5 Å in Arg 245 is observed, leading to the movement of the side chain closer to the filament cavity containing the L2 loop (Figure 6). In other RecA structures, Arg 245, a conserved residue is held by hydrogen bonds with a Glu 261, also a conserved residue. In the present structure, this interaction is disrupted presumably due to a hydrogen bond of Glu 261 with the base amine of the second nucleotide. Such a movement of Arg 245 could be stabilized by a strong interaction such as a salt-bridge with loop L2, the only segment in its neighbourhood. Loop L2 is not visible in the current structure, but in the context of the observed changes in the segments preceding and succeeding L2, either Glu199 or Glu 209 of L2 can be modelled to be positioned appropriately for interaction with Arg245. Such an interaction would help in communicating the information of the binding of second ATP molecule to L2. Both residues are well conserved in bacterial RecAs and mutation of either of them can lead to significant loss of RecA function (1).
A shift of 2.6 Å is observed in the side chain of Asp114 (structurally equivalent to Asp144 of the Walker-B motif), present at the second binding site. Asp114 is also involved in inter-subunit interactions (with residues 30–32 of an adjacent molecule) that form the filament, in all other RecA structures known so far. In this structure however, there is not only a shift in the side chain of Asp114, but there is also a significant shift in the 30–37 segment, that acts as a linker between the N and M domains. In this structure, the side chain of Asp114 is not in a position to hydrogen bond either with the peptidyl nitrogen of residue Gly32, or form a salt-bridge with the side chain of Arg30, leading to weakening of filament interactions. This observation forms the first structural indication of a link between nucleotide binding and filament formation. It also explains an earlier report of a loss of filament formation ability in the absence of DNA, when Arg30 is mutated to an alanine (25).
Additional interactions are observed between N and C domains, made by hydrogen bonds of residues Ile331–Gly332 and 341 of the C domain with Glu37 and Glu34 of the N-terminal helix. These are associated with a 2.0° re-orientation of the N-terminal helix, conformational changes in residues 30–37 and consequently shifts in the range of 1 to 2.7 Å. It must be noted that the citrate-like molecule binds at the N–M interface, involving interaction from the N-terminal helix. It is also noteworthy that a large shift of 5.3 Å is observed in the side chain of Lys-25, moving it closer to the citrate-like molecule. Definite identification of the ligand bound at this region awaits biochemical characterization, which will also help in understanding its influence, if any, on the ordering of the C-terminal domain or the binding of the second nucleotide.
Figure 6.
Movement of Arg245 towards L2 in MsRecA-II (black) as compared with that in MsRecA-I (grey). dATPI, dATPII, Glu261 are shown (see text). N- and C-termini, positions of Glu199 and Glu209 in L2 are also indicated. The regions preceding and succeeding L2, from the same superposition, indicating conformational changes, are shown in another orientation in the inset.
Thermodynamic parameters
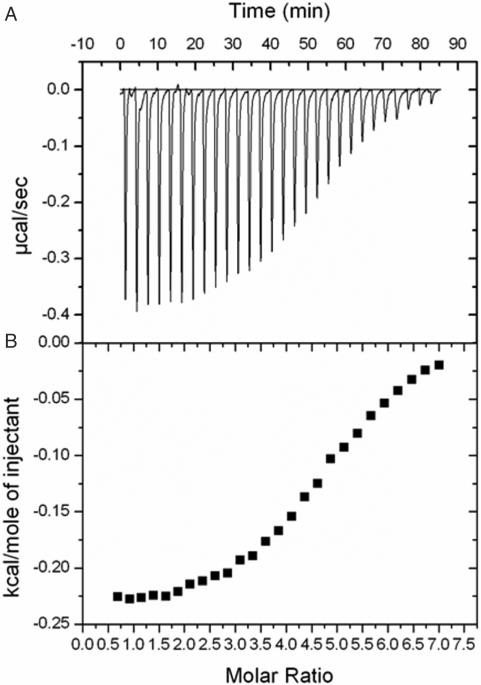
Repeated attempts to measure the thermodynamic parameters of MsRecA-nucleotide interactions in the presence of DNA did not yield interpretable results. This is presumably on account of the complex aggregation properties of the RecA-DNA filament. RecA by itself exhibits unusual aggregation properties and calorimetric experiments involving it is difficult to perform. That must be the reason for the extreme paucity of thermodynamic parameters on the interactions involving the protein. However, we could demonstrate the interactions of RecA with the non-hydrolysable ATP analogue ATPγS using isothermal titration calorimetry (Figure 7). The results of the calorimetric experiments could be best explained in terms of multiple binding sites, each with affinity in the millimolar range. The four binding sites (the actual number provided by the software is between 4 and 5) could possibly be explained in terms of the well-established ATP-binding site in RecA, the two DNA-binding loops and the additional binding site observed in the present structure. However, it should be emphasized that extreme caution needs to be exercised in interpreting the calorimetric measurements involving RecA on account of the complicated aggregation properties of the protein. All the same, the results are compatible with the additional binding site observed in the crystal structure.
Figure 7.
ITC profile of the titration of MsRecA (140 µM) in citrate buffer pH (7.0) with ATPγS (5 mM) in the same buffer. Titration was performed at 27°C with a reference power of 15 µcal/s. Injection volume was 10 µl. (A) Represents the change in reference power (µcal/s) with respect to time. (B) Represents the heat exchange per mole (kcal/mol) of ATPγS added. The curve obtained was fitted using software Origin 7.0 with one set of binding sites. The value of N, K, ΔH and ΔS obtained were 4.78, 4.14 × 104 M−1, −238 kcal mol−1and 20.3 kcal mol−1k−1, respectively.
Biological implications
The C domain has been the focus of recent biochemical studies. Based on studies involving reaction pH profiling of various truncation mutants, Cox and co-workers (14) suggested a potentially complex interaction between the C-terminus and the rest of the protein. The crystal structure reported here, confirms the hypothesis and in fact reveals the exact nature of such interactions. Studies on a set of C-terminal deletion mutants of E.coli RecA, progressively removing 6, 13, 17 and 25 amino acid residues from the C-terminus, have indicated that deletions at the extreme end of C* exhibit enhanced binding to DNA (26), suggesting a regulatory role for the region. Moreover the deletion mutants exhibit dramatic enhancements in the ability of RecA protein to displace single-stranded binding protein (SSB), suggesting that the C* region suppresses RecA's inherent ability to compete with SSB for single-stranded DNA (ssDNA) (26). The regulatory role of the C* region becomes clearer from this observation, especially because the nucleation step of the RecA filament formation on ssDNA is strongly inhibited by prebound SSB, making it essential for RecA to displace SSB. There have been several reports suggesting that, apart from the well-characterized primary and secondary sites formed by the L2 and L1 loops at the centre of the helical groove, a third weak binding site for nucleic acids is present on the C-terminal domain (1,27,28). Based on site-directed mutagenesis studies of conserved amino acid residues in the C-terminal domain, Shibata and co-workers (27,28) have suggested that residues Lys302 and Lys286 have a direct role in double-stranded DNA (dsDNA) binding and that they constitute a part of a gateway for homologous recognition. The second nucleotide-binding site observed in our structure is at the periphery of the suggested site for DNA binding. It is possible that the binding of the second nucleotide through the ordering of the C* facilitates binding of DNA. On the other hand, since we do not have structural data about the weak DNA-binding site and hence cannot define its boundaries, the possibility of dATP occupying part of a large DNA-binding pocket cannot be ruled out. The issue cannot be resolved conclusively on the basis of currently available experimental results. Despite the lack of clarity on this issue, crystallographic definition of a binding site at this region sets the stage for further studies in this regard.
Studies on the magnesium ion dependency for the strand exchange reactions catalysed by RecA have indicated that presence of the RecA C domain imposes a requirement for high concentrations of magnesium for optimal RecA activity (15), whereas the truncation mutants in which segments of the C* have been deleted do not require such high magnesium concentrations. It is also proposed that the C domain acts as a regulatory switch, modulating the access of dsDNA, thereby inhibiting strand exchange at low magnesium levels. The observation of the second dATP and magnesium binding site, formed by the ordering of the C*, could explain such a requirement.
Two types of RecA filaments have been observed by electron microscopy: active filaments with a pitch of 90–110 Å formed in the presence of DNA and nucleotide co-factor (3,29–31) and inactive filaments with a pitch of about 76 Å formed in their absence (30). The pitch of the filaments observed in the crystal structures of different bacterial RecA range from 67.5 to 83 Å. These filaments are considered to correspond to the inactive form. Recently the crystal structures of yeast Rad51 (32) and an archaeal RadA (33), both structural and functional homologues of bacterial RecA, have been determined. The filaments in them have pitches of 130 and 106.7 Å respectively. The main (M) domain has similar structure in RecA, Rad51 and RadA. However, the C-terminal (C) domain is comparatively small in Rad51 and is absent in RadA. On the contrary, the N-terminal (N) domain in them are larger than that in RecA by different extents. Therefore, the results on the aggregation in one cannot be directly extrapolated to those in another. All the same, RecA filaments of the type observed in the Rad51 and the RadA crystal structures were modelled by superposing the M domain of RecA on the M domain of each molecule of Rad51 or RadA in the respective filaments. The resulting RecA filaments are structurally viable. In the RadA type filament, the nucleotide at the second dATP site is at hydrogen bonding distance from the nucleotide in the primary site of an adjacent molecule. In the Rad51 type filament, the two are not in hydrogen bonded contact, but are close to each other with a minimum distance between them of 11.2 Å.
In the meantime, fresh electron microscopic studies on RecA-DNA filaments, with considerable inputs based on the structure of Rad51, have been reported (3,31). In the model of the active filaments with pitch 91 Å (PDB:1NO3), the C domain, sans the C* component which for the first time is reported here, is rotated by about 10° with respect to the M domain compared with its position in the EcRecA crystal structure, leading to an opening between the C and the M domains about the domain junctions. C*, when added to C as in the crystal structure presented here, falls on the periphery of the filament. All the same, it has a few short unfavourable steric contacts with the M domain. These can be relieved by small rotations of the C domain as a whole. Alternatively, the unfavourable contacts may lead to the instability of the active filament or the disordering of C*. This is in consonance with the observed increased action of C-terminal deletion mutants. Furthermore, if a second ATP were to bind in the active state, the position of the second nucleotide-binding site in the model of the active filament is again close to the primary binding site of an adjacent molecule, with a minimum distance of 12.2 Å between ATP molecules located at the two positions. The intervening region between the two sites is a highly conserved loop between the two β-strands (includes 250–259). One lysine residue in this stretch (K258) interacts with the second dATP molecule while another (K250), which is implicated in co-operative DNA binding (34), is positioned close to the first ATP site in the adjacent molecule in the filament. There are also other residues in the stretch which are close to one or the other of the two sites. Thus the stretch can possibly function as a communication channel between the two sites. This possibility needs to be further explored experimentally.
The observation of the full C domain and the second dATP molecule in communication with the first, in the crystal filament, leads us to propose a model for ATP-induced allosteric control in MsRecA, which could be a general model for the RecA family of proteins, given the high degree of sequence, tertiary and quaternary structure conservations among RecAs from different sources. The present structure, suggests that the C domain ordering would lead to the formation of a second nucleotide-binding site, which can communicate with the first ATP molecule situated on an adjacent molecule in the filament thus linking C-terminal ordering and ATP binding. The change in orientation of Asp114 upon the second nucleotide binding, leading to loss of interaction with the N domain of an adjacent molecule, suggests a link between C-terminal ordering and second nucleotide binding with filament assembly/disassembly. Conformational changes implied in the present structure for loop L2, and the possibility of the loop being stabilized by Arg 245, whose conformation has significantly altered in the present structure, hints strongly of a possible link between the second nucleotide binding and the movement in loop L2. Although not characterized in full detail, multiple lines of evidence has led to several reports regarding the influence of the first nucleotide binding and hydrolysis on the conformation of L2 and consequent changes in L1. Taken together, it is possible to envisage a structural basis for the regulatory role of the C domain, owing to its ability to turn on or off the binding of the second nucleotide and its communication with the first, and hence modulate not only conformational changes in the DNA binding loops but also filament assembly. The sequence of events and structural changes, that form the ATP hydrolytic cycle and filament assembly and disassembly are still obscure, but the observation of the ordered C domain and a second ATP-binding site provide new insights to understand allostery in RecA filaments.
Acknowledgments
M.V. is supported by a distinguished biotechnologist award of the DBT, Government of India. Use of X-ray facility for structural biology supported by the Department of science and Technology (DST), and Department of Biotechnology (DBT) at Molecular Biophysics Unit, facilities at the Super Computer Education and Research Centre, DBT supported Bioinformatics Centre and Interactive Graphics facilities at this Institute are gratefully acknowledged. The work forms part of a DBT-sponsored genomics program. Funding to pay the Open Access publication charges for this article was provided by project grants from DBT.
Conflict of interest statement. None declared.
REFERENCES
- 1.McGrew D.A., Knight K.L. Molecular design and functional organization of the RecA protein. Crit. Rev. Biochem. Mol. Biol. 2003;38:385–432. doi: 10.1080/10409230390242489. [DOI] [PubMed] [Google Scholar]
- 2.Flory J., Tsang S.S., Muniyappa K. Isolation and visualization of active presynaptic filaments of recA protein and single-stranded DNA. Proc. Natl Acad. Sci. USA. 1984;81:7026–7030. doi: 10.1073/pnas.81.22.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanloock M.S., Yu X., Yang S., Lai A.L., Low C., Campbell M.J., Egelman E.H. ATP-mediated conformational changes in the RecA filament. Structure. 2003;11:187–196. doi: 10.1016/s0969-2126(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 4.Story R.M., Weber I.T., Steitz T.A. The structure of the E.coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 5.Story R.M., Steitz T.A. The Structure of the recA protein–ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 6.Xing X.U., Bell C.E. Crystal structures of Escherichia coli RecA in a compressed helical filament. J. Mol. Biol. 2004;342:1471–1485. doi: 10.1016/j.jmb.2004.07.091. [DOI] [PubMed] [Google Scholar]
- 7.Xing X.U., Bell C.E. Crystal structures of Escherichia coli RecA in complex with MgADP and MnAMP-PNP. Biochemistry. 2004;43:16142–16152. doi: 10.1021/bi048165y. [DOI] [PubMed] [Google Scholar]
- 8.Datta S., Prabu M.M., Vaze M.B., Ganesh N., Chandra N.R., Muniyappa K., Vijayan M. Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF4: implications for decreased ATPase activity and molecular aggregation. Nucleic Acids Res. 2000;28:4964–4973. doi: 10.1093/nar/28.24.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta S., Ganesh N., Chandra N.R., Muniyappa K., Vijayan M. Structural studies on MtRecA–nucleotide complexes: insights into DNA and nucleotide binding and the structural signature of NTP recognition. Proteins. 2003;50:474–485. doi: 10.1002/prot.10315. [DOI] [PubMed] [Google Scholar]
- 10.Datta S., Krishna R., Ganesh N., Chandra N.R., Muniyappa K., Vijayan M. Crystal structures Mycobacterium smegmatis RecA and its nucleotide complexes. J. Bacteriol. 2003;185:4280–4284. doi: 10.1128/JB.185.14.4280-4284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajan R., Bell C.E. Crystal structure of RecA from Deinococcus radiodurans: insights into the structural basis of extreme radioresistance. J. Mol. Biol. 2004;344:951–963. doi: 10.1016/j.jmb.2004.09.087. [DOI] [PubMed] [Google Scholar]
- 12.Pellegrini L., Yu D.S., Lo T., Anand S., Lee M., Blundell T.L., Venkitaraman A.R. Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 13.Hirshberg M., Stockley R.W., Dodson G., Webb M.R. The crystal Structure of human Rac1, a member of the rho-family complexed with a GTP analogue. Nature Struct. Biol. 1997;4:147–152. doi: 10.1038/nsb0297-147. [DOI] [PubMed] [Google Scholar]
- 14.Lusetti S.L., Wood E.A., Fleming C.D., Modica M.J., Korth J., Abott L., Dwyer D.W., Roca A.I., Inman R.B., Cox M.M. C-terminal deletions of the Escherichia coli RecA protein. J. Biol. Chem. 2003;278:16372–16380. doi: 10.1074/jbc.M212917200. [DOI] [PubMed] [Google Scholar]
- 15.Lusetti S.L., Shaw J.J., Cox M.M. Magnesium ion-dependent activation of the RecA protein involves the C-terminus. J. Biol. chem. 2003;278:16381–16388. doi: 10.1074/jbc.M212916200. [DOI] [PubMed] [Google Scholar]
- 16.Ganesh N., Muniyappa K. Characterization of DNA strand transfer promoted by Mycobacterium smegmatis RecA reveals functional diversity with Mycobacterium tuberculosis RecA. Biochemistry. 2003;42:7216–7225. doi: 10.1021/bi0340548. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;50A:157–163. [Google Scholar]
- 19.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gross P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallographic and NMR system (CNS): a new software for macromolecular structure determination. Acta Crystallogr. 1998;54D:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.Jones T.A. A graphic model building and refinement system for macromolecules. J. Appl. Crystallogr. 1978;11:268–272. [Google Scholar]
- 21.Kleywegt G.J., Jones T.A. Databases in protein crystallography. Acta Crystallogr. 1998;54D:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski R.A., MacArthur M.W., Moss D.S., Thronton J.M. PROCHECK: a program to check the streochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 23.Liang J., Edelsbrunner H., Woodward C. Anatomy of protein packets and cavities: measurements of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly related sequences in the alpha-and beta-subunits of ATP synthase, myosin kinase and other ATP-requiring enzymes and common nucleotide-binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eldin S., Forget A.L., Lindenmuth D.M., Logan K.M., Kinght K.L. Mutations in the N-terminal region of RecA that disrupt the stability of free protein oligomers but not RecA–DNA complexes. J. Mol. Biol. 2000;299:91–101. doi: 10.1006/jmbi.2000.3721. [DOI] [PubMed] [Google Scholar]
- 26.Eggler A.L., Lusetti S.L., Cox M. The C-terminus of the Escherichia coli RecA protein modulates the DNA binding competition with single-stranded DNA-binding protein. J. Biol. chem. 2003;278:16389–16396. doi: 10.1074/jbc.M212920200. [DOI] [PubMed] [Google Scholar]
- 27.Kurumizaka H., Aihara,H, Ikawa S., Kashima T., Bazemore.L.R., Kawasaki K., Sarai A., Radding C.M., Shibata T. A possible role of the C-terminal domain of the RecA protein. J. Biol. Chem. 1996;271:33515–33524. doi: 10.1074/jbc.271.52.33515. [DOI] [PubMed] [Google Scholar]
- 28.Aihara H., Ito Y., Kurumizaka H., Terada T., Yokoyama S., Shibata T. An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing. J. Mol. Biol. 1997;274:213–221. doi: 10.1006/jmbi.1997.1403. [DOI] [PubMed] [Google Scholar]
- 29.Egelman E.H., Stasiak A. Structure helical RecA–DNA complexes. Complexes formed in the presence of ATP-γ-s or ATP. J. Mol. Biol. 1986;191:677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- 30.Egelman E.H., Stasiak A. Electron Microscopy of RecA–DNA complexes: two different states, their functional significance and relation to the solved crystal structure. Micron. 1993;24:309–324. [Google Scholar]
- 31.Yu X., Jacobs S.A., West S.C., Ogawa T., Egelman E.H. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl Acad. Sci. USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conway A.B., Lynch T.W., Zhang Y., Fortin G.S., Fung C.W., Symington L.S., Rice P.A. Crystal structure of a Rad51 filament. Nature Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y., He Y., Moya L.A., Qian X., Luo Y. Crystal structure of archaeal recombinase RadA: a snapshot of its extended conformation. Mol. Cell. 2004;15:423–435. doi: 10.1016/j.molcel.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen T.T., Muench K.A., Bryant F.R. Inactivation of the recA protein by mutation of histidine 97 or lysine 248 at the subunit interface. J. Biol. Chem. 1993;268:3107–3113. [PubMed] [Google Scholar]